Abstract
The outer membrane proteins (OMPs) of Gram-negative bacteria play a crucial role in virulence and pathogenesis. Identification of these proteins represents an important goal for bacterial proteomics, because it aids in vaccine development. Here, we have developed such an approach for Ehrlichia ruminantium, the obligate intracellular bacterium that causes heartwater. A preliminary whole proteome analysis of elementary bodies, the extracellular infectious form of the bacterium, had been performed previously, but information is limited about OMPs in this organism and about their role in the protective immune response. Identification of OMPs is also essential for understanding Ehrlichia’s OM architecture, and how the bacterium interacts with the host cell environment. First, we developed an OMP extraction method using the ionic detergent sarkosyl, which enriched the OM fraction. Second, proteins were separated via one-dimensional electrophoresis, and digested peptides were analyzed via nano-liquid chromatographic separation coupled with mass spectrometry (LC-MALDI-TOF/TOF). Of 46 unique proteins identified in the OM fraction, 18 (39%) were OMPs, including 8 proteins involved in cell structure and biogenesis, 4 in transport/virulence, 1 porin, and 5 proteins of unknown function. These experimental data were compared to the predicted subcellular localization of the entire E. ruminantium proteome, using three different algorithms. This work represents the most complete proteome characterization of the OM fraction in Ehrlichia spp. The study indicates that suitable subcellular fractionation experiments combined with straightforward computational analysis approaches are powerful for determining the predominant subcellular localization of the experimentally observed proteins. We identified proteins potentially involved in E. ruminantium pathogenesis, which are good novel targets for candidate vaccines. Thus, combining bioinformatics and proteomics, we discovered new OMPs for E. ruminantium that are valuable data for those investigating new vaccines against this organism. In summary, we provide both pioneering data and novel insights into the pathogenesis of this obligate intracellular bacterium.
Introduction
The Rickettsiales Ehrlichia ruminantium is an obligate intracellular bacterium that causes heartwater, a fatal tick-borne disease of ruminants, which is found in the islands of the Indian Ocean and the Caribbean, and in Africa [1]. E. ruminantium is transmitted by Amblyomma ticks and infects the endothelium of blood vessels. It has a complex life cycle with two distinct developmental forms found within mammalian host cells [2]. Initially, the infectious forms of the bacterium (elementary bodies, or EBs) adhere to host target cells and are internalized. Then, inside of intracytoplasmic vacuoles, they differentiate into a replicative, non-infectious form, the reticulate body (RB). After 5 to 6 days of intracellular multiplication, disruption of host cells leads to the release of numerous infectious EBs, initiating a new infectious cycle [1,3].
Current control methods for heartwater consist of a combination of vector control, using acaricides, and immunization against E. ruminantium. Different types of vaccines (inactivated, attenuated, recombinant) are currently being tested experimentally, but they have displayed limited efficacy, thus far, due to the genetic and antigenic diversity of E. ruminantium strains [3–8]. At this time, the only commercially available vaccine is based on the administration of infected blood to ruminants, followed by treatment with antibiotics; however, this remains an expensive, high-risk method [3].
Many studies of Gram-negative bacteria, such as Legionella pneumophila, Bartonella henselae, Pseudomonas syringae, Campylobacter jejuni, and Mannheimia haemolytica, have focused on outer membrane proteins (OMPs), because they have proven to be good targets for vaccine development [9–13]. Indeed, the OM of such pathogens represents an important dynamic interface between the bacterium and its environment. It serves as a selective barrier controlling the passage of nutrients and waste products into and out of the cell, and it also creates a chemically distinct periplasmic compartment, where important processes, such as the degradation of harmful substances from the environment or certain types of respiration, can occur [14,15]. OMPs are involved in the integrity and stability of the bacterial envelope, passive and active transport of substrates and nutrients, cell-to-cell communication, adhesion to host cells, and virulence [16].
Prospective proteomic analysis of E. ruminantium, cultivated in host endothelial cells, has already provided information about OMPs that are potentially implicated in bacterial infection and survival, such as members of the major antigenic protein (map) gene cluster [17,18]. Despite significant evidence implicating this gene family in immune protection in Ehrlichia and Anaplasma [19,20] and even strain penetrance in Anaplasma [21], our understanding of the biological role of this gene family is incomplete. However, studies on the differential expression of genes encoding OMPs has permitted us to understand the adaptation of these bacteria to the environment inside their vector, the tick, and to transmission to the mammalian host [22,23].
The aim of this study was to characterize the proteome of the OM fraction from infectious E. ruminantium EBs. To obtain an enriched OM fraction, we optimized a sarkosyl-based enrichment protocol that selectively solubilizes the inner and cytoplasmic membranes of Gram-negative bacteria, with no effect on the OM subcellular fraction [24]. We identified 46 unique proteins in the OM fraction using one-dimensional gel electrophoresis coupled with liquid chromatography-mass spectrometry (1DE-nanoLC-MALDI-TOF/TOF). Of these, 18 were known or predicted prototypical OMPs, while the others were of inner membrane (n = 5) or cytoplasmic (n = 23) origin or were chaperones. We compared our experimental results to the total set of E. ruminantium OMPs by combining results from three subcellular localization prediction algorithms and 34% of the total OMPs predicted from the genome were detected in the obtained OM fraction. We concluded that our method enriched OMPs. These results provide a better understanding of Ehrlichia OM architecture and may lead to the identification of potential vaccine candidates.
Importance
Ehrlichiae are obligate intracellular bacteria with a unique developmental cycle that includes attaching to and entering eukaryotic host cells, a process mediated by proteins in their outer membrane (OM). Thus far, few experimental data on ehrlichial OM proteins are available. To gain insight into the protein composition of the ehrlichial OM, we performed proteome analysis on OM fractions from Ehrlichia ruminantium elementary bodies, the infectious form of this bacterium. We compared our experimental results with an in silico analysis of the E. ruminantium proteome. We identified 18 proteins, whose OM localization was supported by both studies, and were, therefore, very likely to be located in the E. ruminantium OM. Among these proteins, 6 are completely new discovered OMPs and are therefore of importance as potential vaccine antigens. These results provide the first comprehensive overview of OM proteins in an Ehrlichia species and pave the way for developing novel therapeutic strategies to disrupt the OM or processes essential for its function
Materials and Methods
Ehrlichia ruminantium cultivation
E. ruminantium strain Gardel (from Guadeloupe, FWI) was routinely propagated in bovine aorta endothelial cells (BAE) as previously described [25]. One-hundred and twenty hours post-infection, when cell lysis occurs, infectious EBs were harvested and purified using a multistep, 20,000 × g centrifugation protocol, as described elsewhere [26,27]. Purified EBs were stored at -80°C in sucrose-phosphate-glutamate (SPG) buffer, pH 7.4.
Preparation of the OM fraction from E. ruminantium EBs
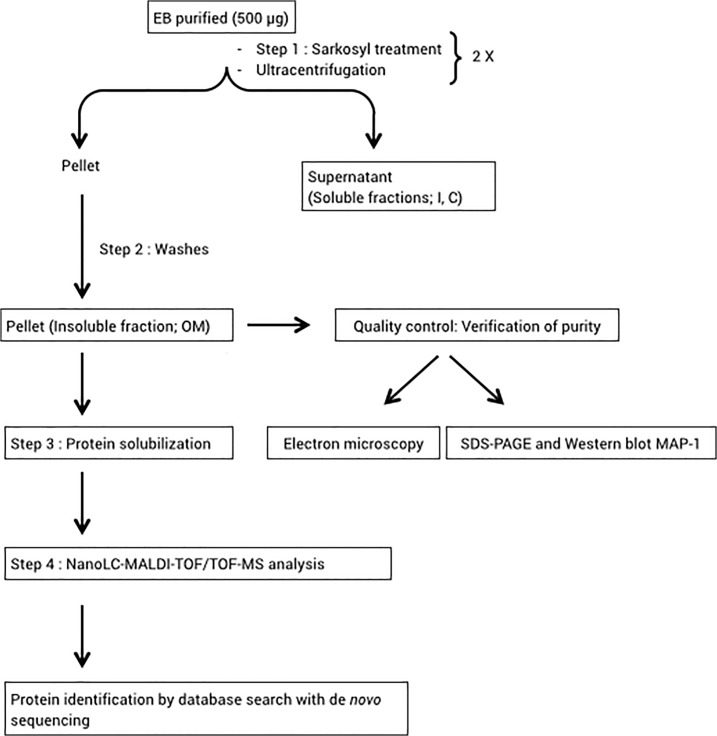
Subcellular fractionation was performed as described by Ohashi et al. [28], modified as follows. Purified EBs stored in SPG were washed in phosphate-buffered saline (PBS, pH 7.4) with a protease inhibitor cocktail (Roche), at 20,000 × g for 30 min at 4°C. Protein content was measured with the microBCA quantification kit (Sigma), according to the manufacturer’s instructions. Five hundred micrograms EBs were pelleted and resuspended in PBS containing 0.1% (v:v) sodium N-laurosyl sarcosine (sarkosyl; Sigma), DNAse (50 μg/mL), RNAse (50 μg/mL), MgCl2 (2.5 mM), and protease inhibitors (Roche), and then incubated for 30 min at 37°C. The sarkosyl treatment was repeated twice, followed by ultracentrifugation at 20,000 × g for 30 min at 4°C (Fig. 1). After the first separation, the insoluble pellet containing the OM fraction was washed twice in PBS and centrifuged at 20,000 × g for 30 min at 4°C to remove residual detergent (Step 2); the final pellet was resuspended in PBS containing protease inhibitors, and then stored at 4°C. Total protein concentration was determined using the 2D Quant Kit (GE Healthcare). Independent biological triplicates were carried out for OMP characterization (Fig. 1).
Fig 1. Experimental workflow for E. ruminantium subcellular fractionation and proteome characterization.
OM, outer membrane; I, inner membrane; C, cytoplasm.
Evaluation of OM enrichment protocol
Transmission Electron Microscopy (TEM). Samples were pre-fixed at 4°C in 2.5% (v/v) glutaraldehyde in PBS (pH 7.2). After a brief rinse with 1 × PBS, samples (intact EBs or OM complex) were fixed for 45 min at 25°C in 1% (w/v) osmium tetroxide in the same buffer, rinsed in distilled water and post-fixed with 2% (w/v) aqueous uranyl acetate for 1 h at 25°C before being embedded in epoxy resin. Two grids containing 4–5 ultrathin sections (60 nm thick) were observed using a Tecnai G2 TEM at 200 kV [29]. The TEM micrographs presented in this study are representative of all samples.
SDS-PAGE and Western blots to monitor OM fraction. Biological samples (15 μg) were precipitated in acetone for 3 h at -20°C and centrifuged at 20,000 × g for 10 min at 4°C. The pellet was solubilized in NuPAGE LDS Sample Buffer loaded on NuPAGE Novex 4–12% Bis-Tris polyacrylamide gels, and electrophoresis was carried out for 40 min at 200 V. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were blocked for 1 h in PBS with 0.05% (v/v) Tween 20 and 5% (w/v) milk, and then incubated with anti-MAP1 mouse monoclonal antibody (mAB) (4F10B4, Abcam) at a dilution of 1:2,000 for 1 h. Anti-Map1 monoclonal antibody was used as a specific OM marker. Membranes were washed three times in PBS with 0.05% (v/v) Tween 20 for 10 min, followed by incubation with the appropriate phosphatase alkaline-conjugated secondary antibodies (Sigma) at a 1:2,000 dilution for 1 h. Finally, membranes were developed using 5-bromo-4-chloro-3’-indolyphosphate/nitro-blue tetrazolium (BCIP/NBT) substrate (Roche) [17].
Proteome Characterization
1D gel electrophoresis for proteomics analysis. Forty μg intact EBs or OM fraction (from ERGp45, p52, and p57) were precipitated in acetone for 3 h at -20°C and centrifuged at 20,000 × g for 10 min at 4°C. Pellets were resuspended in 5 μL solubilization buffer [7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), and 30 mM Tris; Step 3 in Fig. 1]. After protein solubilization, 6 μL loading buffer [0.5 M dithiothreitol (DTT), 10% (w/v) sodium dodecyl sulfate (SDS), 250 mM Tris, 30% (v/v) glycerol, and 0.02% (w/v) bromophenol blue] was added. Samples were vortexed, and 9 μL water was added followed by agitation overnight at room temperature. Finally, samples were centrifuged at 16,000 × g for 2 min, and supernatants were loaded on NuPAGE Novex 4–12% Bis-Tris polycacrylamide gels; electrophoresis was performed for 40 min at 200 V. Gels were stained for 24 h using colloidal Coomassie Blue, and then washed 3 times in double distilled water [17].
In-gel digestion. For the evaluation of the optimized protocol to obtain an OMP enriched fraction, the more intense gel bands were excised. Previously to the NanoLC-MALDI-TOF/TOF analysis and in order to extend the number of proteins identified starting from simpler peptide digests, the OMP enriched fraction was separated by SDS-PAGE and each gel lanes was sliced. For in-gel digestion each band or slice was cut into 1 mm3 gel pieces, and Coomassie Blue was washed off with alternating water and 50% (v/v) acetonitrile (ACN) treatments until the gel pieces were transparent. Proteins were in-gel reduced with 10 mM dithiothreitol (DTT), alkylated with 55 mM iodoacetamide. Next, 6.7 ng/μL modified porcine trypsin (Promega) in 50 mM NH4CO3 was added to each gel band/slice. Digestion was performed at 37°C overnight. Peptides were extracted from the gel by washing it with 5% (v/v) formic acid, followed by two ACN washes. Digestion supernatants and extracted peptides were added, dried in a SpeedVac concentrator, and reconstituted in 5% (v/v) formic acid [30].
NanoLC-MALDI-TOF/TOF analysis. Chromatographic peptide separation was performed on a Thermo EASY-nLC 1000 with a pre-column Acclaim PepMap 100 C18 (75 μm × 2 cm) used as the Peptrap and an Acclaim PepMap RSLC C18 (50 μm × 15 cm) as the chromatographic separation column (Step 4, Fig. 1). A chromatographic gradient was established using mixed volumes of 0.1% (v/v) formic acid in water (buffer A) and 0.1% (v/v) formic acid in acetonitrile (buffer B, all LC-MS grade, from MERCK); peptides were eluted at a constant rate of 2 mL/min for 40 min in 5–40% (v/v) buffer A, according to their hydrophilic/hydrophobic properties. Peptide fractions were spotted onto MALDI plates and co-crystalized with 5 mg/mL alpha-cyano-4-hydroxycinnamic acid using a Micro-Spotter (Sunchrom). Peptide mass spectra were acquired with an Applied Biosystems 4800 Plus MALDI TOF/TOF Analyzer apparatus in both MS and MS/MS mode. Positively charged ions were analyzed in the reflectron mode over an m/z range of 800–3,500 Da. Each MS spectrum was obtained in result-independent acquisition mode with a total of 800 laser shots per spectra and a fixed laser intensity of 3,500 V. Calibration was performed using Des-Arg-bradykinin (904.468 Da), angiotensin 1 (1,296.685 Da), Glu-Fibrinopeptide B (1,570.677 Da), ACTH (1–17 clip) (2,093.087 Da), and ACTH (18–39 clip) (2,465.199 Da) (Calibration Mix from Applied Biosystems). Fifteen s/n best precursors from each MS spectrum were selected for MS/MS analysis. MS/MS analyses were performed using collision-induced dissociation (CID) assisted with air, using collision energy of 1 kV and a gas pressure of 106 Torr. Two thousand laser shots were collected for each MS/MS spectrum using a fixed laser intensity of 4,500 V. Raw data were generated using 4000 Series Explorer Software v3.0 RC1 (Applied Biosystems, Foster City, CA, USA), and all contaminant m/z peaks originating from human keratin, trypsin autodigestion, or matrix were placed on the exclusion list used to generate the peptide mass list used in the database search [17].
Database query. To identify proteins, Mascot generic format files combining MS and MS/MS spectra were used to interrogate a non-redundant protein database using a local Mascot v2.2 license from Matrix Science and the Global Protein Server (GPS) v3.6 (Applied Biosystems). Search parameters for the MS/MS spectra were as follows: i) the Uniprot (2013) sequence database (E. ruminantium with isoforms) was used; ii) taxonomy was set to “all entries” (302,409); iii) variable modifications were considered [i.e., carbamidomethylation (Cys), deamidation (Asn and Gln), and oxidation (Met, Pro, Lys, Arg)]; iv) two missed cleavage sites were allowed; v) precursor tolerance was set to 50 ppm and MS/MS fragment tolerance to 0.5 Da; vi) peptide charge was 1+; and vii) the algorithm used trypsin as the enzyme. A protein candidate provided by this MS/MS search was considered valid if the global Mascot score was >40 at a significance level of p<0.05, if at least one peptide was identified with 95% confidence, and if it was found in at least two of the three biological replicates.
In silico genome analysis
The publicly available proteome of the E. ruminantium strain Gardel, which was extracted from the Uniprot database [31] in FASTA format, was used for bioinformatics studies. The subcellular localization of the 948 E. ruminantium protein-coding genes was predicted using three global programs: PSORTb 3.0 [32], CELLO 2.5 [33], and MetaLocGramN [34]. The predicted utilization locations of each protein were filtered from raw software output using in-house scripts written in the R programming language and exported to Excel. In some cases, CELLO 2.5 predicted multiple localization sites for the same protein. The proteins involved were grouped under the heading “unknown localization.”
As a result of the varying predictions for a given protein, the consensus prediction was calculated using a majority vote procedure. If two of three algorithms agreed on localization, this localization was attributed to the protein. As for the remaining results, when outer or inner membrane localization was predicted by only one program, protein subcellular localization was refined manually, based on the experimental data in the literature, or the presence of signal peptides, transmembrane domains using dedicated algorithms (Table 1; S1 Table).
Table 1. Subcellular localization of E. ruminantium strain Gardel proteins as predicted by PSORTb 3.0, CELLO 2.5, MetaLocGramN, and consensus.
| Subcellular localization | PSORTb 3.0 | CELLO 2.5 | MetaLocGramN | Consensus prediction | ||||
|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | Number | % | |
| Cytoplasmic | 490 | 51.6 | 461 | 48.6 | 526 | 55.4 | 499 | 52.6 |
| Periplasmic | 4 | 0.4 | 9 | 0.9 | 1 | 0.1 | 1 | 0.1 |
| Inner Membrane | 198 | 20.8 | 109 | 11.4 | 192 | 20.2 | 124 | 13.0 |
| Extracellular | 9 | 0.9 | 23 | 2.4 | 158 | 16.6 | 16 | 1.6 |
| Outer membrane | 11 | 1.1 | 90 | 9.4 | 71 | 7.4 | 52 | 5.4 |
| Unknown | 236 | 24.8 | 256 | 27.0 | 0 | 0 | 256 | 27.0 |
| Total | 948 | 948 | 948 | 948 | ||||
Percentages correspond to the number of proteins in each compartment relative to the total number of proteins.
Results
Enrichment of E. ruminantium OM fraction
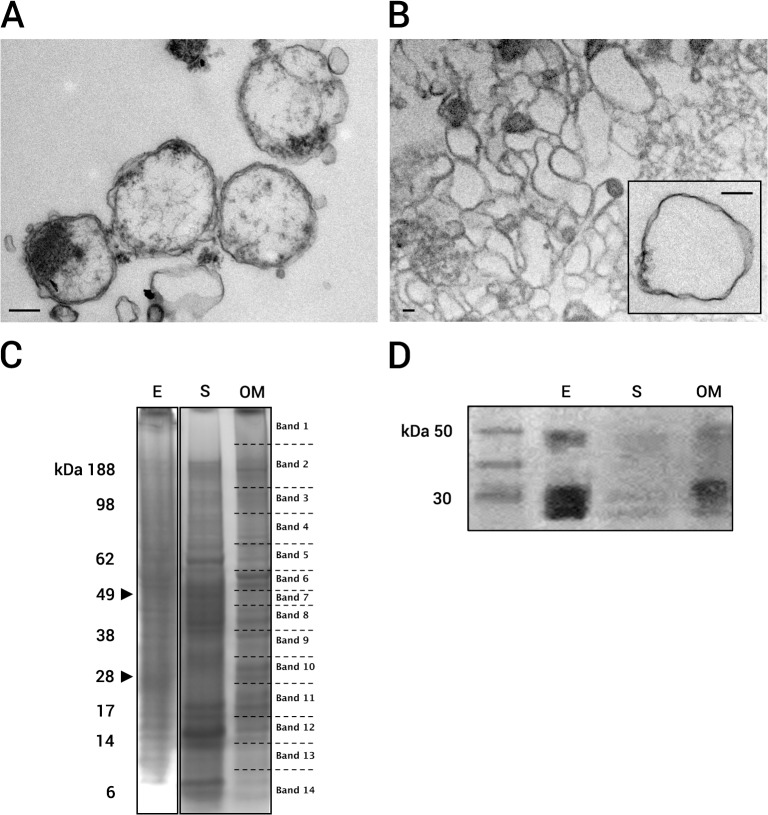
The first step in this study was to recover most of the OM complex with minimal contamination by cytoplasmic and inner membrane fractions. To do this, we used sarkosyl, an ionic detergent commonly used in the purification of OMs in Gram-negative bacteria, because it selectively solubilizes cytoplasmic and inner membranes while conserving the integrity of the OM [24]. Fig. 1 shows the workflow used to obtain the OM fraction. To assess protocol efficacy, samples were harvested at critical time points during the purification process, and their quality was evaluated using TEM, SDS-PAGE to identify proteins in the most intense bands, and Western blotting (Fig. 2). After sarkosyl treatment of intact EBs (Fig. 2A), empty shells with spherical morphology, corresponding to the OM fraction, were observed (Fig. 2B). These OM complexes, with a diameter of approximately 200 nm, appeared to be devoid of inner membrane and cytoplasm components, in contrast to intact EBs (Fig. 2A). Comparative protein migration profiles of the different fractions (intact EBs, E; sarkosyl soluble fractions, S; and outer membrane fractions, OMs) were analyzed using SDS-PAGE (Fig. 2C), and each subcellular fraction displayed a distinct migration pattern. The OM preparation showed prominent bands at approximately 134, 63, 55, 41, 37, and 29 kDa. The most abundant proteins, in the 30 kDa range, may represent Map1 protein family. When the different fractions were analyzed via Western blot using a monoclonal antibody against Map1 (a specific OM marker), intact EBs (the positive control) displayed a strong ~30 kDa band corresponding to Map1 (Fig. 2D). This protein was detected in the OM fraction but not in the soluble fraction, confirming the efficacy of the purification protocol (Fig. 2D). Altogether, these results clearly indicate that the insoluble sarkosyl fraction was strongly enriched with E. ruminantium OM complexes.
Fig 2. Evaluation of OM isolation quality.
Transmission electron microscopy of (A) purified E. ruminantium and (B) the insoluble precipitate after 0.1% sarkosyl treatment; scale bar = 200 nm. (C) SDS-PAGE and (D) Western blot of E (elementary bodies), S (sarkosyl-soluble fraction), and OM (outer membrane fraction) using monoclonal antibodies against Map1. Band 1: Map1-14, X5HG56, GroEL; Band 2: Map1+1, Map1, Map1-6, VirB10, VirB4, GroEL, PyrE, Q5HAR6, X5HG56, 30S-S8; Band 3: Map1+1, Map1-6, Map2, GroEL, PyrE, Q5HAR6, X5HG56, Q5FGC2, Q5HBI2, Q5FHJ9; Band 4: Map1, Map1-6, VirB4, GroEL, DnaK, BamA, FusA, Pnp, Q5HAR6, X5HG56, Q5FH07, Q5HBS6; Band 5: VirB4, VirB10, VirB11, DnaK, HtpG, GroEL, FusA, 30S-S1, Q5FGV5, Q93FS2; Band 6: Map1, Map1-14, VirB10, PleD, GroEL, DnaK, FtsZ, 30S-S1, Q5HB83, Q5FGA7, Q5HBE1; Band 7: Map1-14, GroEL, DnaK, FtsZ, HtpG; Band 8: Map1-6, Map1, GroEL, DnaK, FtsZ, BamA; Band 9: Map1-6, Map1, Map1+1, Map1-14, GroEL, DnaK, BamA, Q5FFE6, Q5HAR6; Band 10: Map1-11, Map1-13, Map1, Map1+1, Map1-6, VirB10, VirB9, Q5FFE6, Q5HAR6, Q5HBI2, Q5HA95; Band 11: Map1, Map2, BamA, DnaK, GroEL, FusA, Def, 50S-L4, PyrE, X5HG56, Q5HBI2; Band 12: 30S-S18, 30S-S12, 50S-L7/L12, 50S-L18, 50S-L24, 50S-L28 X5HG56, Q5HBN6; Band 13: HupB, X5HG56; Band 14: 30S-S12, 50S-L7/L12, 50S-L18, GroEL, YajC, PyrE.
In silico subcellular localization prediction of E. ruminantium proteins
We utilized a combination of three computational prediction tools, CELLO 2.5, PSORTb 3.0, and MetaLocGramN, to predict subcellular localization in the entire E. ruminantium proteome. These programs have been used to identify OMPs in several Gram-negative bacterial species [35–37]. Though the programs made diverse subcellular localization predictions for the same proteins, the combination of different predictors minimizes the risk of false positives for OMP prediction. PSORTb 3.0, CELLO 2.5, and MetaLocGramN predicted 490, 461, and 526 cytoplasmic proteins in E. ruminantium (~50% of total proteins), respectively (Table 1). CELLO 2.5 predicted 11.5% of proteins were inner membrane proteins (IMPs), whereas the two other programs predicted roughly twice as many (20%). CELLO 2.5 identified the highest proportion of OMPs (9.4%, 90/948), followed by MetaLocGramN (7.4%, 71/948) and PSORTb 3.0 (1.1%, 11/948). PSORTb 3.0 could not predict the localization of 236 proteins, while CELLO could not provide predictions for 256.
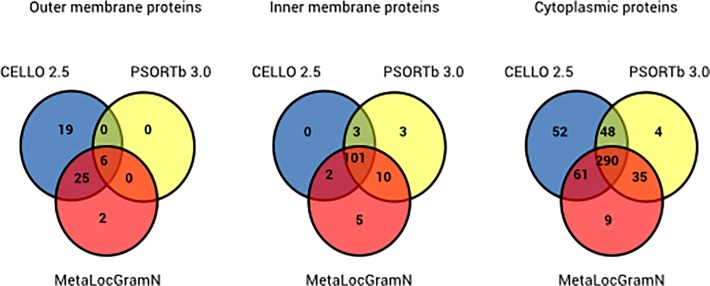
Altogether, we predicted that the total proteome of E. ruminantium (948 proteins) consisted of 53% (499/948) cytoplasmic proteins, 13% (124/948) IMPs, and 5.4% (52/948) OMPs (Table 1). In Fig. 3, the number of proteins in each Venn diagram compartment corresponds the consensus prediction correctly predicted by an algorithm for a given subcellular localization. Of the 52 OMPs identified using consensus predictions, 6 were identified by all three programs. Twenty-one were predicted by only a single program: 19 for CELLO 2.5 and 2 for MetaLocGramN. CELLO 2.5 predicted the highest number of consensus OMPs (50), followed by MetaLocGramN (33) and PSORTb 3.0 (6). All three programs identified two hundred and ninety cytoplasmic proteins. CELLO 2.5 predicted the highest number of cytoplasmic proteins, whereas PSORTb 3.0 predicted the lowest.
Fig 3. Venn diagram representing the predicted subcellular localization of E. ruminantium proteins using PSORTb 3.0, CELLO 2.5, and MetaLocGramN.
The data presented result from consensus prediction of subcellular localization.
Identification of proteins in the E. ruminantium OM fraction
OM fractions prepared from three biological replicates were analyzed individually using 1DE-nanoLC-MALDI-TOF/TOF MS. The proteins identified are presented in Table 2. Of the 46 non-redundant proteins identified in the OM fraction, 41 had known functions (either characterized experimentally or annotated via high sequence similarity), and the remaining five proteins were classified as hypothetical proteins. Several of these proteins (e.g. ERGA_CDS_04510, ERGA_CDS_04580) are conserved among members of Anaplasmataceae. Of the 46 proteins identified, 39% were indeed OMPs (18/46), 11% were IMPs (5/46), and 50% (23/46) were cytoplasmic. These proteins were classified into four functional groups: structural and transport proteins, biogenesis proteins (e.g. BamA, ERGA_CDS_08660), virulence proteins, and proteins involved in metabolic processes (e.g. GroEL, ERGA_CDS_06640 and Ef-Tu, ERGA_CDS_01580). Several ribosomal proteins and chaperones were also identified. Of the 18 OMPs identified, 5 belonged to the well-known MAP1 family (Map1, Map1+1, Map1-6, Map1-13, and Map1-14), 2 comprised β-barrel assembly machinery (BamA and BamD), 3 were components of the type IV secretion system (VirB9-1, VirB9-2, and VirB10), 1 was a porin, and 1 was a major ferric iron-binding protein. The six putative uncharacterized proteins had neither functional annotations in UniProt, nor hits in the Pfam database. Two of these (ERGA_CDS_04580, ERGA_CDS_05150) were predicted by SignalP to contain signal peptides. The first had no homology with known proteins and seemed to be unique in the E. ruminantium genome, whereas the second had similarity to ECH_0525, an ortholog of Esp73, an OMP in Anaplasma phagocytophilum.
Table 2. Proteins identified in the outer membrane fraction of E. ruminantium via 1DE-nanoLC-MALDI-TOF/TOF.
| Locus tag | Protein | Acession Number | Function | Protein MM(kDa) | Number of peptidesa | Protein scoreb | Coverage (%) | PSORTb 3.0 | CELLO 2.5 | MetaLocGramN | Consensus prediction | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Outer membrane proteins (39%) | |||||||||||||
| ERGA_CDS_00150 | VirB10 | Q5HCE9 | Virulence | 48.717 | 2 | 294 | 9 | I | E | P | C | C | OM |
| ERGA_CDS_00160 | VirB9–2 | Q5HCE8 | Virulence | 30.993 | 2 | 86 | 6 | C | OM | C | OM | ||
| ERGA_CDS_01230 | Possible major ferric iron binding protein | Q5FFA9 | Transport/virulence | 41.309 | 2 | 188 | 14 | OM | C | OM | I | OM | |
| ERGA_CDS_02370 | Hypothetical protein | Q5FFH4 | Unknown | 37.402 | 3 | 259 | 26 | U | OM | E | OM | ||
| ERGA_CDS_02510* | Hypothetical protein | Q5HBS6 | Unknown | 90.496 | 1 | 26 | 1 | U | OM | OM | OM | ||
| ERGA_CDS_03960* | Hypothetical protein | Q5HBE1 | Unknown | 55.237 | 1 | 23 | 2 | U | OM | E | OM | ||
| ERGA_CDS_04510 | Hypothetical protein | Q5FGV5 | Unknown | 134.574 | 1 | 124 | 2 | U | C | OM | OM | OM | |
| ERGA_CDS_04580 | Putative exported protein | Q5HB83 | Porin | 41.826 | 9 | 832 | 47 | U | OM | OM | OM | ||
| ERGA_CDS_05150# | Putative exported protein | Q5FH07 | Unknown | 63.139 | 18 | 827 | 27 | OM | OM | OM | OM | ||
| ERGA_CDS_07300 | Hypothetical outer membrane protein | Q93FS2 | Cell struture | 28.127 | 2 | 107 | 18 | U | C | E | OM | ||
| ERGA_CDS_07840 | VirB9–1 | Q5HAC9 | Virulence | 29.489 | 2 | 153 | 10 | I | C | OM | C | OM | |
| ERGA_CDS_08100 | Putative exported lipoprotein | Q5HAA5 | Outer membrane assembly | 29.344 | 1 | 119 | 10 | OM | C | OM | OM | OM | |
| ERGA_CDS_08660 | Outer membrane protein omp1 | Q5FGI9 | Outer membrane assembly | 87.257 | 2 | 173 | 5 | OM | OM | OM | OM | ||
| ERGA_CDS_09000 | Map1-13 | Q4L0D3 | Cell struture | 32.965 | 4 | 419 | 28 | I | OM | P | C | OM | |
| ERGA_CDS_09010 | Map1-14 | Q4W4X7 | Cell struture | 34.186 | 1 | 122 | 7 | U | OM | OM | OM | ||
| ERGA_CDS_09090 | Map1-6 | Q4L0C5 | Cell struture | 33.736 | 4 | 529 | 36 | OM | OM | OM | OM | ||
| ERGA_CDS_09160 | Map1 | Q46330 | Cell structure | 31.204 | 5 | 948 | 38 | OM | E | E | OM | ||
| ERGA_CDS_09170 | Map1+1 | Q4L0B8 | Cell structure | 31.817 | 3 | 136 | 14 | U | OM | OM | OM | ||
| Inner membrane proteins (11%) | |||||||||||||
| ERGA_CDS_01470 | Major antigenic protein 2 SCO2 like-protein | Q9R416 | Cell struture | 23.562 | 1 | 79 | 10 | I | P | C | I | I | |
| ERGA_CDS_03170 | Phosphatidylserine decarboxylase proenzyme | Q5FHJ9 | General metabolism | 25.245 | 1 | 103 | 7 | I | I | I | I | ||
| ERGA_CDS_05400 | VirB4 | Q5FFK8 | Virulence | 90.865 | 1 | 82 | 2 | I | C | C | I | ||
| ERGA_CDS_06350 | Putative Protease IV | Q5HAR6 | General metabolism | 32.263 | 2 | 157 | 14 | I | C | OM | I | I | |
| ERGA_CDS_08130 | Preprotein translocase. YajC subunit | X5HHA7 | Cellular processes and signaling | 13 | 1 | 43 | 15 | I | C | P | I | I | |
| Cytoplasmic proteins (50%) | |||||||||||||
| ERGA_CDS_01570 | Elongation factor G | Q5FFE7 | Protein synthesis | 76.042 | 1 | 40 | 3 | C | C | C | C | ||
| ERGA_CDS_01580 | Elongation factor Tu | Q5FFE6 | Protein synthesis | 43.282 | 5 | 344 | 17 | C | C | C | C | ||
| ERGA_CDS_01760 | Peptide deformylase | Q5HBZ5 | Cell process | 21.926 | 1 | 43 | 9 | C | C | C | C | ||
| ERGA_CDS_02930 | Putative DNA-binding protein HU-beta | Q5HBN6 | Cell process | 10.665 | 1 | 94 | 19 | U | C | P | E | C | |
| ERGA_CDS_03000# | Helix-turn-helix domain protein | X5HG56 | DNA binding | 12250 | 1 | 40 | 6 | U | C | E | C | ||
| ERGA_CDS_03230 | Response regulator pleD | Q5HBK9 | Regulation | 52.358 | 1 | 154 | 5 | C | C | C | C | ||
| ERGA_CDS_03510 | Putative peroxiredoxin | Q5HBI2 | General metabolism | 23.349 | 8 | 760 | 52 | C | C | C | C | ||
| ERGA_CDS_03570 | Polyribonucleotide nucleotidyltransferase | Q5FHK5 | General metabolism | 86.507 | 1 | 77 | 3 | C | OM | C | C | ||
| ERGA_CDS_07810 | Inosine-5'-monophosphate dehydrogenase | Q5FGA7 | General metabolism | 52.348 | 1 | 106 | 6 | C | C | C | C | ||
| ERGA_CDS_08210 | Putative response regulator | Q5HA95 | Regulation | 30.477 | 2 | 118 | 9 | C | C | C | C | ||
| ERGA_CDS_08900 | Orotate phosphoribosyltransferase | Q5FGJ6 | General metabolism | 22.343 | 1 | 57 | 7 | C | C | C | C | ||
| ERGA_CDS_09220 | Cell division protein FtsZ | A1XRC7 | Cell division | 46.126 | 1 | 58 | 4 | I | C | E | I | C | |
| Chaperones | |||||||||||||
| ERGA_CDS_02450 | Chaperone protein HtpG | Q5FHC4 | Chaperone | 72485 | 1 | 60 | 3 | C | C | C | C | ||
| ERGA_CDS_05670 | Chaperone protein DnaK | Q5FFM4 | Chaperone | 69. 957 | 2 | 183 | 6 | C | C | C | C | ||
| ERGA_CDS_06640 | Chaperonin. 60 kDA (GroEL) protein | Q5FFZ1 | Chaperonin | 58. 859 | 16 | 1473 | 43 | C | C | C | C | ||
| Ribosomal proteins | |||||||||||||
| ERGA_CDS_01550 | 30S ribosomal protein S12 | Q5FFE9 | Translation | 13.671 | 1 | 49 | 23 | C | P | C | C | C | |
| ERGA_CDS_01640 | 50S ribosomal protein L7/L12 | Q5HC06 | Translation | 14.269 | 1 | 59 | 15 | C | C | C | C | ||
| ERGA_CDS_05500 | 50S ribosomal protein L28 | Q5FFP1 | Translation | 11.53 | 1 | 63 | 18 | C | C | C | C | ||
| ERGA_CDS_06130 | 50S ribosomal protein L18 | Q5FFR4 | Translation | 14.051 | 1 | 39 | 11 | C | C | C | C | ||
| ERGA_CDS_06150 | 30S ribosomal protein S8 | Q5FFV4 | Translation | 14.668 | 1 | 105 | 11 | C | C | OM | P | C | C |
| ERGA_CDS_06180 | 50S ribosomal protein L24 | Q5FFV1 | Translation | 11.7 | 1 | 57 | 28 | C | P | C | E | C | C |
| ERGA_CDS_06280 | 50S ribosomal protein L4 | Q5FFU1 | Translation | 23.28 | 1 | 55 | 9 | C | C | P | C | C | |
| ERGA_CDS_06340* | 30S ribosomal protein S1 | Q5HAR7 | Translation | 63.425 | 1 | 22 | 2 | C | OM | C | C | ||
Their predicted subcellular localization is shown by U, unknown; C, cytoplasmic; I, inner membrane; O, outer membrane; E, extracellular; P, periplasmic.
aNumber of unique peptides that match the sequence of the identified protein
bMASCOT Score—Identified proteins were only considered if a protein score above 40 was obtained (p<0.05)
#Hypothetical/uncharacterized proteins that had a significant hit on the BLASTp searches. The name of the BLASTp search best hit is here presented.
* proteins identified below the Mascot score (>20) and considered for the study as peptides were checked and interpreted manually to confirm the MASCOT suggestion
In summary, our study increased the number of OMPs experimentally identified accounting for 34% of total predicted OMPs in E. ruminantium (18/52), whereas the total number OMPs account only for 5.5% of E. ruminantium proteome (52/948). Thus, the OM purification process described enriched OMPs.
Discussion
The OM of Gram-negative bacteria is an important interface between the outside and inside of the cell. It protects bacteria against hostile environments. OMPs fulfill a number of crucial functions, such as supporting the biogenesis and integrity of the OM and acting as porins and virulence factors, playing a fundamental role in adherence to host cells, invasion, and evasion of host-defense mechanisms [38].
The purification of OMs is a key step in the identification of OMPs. Several methods, such as isopycnic centrifugation using a sucrose gradient, addition of Triton X-100, and carbonate extraction protocols, have been tested in bacteria [9–11,39]. However, the sarkosyl solubilization strategy, which solubilizes IM proteins and separates IM and OM proteins [24], has become the preferred method for many Gram-negative bacteria, due to the higher purity and better reproducibility of the OM extracts obtained in this manner [13,40,41]. By applying this method to E. ruminantium EBs, we obtained a highly enriched OM fraction. Our proteomic analysis led to the identification of 18 unique OMPs corresponding to 34% of total cell OMPs. The low percentage of sarkosyl-insoluble proteins obtained may be due to excessive washing of the pellets after sarkosyl treatment, resulting in loss of proteins or lysis of cells [10,25]. In addition, OMP extraction was performed on the extracellular, infectious form of Ehrlichia. It is likely that only certain E. ruminantium proteins are expressed at a given life cycle stage [42]. For instance, expression of most E. chaffeensis proteins varies depending on host and vector environments and stage of development [43,44].
We also analyzed the entire E. ruminantium proteome to determine the theoretical subcellular localization of all proteins (OM, IM, cytoplasmic, periplasmic, or extracellular). These in silico predictions allowed us to estimate the quality of the enrichment of OMPs in the OM fraction obtained using our purification protocol. PSORTb 3.0 is one of the most precise subcellular localization predictor for many Gram-negative bacteria [32]. It uses a combination of factors based on motif and profile analyses, e.g. the presence of signal peptides, OM motifs, transmembrane helices, and similarity to proteins with known localization [32]. However, in this study, it returned a high number of proteins with unknown localization (236 or 24.8% of total proteins). This problem may be due to the absence of significant sequence similarity between some E. ruminantium proteins and proteins in the PSORTb 3.0 database. Similar results have been observed in numerous other bacteria [34]. Consequently, we chose two other computational localization predictors to overcome this weakness. CELLO 2.5 has the advantage of using multiple Support Vector Machines (SVMs) to analyze four types of protein descriptors, including amino acid composition, dipeptide composition, partitioned amino acid composition, and frequency of residues with particular physicochemical properties [33], yielding better predictive performance [33]. However, in our study, CELLO 2.5 predicted multiple localization sites for 256 proteins that were subsequently grouped in a “unknown localization” category [35]. Finally, we included MetaLocGramN program, a meta-predictor that combines multiple primary methods, including general subcellular localization, signal peptide predictors, transmembrane helix predictors, and beta barrel OMP predictors [34]. The combination of results from these three programs improved the accuracy of subcellular localization predictions [9,35,45].
Collectively, our bioinformatics analysis predicts that 5.4% of the annotated genes in the E. ruminantium genome are OMPs. Analyses of other Gram-negative bacteria have identified approximately the same percentage of predicted OMPs. For example, an analysis employing 10 different predictors to analyze the Pasteurella multocida genome identified 98 OMPs in an avian strain and 107 in a porcine strain (4.8% and 5.0% of total proteins, respectively) [46]. Similarly, prediction of the subcellular localization of P. syringae Lz4W proteins, performed using PSORTb 3.0, revealed that 148 out of a total of 1,479 proteins (10%) were OMPs [11]. In addition, we compared our results to those obtained experimentally from many other bacteria. In L. pneumophila, OM and surface-exposed proteome analyses using cellular fractionation and fluorescent labeling led to the identification of OMPs accounting for 8.5% of total proteins [12]. These results suggest that our prediction of E. ruminantium OMPs yielded a reasonable identification rate.
We experimentally identified a total of 46 non-redundant proteins in the OM fraction, 18 of which were clearly classified as OMPs. These 18 OMPs correspond to 1.9% of the entire E. ruminantium proteome (18/948) and 34.6% of predicted OMPs in the entire proteome (18/52). Previous studies on the total E. ruminantium proteome have identified 64 non-redundant proteins including 8 OMPs [17]. Thus, as expected, enriching the OM fraction resulted in an increased number of OMPs being identified. Some of these OMPs have known functions and include proteins of the Map1 cluster [47], BamA/D [48], VirB9-1 [49], VirB9-2, VirB10 [50], a porin [51], and major ferric iron-binding protein [52]. We also characterized five proteins classified as hypothetical but predicted to be OMPs, including ERGA_CDS_04510, 03960, 02510, 02370, and 05150. BLAST search on ERGA_CDS_05150 revealed an ortholog in Ehrlichia chaffeensis, Esp73; an ortholog to A. phagocytophilum Asp55 and Asp62, that is predicted to contain 22 transmembrane β-strands forming a β-barrel and, thus, may be involved in membrane transport [53]. Further functional characterization of these newly discovered OMPs should be carried out to evaluate their potential as protective antigens.
Map1, the immunodominant, major OMP expressed by E. ruminantium in the mammalian host, is encoded by a member of a multigene family comprising 16 paralogs [54]. The number of Map1 family proteins detected in this study (n = 5: Map1, Map1+1, Map1-6, Map1-14, and Map1-13) was greater than that detected in a previous proteomic analysis [17]. These proteins are known to be differentially transcribed in vitro in endothelial and tick cell cultures [54,55] and are well conserved, since omp-1, msp2, p44, p30, and map-1 belong to a superfamily harboring the PF01617 Pfam domain [1]. Map1 family proteins are considered priority targets for candidate vaccines [56], as they are potentially involved in E. ruminantium adaptation to the mammalian host and its vector, the tick [18]. However, few data are currently available on the expression and characterization of Map1 family proteins throughout the bacterial life cycle [17].
Proteins of the β-barrel Assembly Machinery (BAM) complex are involved in diverse cellular functions, including solute transport, protein secretion, and assembly of protein and lipid components of the OM [57]. They account for the vast majority of bacterial OMPs and are essential for bacterial viability and function [58]. The insertion of proteins in the OM depends on a protein complex that contains the OMP BamA and four associated lipoproteins (BamB, C, D, and E) [59]. BamA (ERGA_CDS_08660) and BamD (ERGA_CDS_08100) were identified in our experimental analysis. BamA proteins are essential for the biogenesis of β-barrel OMPs and play a central part in OMP assembly [60–62]. It has been observed that reducing the levels of BamA significantly affects the ability of the β-barrel membrane protein OprF to localize to the OM, showing its essential role in OM biogenesis [61]. BamD is the only essential lipoprotein in the BAM complex [63], and it is highly conserved in Gram-negative bacteria as well [64].
Many bacterial species use specialized secretion systems to transfer macromolecules across membranes [65]. The type IV secretion system (T4SS) translocates DNA or proteins across membranes directly into eukaryotic host cells to subvert host cellular functions. Consequently, the proteins that make up this system represent crucial bacterial virulence determinants in important human pathogens such as B. henselae, Helicobacter pylori, L. pneumophila, Bordetella pertussis, and Brucella melitensis [66,67]. In this study, we identified three conserved pathogenesis-associated proteins: VirB4, VirB9, and VirB10. VirB9 is an OM component of the T4SS and is hypothesized to be a translocation pore [68,69]. It is essential for the stability of the translocation machinery and substrate selection [69]. It interacts with VirB10, which bridges the IM and OM protein subcomplexes, and actively participates in T4SS substrate transfer across the bacterial envelope [12,70–72]. VirB4 is an ATPase, providing energy for substrate export and pilus biogenesis, and it interacts with several other VirB proteins, such as VirB10 [50]. It is not surprising, then, to identify such proteins in the E. ruminantium OM fraction. Moreover, a recent study showed that some T4SS components could be potential vaccine candidate for pathogenic bacteria [49].
We also identified a porin (ERGA_CDS_04580) that has no homology to other proteins and that seems to be unique to E. ruminantium. Porins play a fundamental role in pathogenicity [51], participating in adhesion to and invasion of host cells and evasion of host defense mechanisms [73]. They represent good targets for therapeutic development. Some porins activate immunological responses, induce signaling pathways, and modify the properties of the OM lipid barrier [73]. It would be interesting to further investigate the role of this porin with functional studies.
The periplasmic major ferric iron binding protein of Gram-negative bacteria (ERGA_CDS_01230), which has homologous counterparts in many other pathogenic species, plays a key role in the acquisition of iron from mammalian host serum iron transport proteins; thus, it is essential for the survival of the pathogen within the host [40,74].
Within the cell, the full-length protease (ERGA_CDS_06350), may be processed into the intermediate 45 kDa form, which represents a form of protease IV that lacks the signal sequence. This 45 kDa intermediate may undergo a conformational change that activates its protease activity, triggering the cleavage of the propeptide from the mature protease domain. The mature protease IV may be secreted through the OM, functioning in the developmental cycle [75,76] and as an important virulence factor [77].
In this study, we detected the chaperones DnaK and GroEL in the OM fraction, though they are depicted as cytoplasmic proteins. These results are not surprising, as these proteins are often membrane-associated [13,78]. In many bacteria, such as L. pneumophila and Borrelia burgdorferi [12,79], GroEL (Hsp60) is found in the OM and plays a role in the folding of a large number of proteins; in other bacteria, this protein is active in bacterial adhesion [80,81]. Similarly, in E. chaffeensis, the chaperone proteins GroEL and DnaK, and the translation elongation factor G, are localized to the membrane surface [82]. GroEL has also been detected on the surfaces of H. pylori [83], L. pneumophila [84], Haemophilus ducreyi [85], and Clostridium difficile [80] via immunofluorescence or immunoelectron microscopy. Finally, DnaK has been detected on the surface of H. pylori [83]. Other important cytoplasmic proteins identified in our study (FusA, TypA, EF-Tu, and Tig) are associated with ribosomes but can be membrane-associated during the transport of nascent OMPs across the periplasmic space to the OM [86]. Recently, EF-Tu was shown to be membrane-associated, secreted in outer membrane vesicles (OMVs), and immunogenic during Burkholderia infection in a murine model of melioidosis [87]. Therefore, we cannot deny the possibility that these proteins with well-known functions in the cytoplasmic, periplasmic, or inner membrane are present in the OM of E. ruminantium and play unexpected roles in E. ruminantium -host interaction.
Surprisingly, we also detected ribosomal proteins with a predicted cytoplasmic localization. These proteins may represent a contamination with cytoplasmic proteins. Such proteins have also been identified in OM fractions of Pseudomonas and Yersinia strains, however [88,89]. Moreover, it should be noted that among these ribosomal proteins, we obtained a majority of 50S ribosomal subunits, as has been shown in Legionella [12]. Interestingly, one ribosomal protein we found in the OM fraction (ERGA_CDS_01640) has been predicted by S4TE software as a putative type IV effector [27]. Type IV effectors are proteins produced by pathogenic bacteria to manipulate host cell gene expression and other processes and have been shown to be critical for pathogenicity, making them salient targets for understanding bacterial virulence [90]. The function of this particular protein and its role in E. ruminantium pathogenicity is currently under investigation.
Conclusion
This study provides the first proteomic profile of the Ehrlichia ruminantium OM. The combination of subcellular fractionation via sarkosyl solubilization and a high degree of accuracy in predicting OMP status allowed us to generate a high-resolution OM proteome comprised of 46 proteins identified in the OM fraction. We identified OMPs involved in cell wall structure, i.e. at the interface between bacteria and host cells, and proteins known to be virulence factors. Moreover, we identified new OMPs by our approach coupling a consensus of computer algorithms, manual sequence analysis and experimental proteomics. In the future, functional studies should explore the potential of using these OMPs as vaccine candidates against E. ruminantium.
Supporting Information
948 proteins were analyzed using 3 bioinformatic predictors and the resulting consensus prediction is indicated.
(XLSX)
Acknowledgments
We are grateful to Elisabete Pires and Renata Soares for technical assistance with MS/MS analysis, and David Pleydell for help with bioinformatics analysis. We thank C3MAG platform for assistance with Transmission Electron Microscopy.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors acknowledge financial support from FEDER grant FED 1/1.4-30305, 2007–2013, “Risque en santé animale et végétale” (PhD grant to AM), a PAUILF grant (Programme des Actions Universitaires Intégrées Luso-Françaises), 2011–2012, “Identification des protéines de la membrane externe de la Rickettsiales Ehrlichia ruminantium par une approche de protéomique,” and the Fundação para a Ciência e Tecnologia (FCT, Lisbon, Portugal; research project PTDC/CVT/114118/2009, research grant PEst-OE/EQB/LA0004/2011, and Post-doc grant SFRH/ BPD/ 45978/ 2008 to IM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Allsopp BA (2010) Natural history of Ehrlichia ruminantium . Vet Parasitol 167: 123–135. 10.1016/j.vetpar.2009.09.014 [DOI] [PubMed] [Google Scholar]
- 2. Jongejan F, Zandbergen TA, van de Wiel PA, de Groot M, Uilenberg G (1991) The tick-borne rickettsia Cowdria ruminantium has a Chlamydia-like developmental cycle. Onderstepoort J Vet Res 58: 227–237. [PubMed] [Google Scholar]
- 3. Vachiery N, Marcelino I, Martinez D, Lefrancois T (2013) Opportunities in diagnostic and vaccine approaches to mitigate potential heartwater spreading and impact on the American mainland. Dev Biol (Basel) 135: 191–200. 10.1159/000190050 [DOI] [PubMed] [Google Scholar]
- 4. Faburay B, Geysen D, Ceesay A, Marcelino I, Alves PM, et al. (2007) Immunisation of sheep against heartwater in The Gambia using inactivated and attenuated Ehrlichia ruminantium vaccines. Vaccine 25: 7939–7947. [DOI] [PubMed] [Google Scholar]
- 5. Jongejan F, Vogel SW, Gueye A, Uilenberg G (1993) Vaccination against heartwater using in vitro attenuated Cowdria ruminantium organisms. Rev Elev Med Vet Pays Trop 46: 223–227. [PubMed] [Google Scholar]
- 6. Mahan SM, Smith GE, Kumbula D, Burridge MJ, Barbet AF (2001) Reduction in mortality from heartwater in cattle, sheep and goats exposed to field challenge using an inactivated vaccine. Vet Parasitol 97: 295–308. [DOI] [PubMed] [Google Scholar]
- 7. Pretorius A, Collins NE, Steyn HC, van Strijp F, van Kleef M, et al. (2007) Protection against heartwater by DNA immunisation with four Ehrlichia ruminantium open reading frames. Vaccine 25: 2316–2324. [DOI] [PubMed] [Google Scholar]
- 8. Raliniaina M, Meyer DF, Pinarello V, Sheikboudou C, Emboule L, et al. (2010) Mining the genetic diversity of Ehrlichia ruminantium using map genes family. Vet Parasitol 167: 187–195. 10.1016/j.vetpar.2009.09.020 [DOI] [PubMed] [Google Scholar]
- 9. Ayalew S, Confer AW, Hartson SD, Shrestha B (2010) Immunoproteomic analyses of outer membrane proteins of Mannheimia haemolytica and identification of potential vaccine candidates. Proteomics 10: 2151–2164. 10.1002/pmic.200900557 [DOI] [PubMed] [Google Scholar]
- 10. Hobb RI, Fields JA, Burns CM, Thompson SA (2009) Evaluation of procedures for outer membrane isolation from Campylobacter jejuni . Microbiology 155: 979–988. 10.1099/mic.0.024539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jagannadham MV, Abou-Eladab EF, Kulkarni HM (2011) Identification of outer membrane proteins from an Antarctic bacterium Pseudomonas syringae Lz4W. Mol Cell Proteomics 10: M110 004549. 10.1074/mcp.O111.015446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khemiri A, Galland A, Vaudry D, Chan Tchi Song P, Vaudry H, et al. (2008) Outer-membrane proteomic maps and surface-exposed proteins of Legionella pneumophila using cellular fractionation and fluorescent labelling. Anal Bioanal Chem 390: 1861–1871. 10.1007/s00216-008-1923-1 [DOI] [PubMed] [Google Scholar]
- 13. Rhomberg TA, Karlberg O, Mini T, Zimny-Arndt U, Wickenberg U, et al. (2004) Proteomic analysis of the sarcosine-insoluble outer membrane fraction of the bacterial pathogen Bartonella henselae . Proteomics 4: 3021–3033. [DOI] [PubMed] [Google Scholar]
- 14. Shevchuk O, Jager J, Steinert M (2011) Virulence properties of the Legionella pneumophila cell envelope. Front Microbiol 2: 74 10.3389/fmicb.2011.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weiner JH, Li L (2008) Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim Biophys Acta 1778: 1698–1713. [DOI] [PubMed] [Google Scholar]
- 16. Oleastro M, Menard A (2013) The Role of Helicobacter pylori Outer Membrane Proteins in Adherence and Pathogenesis. Biology (Basel) 2: 1110–1134. 10.3390/biology2031110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marcelino I, de Almeida AM, Brito C, Meyer DF, Barreto M, et al. (2012) Proteomic analyses of Ehrlichia ruminantium highlight differential expression of MAP1-family proteins. Vet Microbiol 156: 305–314. 10.1016/j.vetmic.2011.11.022 [DOI] [PubMed] [Google Scholar]
- 18. Postigo M, Taoufik A, Bell-Sakyi L, Bekker CP, de Vries E, et al. (2008) Host cell-specific protein expression in vitro in Ehrlichia ruminantium . Vet Microbiol 128: 136–147. [DOI] [PubMed] [Google Scholar]
- 19. Lopez JE, Palmer GH, Brayton KA, Dark MJ, Leach SE, et al. (2007) Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect Immun 75: 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohashi N, Zhi N, Zhang Y, Rikihisa Y (1998) Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun 66: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Futse JE, Brayton KA, Dark MJ, Knowles DP Jr., Palmer GH (2008) Superinfection as a driver of genomic diversification in antigenically variant pathogens. Proc Natl Acad Sci U S A 105: 2123–2127. 10.1073/pnas.0710333105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Felek S, Greene R, Rikihisa Y (2003) Transcriptional analysis of p30 major outer membrane protein genes of Ehrlichia canis in naturally infected ticks and sequence analysis of p30-10 of E. canis from diverse geographic regions. J Clin Microbiol 41: 886–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Unver A, Rikihisa Y, Stich RW, Ohashi N, Felek S (2002) The omp-1 major outer membrane multigene family of Ehrlichia chaffeensis is differentially expressed in canine and tick hosts. Infect Immun 70: 4701–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Filip C, Fletcher G, Wulff JL, Earhart CF (1973) Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol 115: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marcelino I, Verissimo C, Sousa MF, Carrondo MJ, Alves PM (2005) Characterization of Ehrlichia ruminantium replication and release kinetics in endothelial cell cultures. Vet Microbiol 110: 87–96. [DOI] [PubMed] [Google Scholar]
- 26. Marcelino I, Vachiery N, Amaral AI, Roldao A, Lefrancois T, et al. (2007) Effect of the purification process and the storage conditions on the efficacy of an inactivated vaccine against heartwater. Vaccine 25: 4903–4913. [DOI] [PubMed] [Google Scholar]
- 27. Meyer DF, Noroy C, Moumene A, Raffaele S, Albina E, et al. (2013) Searching algorithm for type IV secretion system effectors 1.0: a tool for predicting type IV effectors and exploring their genomic context. Nucleic Acids Res 41: 9218–9229. 10.1093/nar/gkt718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohashi N, Unver A, Zhi N, Rikihisa Y (1998) Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J Clin Microbiol 36: 2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gros O, Elisabeth NH, Gustave SD, Caro A, Dubilier N (2012) Plasticity of symbiont acquisition throughout the life cycle of the shallow-water tropical lucinid Codakia orbiculata (Mollusca: Bivalvia). Environ Microbiol 14: 1584–1595. 10.1111/j.1462-2920.2012.02748.x [DOI] [PubMed] [Google Scholar]
- 30. Franco CF, Santos R, Coelho AV (2011) Proteome characterization of sea star coelomocytes—the innate immune effector cells of echinoderms. Proteomics 11: 3587–3592. 10.1002/pmic.201000745 [DOI] [PubMed] [Google Scholar]
- 31. Consortium U (2011) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–75. 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu CS, Lin CJ, Hwang JK (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13: 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Magnus M, Pawlowski M, Bujnicki JM (2012) MetaLocGramN: A meta-predictor of protein subcellular localization for Gram-negative bacteria. Biochim Biophys Acta 1824: 1425–1433. 10.1016/j.bbapap.2012.05.018 [DOI] [PubMed] [Google Scholar]
- 35. Kulkarni HM, Swamy Ch V, Jagannadham MV (2014) Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res 13: 1345–1358. 10.1021/pr4009223 [DOI] [PubMed] [Google Scholar]
- 36. Roszczenko P, Radomska KA, Wywial E, Collet JF, Jagusztyn-Krynicka EK (2012) A novel insight into the oxidoreductase activity of Helicobacter pylori HP0231 protein. PLoS One 7: e46563 10.1371/journal.pone.0046563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stekhoven DJ, Omasits U, Quebatte M, Dehio C, Ahrens CH (2014) Proteome-wide identification of predominant subcellular protein localizations in a bacterial model organism. J Proteomics 99: 123–137. 10.1016/j.jprot.2014.01.015 [DOI] [PubMed] [Google Scholar]
- 38. Lin J, Huang S, Zhang Q (2002) Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes Infect 4: 325–331. [DOI] [PubMed] [Google Scholar]
- 39. Wolff S, Hahne H, Hecker M, Becher D (2008) Complementary analysis of the vegetative membrane proteome of the human pathogen Staphylococcus aureus . Mol Cell Proteomics 7: 1460–1468. 10.1074/mcp.M700554-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown RN, Romine MF, Schepmoes AA, Smith RD, Lipton MS (2010) Mapping the subcellular proteome of Shewanella oneidensis MR-1 using sarkosyl-based fractionation and LC-MS/MS protein identification. J Proteome Res 9: 4454–4463. 10.1021/pr100215h [DOI] [PubMed] [Google Scholar]
- 41. Papanastasiou M, Orfanoudaki G, Koukaki M, Kountourakis N, Sardis MF, et al. (2013) The Escherichia coli peripheral inner membrane proteome. Mol Cell Proteomics 12: 599–610. 10.1074/mcp.M112.024711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seo GM, Cheng C, Tomich J, Ganta RR (2008) Total, membrane, and immunogenic proteomes of macrophage- and tick cell-derived Ehrlichia chaffeensis evaluated by liquid chromatography-tandem mass spectrometry and MALDI-TOF methods. Infect Immun 76: 4823–4832. 10.1128/IAI.00484-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singu V, Liu H, Cheng C, Ganta RR (2005) Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infect Immun 73: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singu V, Peddireddi L, Sirigireddy KR, Cheng C, Munderloh U, et al. (2006) Unique macrophage and tick cell-specific protein expression from the p28/p30-outer membrane protein multigene locus in Ehrlichia chaffeensis and Ehrlichia canis . Cell Microbiol 8: 1475–1487. [DOI] [PubMed] [Google Scholar]
- 45. Kossmehl S, Wohlbrand L, Druppel K, Feenders C, Blasius B, et al. (2013) Subcellular protein localization (cell envelope) in Phaeobacter inhibens DSM 17395. Proteomics 13: 2743–2760. 10.1002/pmic.201300112 [DOI] [PubMed] [Google Scholar]
- 46. E-komon T, Burchmore R, Herzyk P, Davies R (2012) Predicting the outer membrane proteome of Pasteurella multocida based on consensus prediction enhanced by results integration and manual confirmation. BMC Bioinformatics 13: 63 10.1186/1471-2105-13-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peter TF, Burridge MJ, Mahan SM (2002) Ehrlichia ruminantium infection (heartwater) in wild animals. Trends Parasitol 18: 214–218. [DOI] [PubMed] [Google Scholar]
- 48. van den Berg B (2013) Lateral gates: beta-barrels get in on the act. Nat Struct Mol Biol 20: 1237–1239. 10.1038/nsmb.2709 [DOI] [PubMed] [Google Scholar]
- 49. Morse K, Norimine J, Palmer GH, Sutten EL, Baszler TV, et al. (2011) Association and evidence for linked recognition of type IV secretion system proteins VirB9–1, VirB9–2, and VirB10 in Anaplasma marginale . Infect Immun 80: 215–227. 10.1128/IAI.05798-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward DV, Draper O, Zupan JR, Zambryski PC (2002) Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci U S A 99: 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Achouak W, Heulin T, Pages JM (2001) Multiple facets of bacterial porins. FEMS Microbiol Lett 199: 1–7. [DOI] [PubMed] [Google Scholar]
- 52. Nowalk AJ, Tencza SB, Mietzner TA (1994) Coordination of iron by the ferric iron-binding protein of pathogenic Neisseria is homologous to the transferrins. Biochemistry 33: 12769–12775. [DOI] [PubMed] [Google Scholar]
- 53. Ge Y, Rikihisa Y (2007) Surface-exposed proteins of Ehrlichia chaffeensis . Infect Immun 75: 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Heerden H, Collins NE, Brayton KA, Rademeyer C, Allsopp BA (2004) Characterization of a major outer membrane protein multigene family in Ehrlichia ruminantium . Gene 330: 159–168. [DOI] [PubMed] [Google Scholar]
- 55. Bekker CP, Postigo M, Taoufik A, Bell-Sakyi L, Ferraz C, et al. (2005) Transcription analysis of the major antigenic protein 1 multigene family of three in vitro-cultured Ehrlichia ruminantium isolates. J Bacteriol 187: 4782–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frutos R, Viari A, Ferraz C, Morgat A, Eychenie S, et al. (2006) Comparative genomic analysis of three strains of Ehrlichia ruminantium reveals an active process of genome size plasticity. J Bacteriol 188: 2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tellez R Jr., Misra R (2012) Substitutions in the BamA beta-barrel domain overcome the conditional lethal phenotype of a DeltabamB DeltabamE strain of Escherichia coli . J Bacteriol 194: 317–324. 10.1128/JB.06192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67: 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rigel NW, Ricci DP, Silhavy TJ (2013) Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for beta-barrel assembly in Escherichia coli . Proc Natl Acad Sci U S A 110: 5151–5156. 10.1073/pnas.1302662110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Doerrler WT, Raetz CR (2005) Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J Biol Chem 280: 27679–27687. [DOI] [PubMed] [Google Scholar]
- 61. Hoang HH, Nickerson NN, Lee VT, Kazimirova A, Chami M, et al. (2011) Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. MBio 2: 00246–00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Werner J, Misra R (2005) YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli . Mol Microbiol 57: 1450–1459. [DOI] [PubMed] [Google Scholar]
- 63. Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, et al. (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli . Mol Microbiol 61: 151–164. [DOI] [PubMed] [Google Scholar]
- 64. Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, et al. (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli . Proc Natl Acad Sci U S A 104: 6400–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Voth DE, Broederdorf LJ, Graham JG (2012) Bacterial Type IV secretion systems: versatile virulence machines. Future Microbiol 7: 241–257. 10.2217/fmb.11.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Christie PJ (2009) Structural biology: Translocation chamber's secrets. Nature 462: 992–994. 10.1038/462992a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dehio C (2008) Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell Microbiol 10: 1591–1598. 10.1111/j.1462-5822.2008.01171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cascales E, Christie PJ (2003) The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ (2005) Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol 187: 3486–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alvarez-Martinez CE, Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73: 775–808. 10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cascales E, Christie PJ (2004) Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304: 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fronzes R, Christie PJ, Waksman G (2009) The structural biology of type IV secretion systems. Nat Rev Microbiol 7: 703–714. 10.1038/nrmicro2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Galdiero S, Falanga A, Cantisani M, Tarallo R, Della Pepa ME, et al. (2012) Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr Protein Pept Sci 13: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shouldice SR, Dougan DR, Skene RJ, Tari LW, McRee DE, et al. (2003) High resolution structure of an alternate form of the ferric ion binding protein from Haemophilus influenzae . J Biol Chem 278: 11513–11519. [DOI] [PubMed] [Google Scholar]
- 75.Hoge R, Pelzer A, Rosenau F, Wilhelm S (2010) Weapons of a pathogen: proteases and their role in virulence of Pseudomonas aeruginosa. In current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology FORMATEX. pp. 383–395.
- 76. Traidej M, Caballero AR, Marquart ME, Thibodeaux BA, O'Callaghan RJ (2003) Molecular analysis of Pseudomonas aeruginosa protease IV expressed in Pseudomonas putida . Invest Ophthalmol Vis Sci 44: 190–196. [DOI] [PubMed] [Google Scholar]
- 77. Engel LS, Hill JM, Caballero AR, Green LC, O'Callaghan RJ (1998) Protease IV, a unique extracellular protease and virulence factor from Pseudomonas aeruginosa . J Biol Chem 273: 16792–16797. [DOI] [PubMed] [Google Scholar]
- 78. Boonjakuakul JK, Gerns HL, Chen YT, Hicks LD, Minnick MF, et al. (2007) Proteomic and immunoblot analyses of Bartonella quintana total membrane proteins identify antigens recognized by sera from infected patients. Infect Immun 75: 2548–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Scopio A, Johnson P, Laquerre A, Nelson DR (1994) Subcellular localization and chaperone activities of Borrelia burgdorferi Hsp60 and Hsp70. J Bacteriol 176: 6449–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hennequin C, Collignon A, Karjalainen T (2001) Analysis of expression of GroEL (Hsp60) of Clostridium difficile in response to stress. Microb Pathog 31: 255–260. [DOI] [PubMed] [Google Scholar]
- 81. Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K (1989) Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli . Embo J 8: 3517–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ge Y, Rikihisa Y (2007) Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J Bacteriol 189: 7819–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Huesca M, Borgia S, Hoffman P, Lingwood CA (1996) Acidic pH changes receptor binding specificity of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect Immun 64: 2643–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Garduno RA, Garduno E, Hoffman PS (1998) Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect Immun 66: 4602–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Frisk A, Ison CA, Lagergard T (1998) GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect Immun 66: 1252–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Herskovits AA, Shimoni E, Minsky A, Bibi E (2002) Accumulation of endoplasmic membranes and novel membrane-bound ribosome-signal recognition particle receptor complexes in Escherichia coli . J Cell Biol 159: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nieves W, Heang J, Asakrah S, Honer zu Bentrup K, Roy CJ, et al. (2010) Immunospecific responses to bacterial elongation factor Tu during Burkholderia infection and immunization. PLoS One 5: e14361 10.1371/journal.pone.0014361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Coquet L, Cosette P, De E, Galas L, Vaudry H, et al. (2005) Immobilization induces alterations in the outer membrane protein pattern of Yersinia ruckeri . J Proteome Res 4: 1988–1998. [DOI] [PubMed] [Google Scholar]
- 89. Seyer D, Cosette P, Siroy A, Dé E, Lenz C, et al. (2005) Proteomic comparison of outer membrane proteins patterns of sessile and planktonic Pseudomonas aeruginosa cells. Biofilms 2: 27–36. [Google Scholar]
- 90. Hicks SW, Galan JE (2013) Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol 11: 316–326. 10.1038/nrmicro3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
948 proteins were analyzed using 3 bioinformatic predictors and the resulting consensus prediction is indicated.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.