Abstract
The ERECT PANICLE 3 gene of rice encodes a peptide that exhibits more than 50% sequence identity with the Arabidopsis F-box protein HAWAIIAN SKIRT (HWS). Ectopic expression of the Os02g15950 coding sequence, driven by the HWS (At3g61950) promoter, rescued the hws-1 flower phenotype in Arabidopsis confirming that EP3 is a functional orthologue of HWS. In addition to displaying an erect inflorescence phenotype, loss-of-function mutants of Os02g15950 exhibited a decrease in leaf photosynthetic capacity and stomatal conductance. Analysis of a range of physiological and anatomical features related to leaf photosynthesis revealed no alteration in Rubisco content and no notable changes in mesophyll size or arrangement. However, both ep3 mutant plants and transgenic lines that have a T-DNA insertion within the Os02g15950 (EP3) gene exhibit smaller stomatal guard cells compared with their wild-type controls. This anatomical characteristic may account for the observed decrease in leaf photosynthesis and provides evidence that EP3 plays a role in regulating stomatal guard cell development.
Key words: Arabidopsis, ERECT PANICLE3, F-box protein, guard cell, HAWAIIAN SKIRT, photosynthesis, rice, stomatal conductance.
Introduction
Inflorescences of the erect panicle 3 (ep3) mutant in rice remain upright throughout grain filling. The phenotype is associated with an elevation in the number of small vascular bundles and an increase in the thickness of the parenchyma in the peduncle and a reduction in the number of spikelets per panicle. The EP3 gene has been cloned and the ep3 mutation attributed to a single base pair change in Os02g15950 that leads to the introduction of a premature termination codon (Piao et al., 2009).
A potential orthologue of EP3 in Arabidopsis is the gene HAWAIIAN SKIRT (At3g61950). The protein encoded by EP3 shares in excess of 50% sequence homology with HWS at the amino acid level. The hws-1 mutant is distinctive as its floral organs look as if they are attached to the developing silique throughout maturation and senescence. A detailed study of flower development has revealed that this is the consequence of the sepals being fused into a single whorl trapping the separated petals and anther filaments and preventing them from being shed (González-Carranza et al., 2007). Identification and characterization of the gene responsible for the mutant phenotype has revealed a 28bp deletion in a gene encoding an F-box protein leading to the synthesis of a truncated HWS peptide (González-Carranza et al., 2007). Phenotypic studies of hws-1, and transgenic lines ectopically expressing HWS, have shown that the gene may also regulate both the size of aerial organs and seeds (González-Carranza et al., 2007) and play a role in regulating stomatal distribution and aspects of chloroplast assembly (Z González-Carranza et al., unpublished data).
There is, therefore, reason to believe that the EP3 gene in rice (Oryza sativa) may have a role in determining anatomical, stomatal, and photosynthetic properties. There are known relationships in many species between photosynthetic rate and features of leaf anatomy such as vein density and mesophyll cell properties (McKown and Dengler, 2007; Flexas et al., 2012; Feldman et al., 2014). Stomatal properties are especially important in the context of balancing CO2 uptake with transpirational water loss and gaseous exchange through stomatal pores is optimized via a series of signalling pathways which have been well studied in Arabidopsis and other species (Raven, 2002; Bergmann and Sack, 2007; Kim et al., 2010). Over 60 genes have been reported to regulate stomatal development in Arabidopsis and detailed functional analysis has indicated that these contribute to cell-fate specification, cell polarity, cell division, and cell–cell communication (MacAlister et al., 2006; Ohashi-Ito and Bergmann, 2006; Pillitteri et al., 2006; Pillitteri and Torii, 2012). The mechanisms responsible for stomatal development and signal transduction in rice have been less well studied. However, it has been highlighted recently that both stomatal and mesophyll conductance have an effect on photosynthetic potential in this crop species (Kusumi et al., 2012; Adachi et al., 2013).
In this study, our attention was focused on the physiological impact of the EP3 gene on photosynthesis in rice as there is evidence that its putative orthologue in Arabidopsis can influence stomatal development. Our results show that EP3 is a functional orthologue of HWS and that the gene has a role in regulating rice photosynthesis via stomatal development. This study has practical as well as fundamental implications as rice is one of the most important crops in the world and feeds over half of the world’s population. Further improvement of potential yield in rice is considered to require an increase in photosynthetic efficiency (Zhu et al., 2010). However, the improvement of rice photosynthesis and water use efficiency is a major challenge for crop physiologists and an understanding of how leaf morphology, stomatal development, and guard cell aperture is controlled is integral to this goal (Zhang, 2007; Xing and Zhang, 2010; Zhu et al., 2010; Sharma et al., 2013). Detailed gene function studies are necessary to understand further the regulation of rice leaf photosynthesis and the impact on final yield.
Materials and methods
Plant materials and growth conditions
Seeds of the rice mutant erect panicle3 (ep3), and a T-DNA insertion line 1C-03432 were kindly supplied by Dr Hee-Jong Koh (Seoul National University) together with seeds of their wild-type controls. The ep3 mutant was generated by N-methyl-N-nitrosourea mutagenesis of Hwasunchalbyeo seed resulting in the introduction of a single base-pair change in the second exon (G/C–A/T) leading to the conversion of a codon for tryptophan into a premature stop (Piao et al., 2009). Hwayoungbyeo plants contained a T-DNA insertion, sited between 1921bp and 1922bp downstream of the ATG, that disrupted the putative second exon of Os02g15950.
Rice plants were grown in a growth room at 28 °C with a 12/12h light/dark photoperiod and illuminated by a bank of 400W metal halide lamps, a light intensity at plant height of 400 µmol m–2 s–1 and a relative humidity of approximately 50%. Rice seeds were placed on filter papers moistened with distilled water in Petri dishes to promote germination and were then transferred to 1.5ml Eppendorf tubes with their bottoms removed and floated on a hydroponic nutrient solution (Murchie et al., 2005) until the emergence of the second leaf. They were then transferred to a hydroponic tank system which was used for the further growth of rice plants and the nutrient compositions were as described in Murchie et al. (2005). Plants were held in place via sponges through 3cm diameter holes. Light-resistant materials were used to inhibit the growth of algae. Tap water was used to make up the nutrient solution and the final pH was adjusted to between 5.0 and 5.5 using HCl. Fully expanded leaf 6 was used throughout for the measurements.
Plants of Arabidopsis including Col-0 and the hawaiian skirt-1 (hws-1) mutant were grown in plastic pots or plastic trays containing Levington M3 compost under growth-room conditions of 20/4h light/dark at 23±1 °C. Intercept at 0.2g l–1 was added to the soil to prevent any attack of the plants by insect larvae.
Genomic PCR
The primers FCAPsEP3 (see Supplementary Fig. S1 at JXB online) were modified from Piao et al. (2009) to improve amplification efficiency and used to genotype the ep3 MNU mutant. Primers F2EP3, R2EP3, and FLBC 5′ were designed to genotype the T-DNA insertion line (see Supplementary Fig. S1 at JXB online). A Polymerase Chain Reaction (PCR) was performed using MangoTaq™ DNA polymerase (BiolineTM) according to the manufacturer’s instructions and programmes were run using a GeneAmp PCR system 9700 (Applied Biosystems). The PCR parameters used were: 94 °C for 5min; 32 cycles of 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 1min. A final elongation step was performed at 72 °C for 7min.
Plasmid construction and plant transformation
Rice Hwasunchalbyeo genomic DNA was extracted using the Qiagen DNAeasy Plant Mini kit (Qiagen) following the manufacturer’s instructions and a genomic region of 1227bp containing an open reading frame beginning in the second predicted exon 2 was amplified using PhusionTM High-Fidelity DNA Polymerase (New England BioLabs) and sub-cloned into the vector pBI101.2:HWS pro (González-Carranza et al., 2007) to perform a complementation test in Arabidopsis hws-1 mutant plants. The first predicted exon 1 was not included as all other putative HWS orthologues have only a single exon. The primers used were: ForEP3 and RevEp3 (see Supplementary Fig. S1 at JXB online); a second set of primers was designed to sub-clone this genomic region containing the restriction sites BamHI and SmaI in the forward and reverse primers, respectively: ForEP3BamHI and RevEP3SmaI (see Supplementary Fig. S1 at JXB online) The E.coli DH5α competent cells were transformed and positive colonies from kanamycin selective plates were tested by PCR. Plasmids were extracted from the PCR-confirmed colonies using QIAprep® Miniprep Kit (Qiagen) and sequenced with primers ForHWS5UTR and RevGUS90 (Eurofin).
Positive plasmids were electroporated into Agrobacterium tumefaciens strain C58 and transformed into Arabidopsis plants hws-1 and Col-0 using the floral dip method (Clough and Bent, 1998). T1 plants were screened on MS plates containing kanamycin at a final concentration 50ng ml–1 and were grown at 22 °C with 24h daylight. Antibiotic-resistant plants were transplanted into soil under the Arabidopsis growth conditions described in the Pplant materials and growth conditions section. Genotyping was carried out using primers SSLPHSfor and SSLPHSrev (see Supplementary Fig. S1 at JXB online) to test the hws-1 background and F2EP3 and RevGUS90 (see Supplementary Fig. S1 at JXB online) to test the cloned ORF from the rice EP3 gene.
Gas-exchange and chlorophyll measurements
The fully expanded leaf 6 of rice plants was used for gas-exchange measurements using a Li-Cor 6400 XT Portable Photosynthesis System with chlorophyll fluorescence attachment (Li-Cor Biosciences, USA). Measurements were taken in the growth room between 10.00h and 15.00h. The settings were as follows: flow rate 500 µmol s–1, block temperature 30 °C with ambient relative humidity. To determine the light-response curve, the sample (cuvette) CO2 concentration was set to 400 µmol mol–1 and 10 points of lamp PAR were used varying between 0 and 2000 µmol m–2 s–1 (10% blue). Apparent quantum yield, light compensation points, light-saturated photosynthetic capacity, and dark respiration were measured using the resulting light-response curve.
Assimilation versus internal leaf CO2 concentration (A/C i) curves were performed to gain information on the carboxylation capacity (V cmax), electron transport capacity (J max), and the proportion of photosynthesis limited by stomatal conductance (l). The flow rate was set to 500 µmol s–1, the block temperature was set to 30 °C, and lamp PAR was set to 500 µmol m–2 s–1 (10% blue). A series of reference cuvette CO2 concentrations was used.
To explore stomatal conductance (g s) responses to changes in relative humidity, gas exchange measurements were performed using manually set levels of cuvette humidity. The setting was as above for the light-response curves but with a PAR of 500 µmol m–2 s–1 (10% blue). The relative humidity (RH) was set to 60%, 55%, 50%, 45%, and 40% by manually adjusting the desiccant bypass/scrub when measurements were taken. Leaves used for measurements were left in the chamber under the respective RHs until the values of photosynthesis and stomatal conductance stabilized, usually about 2–3min. Leaf temperature was set to 25 °C. The PAR was first set to 2000 µmol m–2 s–1 (10% blue) for about 1min to make stomata open fully, and then set to 500 µmol m–2 s–1 (10% blue) for taking measurements. Manual change of humidity was performed when the g s value stabilized at a PAR of 500 µmol m–2 s–1.
Leaf chlorophyll content was estimated using a SPAD-502 chlorophyll meter (Konica, Minolta) before the gas-exchange measurement (Markwell et al., 1995). Five points on each leaf were used to calculate a mean SPAD value for each biological replicate.
Dark adapted F v/F m was determined using a Mini-PAM (Heinz Walz GmbH) at the widest region through the leaf (the middle of the leaf). A 1h dark adaption was performed for the plant leaves using a dark leaf clip DLC-8 (Heinz Walz GmbH).
Leaf anatomical analysis
A silicon based impression resin Coltene President Plus Jet (Colténe Whaledent, Switzerland) was used for leaf surface impression to determine stomatal density and stomatal size (Smillie et al., 2012). The two components of the resin were mixed in a Petri dish just before application on the leaf surface using a plastic stick. The resin impression was removed from the leaf surface after 10min and then painted with a layer of clear nail varnish. The nail varnish was then peeled off from the resin and put onto a glass slide for further imaging under a light microscope (Nikon optiphot-2) with a Leica DFC 320 camera. Stomatal guard cell length and width were measured using the straight line in Image J software while stomatal guard cell area (the area of two stomatal guard cells when stomata were fully shut so that the influence of stomatal aperture was avoided) was measured using the freehand selection area in Image J (Abràmoff et al., 2004). Subsidiary cells were not included in these measurements. All stomata were closed when measurements were taken to avoid an influence of aperture on the measurement.
Anatomical characters were determined from leaf transverse sections. The leaf sample was positioned in a raw potato cube and a razor blade was used for hand cutting. Sections were then mounted on a microscope slide and then cleared using a solution of 85% (w/v) lactic acid in chloral hydrate. Slides were put into Petri dishes in a 70 °C water bath for 1h. Distilled water was used to wash the sections after incubation and a solution containing 1% (w/v) toluidine blue in 1% (w/v) disodium tetraborate was used for staining. Sections on the slide were placed in dye solutions for 15 s before washing several times with distilled water until they were ready to be observed under a microscope (Smillie et al., 2012). A Leica DMRB microscope with a Leica EC3 camera was used to take photographs. The photographs of the leaf sections were analysed and measured using Image J software.
Separated single cell preparation was generated (Pyke and Leech, 1987; Smillie et al., 2012). A 1mm wide piece of the leaf samples was fixed in a 3.5% (w/v) glutaraldehyde solution for 1h in the dark and then transferred to a solution of 0.1M NaEDTA (pH 9.0) before incubating at 60 °C for at least 6h. These samples were then left at 4 °C overnight and mounted on a microscope slide to produce single cells by slightly hitting. Images were captured using a Leica DM5000B microscope with a DFC camera for further analysis in Image J software (Abràmoff et al., 2004).
Rubisco quantitation
Leaf samples of 1cm were collected from the widest part of a fully expanded leaf 6 after the gas exchange measurements had been taken and then immediately immersed in liquid nitrogen and stored at –80 °C. The leaf area (1cm × leaf width) was calculated and used to determine the quantitation in a unit area. Rubisco extraction was performed by adding 1ml extraction buffer (100mM HEPES, 1mM EDTA, 20mM MgCl2, 0.1% (v/v) Triton, 20% (v/v) glycerol, 0.12% (w/w) DTT, pH 7.5) to lysing matrix tubes with additional 1/4″ ceramic spheres (supplied by MP Biomedical). The tubes were put in MPTM FAST PREP®-24 (MP Biomedical) under a maximum vortex speed for 1min to grind the sample. Samples were centrifuged at 13 000×g for 10min at 4 °C and the supernatant was then transferred to another Eppendorf tube and stored at –80 °C. Total protein calculation and the SDS-PAGE for separating the large subunit of Rubisco was performed as described by Murchie et al. (2002) and pure wheat Rubisco protein was kindly supplied by Martin Parry, Rothamsted Research, UK. The gel was stained in QC Colloidal Coomassie Stain (Bio-Rad) for 1h and destained in water to remove background colour. Gel densitometry was performed using a GS-800 Calibrated Densitometer (Bio-Rad) and Quantity One version 4.4.1 software. The density of Rubisco in 1 µg total protein was calculated from SDS-PAGE densitometry compared with the pure Rubisco reference.
Statistical analysis
Values were used to perform statistical analysis in GraphPad Prism 6 software. Unpaired t-tests were performed to compare two sets of values between mutant plants and their control plants (for instance, the comparison of anatomical structure). Paired t-tests were performed for the analysis of matched pairs of values between mutant plants and their control plants (for instance, stomatal conductance under a series of changing APR).
Results
Complementation of hws-1 using OsEP3
To determine whether the OsEP3 is a functional orthologue of HWS the predicted EP3 exon 2 was cloned using genomic DNA from the Hwasunchalbyeo ecotype and sub-cloned into the vector pBI101.2:HWS pro (González-Carranza et al., 2007). A phenotypic comparison of flowers was performed between transgenic plants and their controls. Sepals of hws-1 flowers were fused into a single whorl and remained attached to siliques throughout maturity and dehiscence (Fig. 1A). Floral organs from hws-1 plants containing the construct pBI101.2: HWS pro:EP3 (Fig. 1C) resembled wild-type plants (Fig. 1B) and exhibited normal floral organ shedding. Similarly, Col-O plants transformed with the pBI101.2: HWS pro:EP3 construct were indistinguishable from the wild type (Fig. 1D).
Fig. 1.

Phenotypic analysis of flowers from hws-1 and Col-0 plants containing pBI101:HWSpro:EP3. Green siliques from hws-1 (A), Col-0 WT (B), HWS:EP3/hws-1 (C), and HWS:EP3/Col-0 WT (D). Sepal fusion in hws-1(A) was rescued by expressing EP3 driven by the HWS promoter (C). Scale bar=0.5cm.
Photosynthetic assimilation and stomatal conductance in the ep3 mutant
Gas-exchange measurements were performed on the fully expanded leaf 6 from the ep3 NMU mutant, Hwasunchalbyeo, the ep3 T-DNA insertion 1C-03432.L, and Hwayoungbyeo grown in a hydroponic system (Fig. 2A–D).
Fig. 2.

Comparison of aerial tissues (A–D) at the leaf 6 stage and mature panicles (E–H) at the harvest stage from two rice lines carrying a mutation in the EP3 gene grown in hydroponics. (A, E) ep3 NMU mutant; (B, F) Hwasunchalbyeo, from which the NMU mutant line was generated; (C, G) ep3 T-DNA insertion 1C-03432.L; (D, H) Hwayoungbyeo, from which the ep3 T-DNA insertion 1C-03432.L was generated. Scale bars: (A–D) 10cm; (E–H) 1cm.
The light-response curves from all four lines showed typical responses but the net photosynthetic assimilation at higher light levels, including the light-saturated rate (A max) was observed to be lower in the ep3 NMU mutant and the T-DNA insertion line when compared with Hwasunchalbyeo (unpaired t-test, P <0.001) and Hwayoungbyeo (unpaired t-test, P <0.001), respectively (Fig. 3A, B). Apparent quantum yield and light compensation point were calculated from the linear portion of the light-response curve at lower PAR but no significant difference between each gene function disruption line and their WT plants was seen (Fig. 3C). Analysis of the light compensation point also indicated that there was no significant difference between each gene function disruption line and their controls (Fig. 3D). Dark respiration (R d, µmol m–2 s–1) was calculated from the light-response curve and there was no significant difference between the ep3 NMU mutant (1.89±0.40) and Hwasunchalbyeo (2.01±0.26) and between the ep3 T-DNA insertion (2.18±0.18) and Hwayoungbyeo (1.86±0.34). These results suggest that the lower photosynthetic assimilation observed in the ep3 NMU mutant and the T-DNA insertion line is neither the consequence of a defect in photosynthetic (quantum) efficiency nor the higher demand of cellular respiration. Stomatal conductance (g s) calculated from the light-response curve is shown in Fig. 3E, F). g s was significantly lower in the ep3 mutants when compared with their WT plants at a PAR of 500 µmol m–2 s–1 and above (paired t-test, ep3 NMU mutant versus Hwasunchalbyeo P <0.05; paired t-test, ep3 T-DNA insertion versus Hwayoungbyeo P <0.001) (Fig. 3E, F).
Fig. 3.
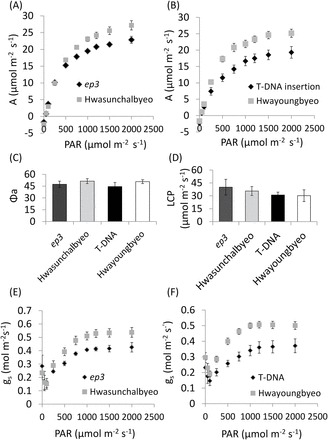
Light-response curve from fully expanded leaf 6 of ep3 mutants and their WT control plants. (A) A significant decrease of photosynthetic assimilation was found in ep3 NMU mutant plants compared with Hwasunchalbyeo over 500 µmol m–2 s–1 PAR, P <0.001 (paired t-test). (B) Significantly lower photosynthesis was also observed in the ep3 T-DNA insertion line compared with Hwayoungbyeo over 500 µmol m–2 s–1 PAR, P <0.001 (paired t-test). Analysis of apparent quantum yield (C) and light compensation point (D) indicate no significant change. (E, F) Stomatal conductance (g s) in response to PAR. Error bars in this figure show the SD, n=5.
A/C i curves were generated to identify the component responsible for the decline in photosynthesis (curves shown in Supplementary Fig. S2 at JXB online). Maximum RuBP saturated rate of carboxylation (V cmax, µmol m–2 s–1), CO2 compensation point (Γ, µmol mol–1), maximum rate of electron transport (J max, µmol m–2 s–1), and stomatal limitation (l) were calculated from the fitted A/C i curve by the A/C i Response Curve Fitting 10.0 suite (http://landflux.org/Tools.php) following the Farquhar–von Caemmerer–Berry leaf photosynthesis model (Ethier and Livingston, 2004). There was no significant difference in J max between the mutants and their wild-type control plants (Table 1). However, V cmax which shows the level of active Rubisco in the leaf and the CO2 compensation point (Γ), was significantly lower only in the ep3 T-DNA insertion when compared with the control (unpaired t-test, P <0.01 and P <0.001, respectively. The proportion of photosynthesis limited by stomatal conductance (l) was calculated according to Long and Bernacchi (2003) and was significantly higher in both the ep3 NMU mutant and the T-DNA insertion line when compared with their control plants (unpaired t-test, ep3 NMU mutant versus Hwasunchalbyeo P=0.024; ep3 T-DNA insertion versus Hwayoungbyeo P=0.018). As a lower g s was the most consistent effect of EP3 disruption, this parameter was focused on in more detail.
Table 1.
Comparison of V cmax, CO2 compensation point, J max, and stomatal limitationCO2 compensation point (Γ, µmol mol–1),maximum rate of electron transport (J max, µmol m–2 s–1), stomatal limitation (l), and maximum RuBP saturated rate of carboxylation (V cmax, µmol m–2 s–1) were calculated from a fitted A/C i curve by the A/C i Response Curve Fitting 10.0 suite (available on http://landflux.org/Tools.php). Stomatal limitation (l) was calculated using l=(A 0–A)/A 0 (A 0 at C i=400 µmol mol–1). An unpaired t-test was performed for statistics: * significant at the P <0.05 level; ** significant at the P <0.01 level; *** significant at the P <0.001 level.
| ep3 NMU mutant | Hwasunchalbyeo | ep3 T-DNA insertion | Hwayoungbyeo | |
|---|---|---|---|---|
| V cmax (µmol m–2 s–1) | 71.18±11.34 | 81.78±8.68 | 57.70±7.58** | 86.22±11.95 |
| Γ (µmol mol–1) | 61.98±5.24 | 60.50±4.36 | 75.78±5.70*** | 55.64±3.02 |
| J max (µmol m–2 s–1) | 106.5±5.57 | 112.6±4.32 | 105.2±8.64 | 110.4±5.84 |
| Stomatal limitation (l) (%) | 10.23±5. 42* | 4.790±2.94 | 18.53±2.61* | 12.82±5.64 |
Maximum quantum yield was also measured as dark adapted F v/F m and no differences were seen between the ep3 NMU mutant and the T-DNA insertion line and their controls, indicating that lowered photosynthesis was not the consequence of a defect in PSII (see Supplementary Fig. S3 at JXB online).
To confirm that the ep3 mutant plants have a lower g s and to explore this property in more detail, the gas-exchange measurement was performed using manually altered humidity levels. g s was determined under changing humidity conditions while the leaf temperature, CO2, and PAR levels were fixed. The g s declined in all four plant lines when the humidity decreased (see Supplementary Fig. S4 at JXB online). The analysis of g s indicated that there were significant decreases in the ep3 mutant lines when compared with their WT controls (paired t-test, ep3 NMU mutant versus Hwasunchalbyeo P<0.05; ep3 T-DNA insertion versus Hwayoungbyeo P<0.01; see Supplementary Fig. S4 at JXB online).The data show consistency in the direction of the response for stomatal conductance in both the humidity and the light response.
Analysis of stomatal density and stomatal area
To determine any morphological origin of the decline in g s, stomatal density and stomatal area were determined (see Supplementary Fig. S5 at JXB online). Both the adaxial and abaxial surfaces of the middle portion of leaf 6 were investigated. Statistical analysis showed that there was no significant difference in stomatal density between ep3 functional disruption lines and their WT plants on either the adaxial or abaxial surfaces (Table 2). However, there was a consistent decrease in guard cell area in both the lines compared with the controls. The stomatal guard cell area was significantly smaller in the ep3 NMU mutant and the T-DNA insertion line on the leaf abaxial surface when compared with Hwasunchalbyeo (unpaired t-test, P <0.01) and Hwayoungbyeo (unpaired t-test, P <0.05), respectively (Table 2). On the adaxial surface, a reduction in stomatal guard cell area was only observed in the T-DNA insertion line (unpaired t-test, P <0.05). The reduced abaxial stomatal guard cells area in the ep3 NMU mutant is the consequence of a reduction in stomatal width (unpaired t-test, P <0.001) while a reduced stomatal guard cell length in the T-DNA insertion line (unpaired t-test, P <0.001) was responsible for the area reduction (Table 2). A decrease in stomatal guard cell length from the adaxial surface was observed in the T-DNA insertion line (unpaired t-test, P <0.001) (Table 2) and this reduction was responsible for a smaller adaxial guard cell area in the T-DNA insertion line (Table 2). Abaxial- and adaxial-specific responses of stomatal density to alterations in growth irradiance and CO2 were observed in a previous study (Hubbart et al., 2013).
Table 2.
Analysis of stomatal density, stomatal (guard cell) area, length, and widthThe table shows measurements of stomatal distribution and stomatal area, length, and width in the form of ‘mean±SD’ for n=40. An unpaired t-test was performed for statistics: * significant at the P <0.05 level; ** significant at the P <0.01 level; *** significant at the P <0.001 level.
| Lines | Surface | Stomatal number(mm–2) | Guard cell length(µm) | Guard cell width(µm) | Guard cell area(µm2) |
|---|---|---|---|---|---|
| ep3 NMU mutant | Adaxial | 182.5±6.29 | 20.50±0.41 | 8.05±0.15 | 143.53±4.89 |
| Hwasunchalbyeo | Adaxial | 190.27±3.93 | 21.12±0.31 | 7.67±0.16 | 137.59±3.46 |
| T-DNA insertion | Adaxial | 200.55±5.39 | 20.72±0.46*** | 7.82±0.14 | 143.02±5.34* |
| Hwayoungbyeo | Adaxial | 197.22±5.47 | 23.35±0.34 | 8.20±0.17 | 158.60±5.84 |
| ep3 NMU mutant | Abaxial | 209.72±7.34 | 19.26±0.41 | 6.37±0.16*** | 116.68±3.53** |
| Hwasunchalbyeo | Abaxial | 215.83±7.72 | 19.26±0.32 | 7.49±0.13 | 129.88±3.29 |
| T-DNA insertion | Abaxial | 222.22±4.01 | 19.19±0.31*** | 7.87±0.14* | 133.68±2.94* |
| Hwayoungbyeo | Abaxial | 238.05±8.48 | 20.24±0.51 | 7.43±0.12 | 143.80±3.51 |
Correlation analysis between stomatal parameters and stomatal conductance was also performed but no significant correlation was observed (see Supplementary Fig. S6 at JXB online).
Leaf anatomy
Alterations in stomatal guard cells area could be indicative of other leaf developmental effects (Hubbart et al., 2013). Therefore, the leaf anatomical structure was analysed using transverse sections taken from the middle area of fully expanded leaf 6 (see Supplementary Fig. S7 at JXB online). Statistical analysis showed that there was no significant difference in the following parameters: leaf thickness at minor veins; interveinal distance between the major vein and minor veins; and leaf thickness at the major vein and the width of the major vein (Table 3). Significant changes were observed in the following parameters: the interveinal distance between minor veins was significantly reduced in the ep3 NMU mutant when compared with Hwasunchalbyeo (unpaired t-test, P <0.05) (Table 3); the leaf thickness at a bulliform cell position was significantly smaller in the T-DNA insertion line when compared with Hwayoungbyeo (unpaired t-test, P <0.01) (Table 3); and the width of the minor vein was also significantly reduced in the ep3 mutant (unpaired t-test, P <0.01) (Table 3). In all of these changed parameters, the significant changes were only observed in one of the ep3 functional disruption lines (ep3 NMU mutant or the T-DNA insertion line).
Table 3.
Analysis of leaf anatomical structuresThe table shows measurements of leaf anatomical structures from leaf sections and single mesophyll cell preparation in the form of ‘mean±SD’ for n=5–40. An unpaired t-test was performed for statistics: * significant at the P <0.05 level; ** significant at the P <0.01 level.
| ep3 NMU mutant | Hwasunchalbyeo | ep3 T-DNA line | Hwayoungbyeo | |
|---|---|---|---|---|
| Major vein width (µm) | 114.21±5.44 | 119.05±2.59 | 115.96±5.88 | 118.66±3.37 |
| Minor vein width (µm) | 46.75±1.39** | 41.44±1.13 | 44.21±1.91 | 43.19±1.39 |
| Leaf thickness at | 174.34±5.44 | 182.97±2.59 | 179.80±5.88 | 176.11±3.37 |
| major vein (µm) | ||||
| Leaf thickness at | 85.39±1.64 | 85.33±1.85 | 87.07±1.80 | 91.52±1.49 |
| minor vein (µm) | ||||
| Leaf thickness at | 86.11±2.18 | 83.07±2.07 | 80.52±1.69** | 88.13±1.64 |
| bulliform cells (µm) | ||||
| Interveinal distance between | 137.01±4.09 | 143.16±4.09 | 122.12±5.08 | 124.76±2.02 |
| major vein and minor vein (µm) | ||||
| Interveinal distance between | 139.99±3.03* | 150.73±3.19 | 128.35±4.40 | 129.57±3.52 |
| minor veins (µm) | ||||
| Mesophyll cell area (µm2) | 279.62±8.35 | 296.91±8.99 | 331.40±10.83 | 347.18±11.73 |
| Mesophyll cell lobe number | 7.08±0.28 | 7.44±0.26 | 7.36±0.23 | 7.68±0.243 |
Measurements of mesophyll cell area and cell shape were performed on single mesophyll cells separated from the fully expanded leaf 6 tissues (see Supplementary Fig. S8 at JXB online). The analysis of mesophyll cell area showed that there was no significant difference between the ep3 functional disruption lines and their WT plants (Table 3). The number of mesophyll cell lobes was also analysed, and no significant difference was observed between the ep3 functional disruption lines and their WT plants (Table 3).
A significant increase in leaf width on leaf 6 from the ep3 NMU mutant was observed when compared with Hwasunchalbyeo (unpaired t-test, P <0.05), however, no significant difference was observed between the T-DNA insertion line and its Hwayoungbyeo control (data not shown).
Leaf chlorophyll content taken as SPAD values were measured on leaf 6 and statistical analysis indicated that the SPAD value was significantly higher in the ep3 mutant lines when compared witho their WT plants (unpaired t-test, ep3 NMU mutant versus Hwasunchalbyeo, P <0.05; ep3 T-DNA insertion line versus Hwayoungbyeo, P <0.001) (Fig. 4A).
Fig. 4.
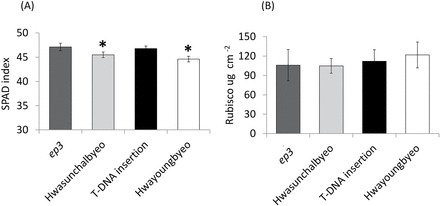
Analysis of SPAD index and Rubisco. Fully expanded leaf 6 from rice plants were used for these measurements. Error bars indicate SD value for n=3 in Rubisco and n=18 in SPAD. (A) SPAD index of mutant plants and WT control plants, indicating a proxy for chlorophyll content. (B) Analysis of Rubisco concentration per unit leaf area (µg cm–2).
Under optimal conditions, saturating light and ambient CO2 concentrations, the content of active Rubisco protein commonly limits a large proportion of photosynthetic rate (Makino, 2011). Our data did not show a consistent lowering of Rubisco activity in both lines, as measured as V cmax. Hence the Rubisco protein concentration per unit leaf area was measured using a SDS-PAGE gel (see Supplementary Fig. S9 at JXB online). There was no significant difference between the EP3 functional disruption line and their WT plants (Fig. 4B). This result suggests that the reduction in photosynthesis of the ep3 mutant and the T-DNA insertion line is not caused by differences in the quantity of Rubisco present but may represent a down-regulation of Rubisco activity.
Discussion
OsEP3 is a functional orthologue of the Arabidopsis HWS gene
The fused sepal phenotype of the hws-1 mutant was rescued by expressing the OsEP3 exon 2 driven by the HWS promoter (Fig. 1B). This observation indicates that the exon 2 of OsEP3 is able to substitute for the function of HWS during floral development and confirms that OsEP3 is a functional orthologue of At3g61590. HWS has two potential orthologues in rice according to protein sequence similarity, these are Os01g47050 and Os02g15950 (ERECT PANICLE3). A previous study confirmed that Os01g47050 is able to complement hws-1 mutant plants (Z González-Carranza, unpublished data) which indicates that the HWS orthologue in rice is duplicated. The phenotypic characteristics of silencing the Os01g47050 gene are unknown and it would be intriguing to determine the features of a double knockout of both this gene and Os02g15950 (ERECT PANICLE3) to ascertain whether organ fusion and growth are also compromised as observed in hws-1 (González-Carranza et al., 2007) and in Fig. 2. The protein sequence alignment reported by Piao et al. (2009) indicated that there are two exons in Os02g15950 but only one exon in HWS and its orthologues. Interestingly, the second exon of OsEP3 also starts with an ATG and our study has shown that the second exon of EP3 is able to rescue the fused sepal phenotype in hws-1. Thus, it is possible that the first predicted exon of EP3 might not be an accurate interpretation of the structure of the gene. However, it is also possible that the EP3 RNA might undergo alternative splicing. Further analyses need to be carried out to determine which of these alternatives is correct.
Reduced stomatal guard cell area is associated with a lowered stomatal conductance and photosynthetic capacity in ep3 mutants
A significantly lower photosynthetic capacity and g s was observed in both the ep3 NMU mutant and the T-DNA insertion line from light-response curves when compared with their WT plants, Hwasunchalbyeo and Hwayoungbyeo, respectively. No consistent change in apparent maximum quantum yield of CO2 uptake (Φa), light compensation point (LCP), quantity of Rubisco, F v/F m, mesophyll cell size, and lobes were found. However, significant differences were observed in guard cell (stomatal) length and guard cell (stomatal) area (Table 2) and the proportion of photosynthesis limited by stomatal conductance between both the ep3 functional disruption lines and their WT plants. In this section, the term stomatal area is used to reflect the changes seen in both the length and width of the guard cells (Table 2). Significant differences in some anatomical changes were only observed in the ep3 NMU mutant or the DNA insertion line (Table 3). The changes only observed in one mutant line may be the consequence of some other mutations in the genome or because the truncated peptides generated in the ep3 mutant retain some function when only the F-box domain of the protein was translated.
The stomatal densities and area observed here are in line with published values for rice (Ohsumi et al., 2007; Hubbart et al., 2013). It is proposed that the smaller area of guard cells in the mutants with no alteration in stomatal density is, at least partly, responsible for the lowered stomatal conductance which, in turn, caused the lower values of photosynthesis. However, an additional change in stomatal aperture cannot be ruled out. There were no consistent changes in other leaf anatomical changes or in Rubisco protein content, suggesting that the reduction of stomatal area is not the consequence of the general cell size reduction throughout the rice leaves. Stomatal area, density of stomata, and depth of the stomatal pore all have a role in determining conductance values, although the relationships can be complex. For example, stomatal size has been reported to have an inverse relationship with stomatal density in rice (Ohsumi et al., 2007) and across species and genera, including fossil plants (Hetherington and Woodward, 2003). Importantly, stomatal density does not correlate with conductance across a range of rice genotypes but a significant positive correlation between stomatal length and stomatal conductance can be observed (Ohsumi et al., 2007) which support our hypothesis. However, a significant change in length was only observed in the Hwayoungbyeo mutant. An inverse relationship between guard cell size and conductance has been observed in other studies and other species (Lawson and Blatt, 2014; Raven, 2014), but not in this study (see Supplementary Fig. S6 at JXB online) or in a survey of rice genotypes (Ohsumi et al., 2007). Rice stomata are small in comparison with other species so it is possible that other factors, including area, density, and depth of the pore are more significant. It is proposed here that the alteration of area with no change in density will reduce the maximum possible pore area per unit leaf area for gas exchange.
To improve photosynthesis and yield under natural environments the speed of stomatal movements is considered to be important especially in fluctuating light (Lawson et al., 2012). Quick responses in terms of stomatal opening and closing were reported to be essential for optimizing water loss and CO2 gain (Vico et al., 2011; Lawson et al., 2012). In the ep3 mutant, smaller stomata may be a benefit in terms of speed of response and reducing the expense of stomatal movement. In the gas exchange measurements the temporal pattern of response (the kinetics of the light-response curve, for example) of the mutant and the wild type was similar in each case: it was only the magnitude of the response that differed, suggesting that there was no substantial change in stomatal movement dynamics over the time-scale used.
Stomatal conductance, Rubisco, and photosynthesis
A correlation between stomatal conductance and photosynthesis is common (Wong et al., 1979; Farquhar and Sharkey, 1982; Leuning, 1995), however, many previous studies have shown that, under optimal conditions and ambient CO2, Rubisco quantity and activation state dominate in the determination of photosynthetic capacity in rice (Makino, 2011; Parry et al., 2013). Rubisco quantity did not show significant differences (Fig. 4B) in this study and it is proposed here that the limitations on CO2 diffusion caused by lowered stomatal conductance resulted in a lowered Rubisco activation state in the ep3 mutant as shown by the V cmax. This is in accord with previous studies where stomatal conductance was shown to place a limitation on Rubisco activity (Flexas et al., 2006). It is interesting that the V cmax was only significantly lower in the T-DNA insertion line (Table 1), indicating that, although stomatal conductance is still the original limiting factor, there may be small differences in the way Rubisco activity is regulated between Hwasunchalbyeo and Hwayoungbyeo.
F-box protein EP3 and stomatal area
In this study, it is proposed that the reduced stomatal area in ep3 functional disruption lines leads to a lower photosynthetic assimilation via a reduction in stomatal conductance while no difference was found in stomatal density and the mesophyll cell size (Table 3). Therefore, the EP3 gene was inferred to play a role in regulating stomatal area. As an F-box protein, EP3 may function as a subunit of ubiquitin E3 ligase to recognize and degrade a certain substrate (Petroski and Deshaies, 2005; Lechner et al., 2006) that is a determinant of stomatal area. Stomatal development has been well studied in Arabidopsis and a number of genes have been reported to be involved in the process (Pillitteri and Torii, 2012). Two F-box genes DROUGHT TOLERANCE REPRESSOR (DOR) (Zhang et al., 2008) and CONSTITUTIVE PHOTOMORPHOGENIC (COP1) (Kang et al., 2009) were reported to regulate the stomatal ABA response and light-controlled stomatal development, respectively. This also provides evidence that F-box proteins are closely related to stomatal regulatory networks. The pathway controlling stomatal size (S) and density (D) seems to be linked by observing an inverse relationship between the S and D under different CO2 levels (Franks and Beerling, 2009; Doheny-Adams et al., 2012). The gene HIGH CARBON DIOXIDE (HIC) was reported to modulate a change in stomatal density in response to elevated CO2 (Gray et al., 2000). In this study, a stomatal area decrease has been observed in the ep3 mutant without disrupting stomatal density, which did not show an inverse relationship. Expression analysis of EP3 RT-PCR was performed to detect the expression pattern of EP3. Expression of EP3 was detected in Hwayoungbyeo, the ep3 NMU mutant, and Hwasunchalbyeo leaf tissues (see Supplementary Fig. S10 at JXB online). Studies on the HAWAIIAN SKIRT (HWS) gene in Arabidopsis also identified expression in leaf tissue (González-Carranza et al., 2007) and an abnormal distribution of stomata took place in the hws-1 mutant (Z González-Carranza et al., unpublished data), suggesting that the gene may be involved in stomatal lineage. EP3, the functional orthologue, might play a similar function as HWS in the regulation of stomatal development. In order to understand how EP3 may be involved in stomatal area regulation, it is important to identify the substrate for this F-box protein and our current work on HWS should prove valuable in this regard.
Summary
EP3 was confirmed to be a functional orthologue of HWS via a complementation test. Observations from gas exchange indicated that a decrease in photosynthesis in the ep3 mutant was the consequence of a decline in stomatal conductance linked to a reduced guard cell area. EP3 is likely to play a role in regulating stomatal guard cell area as an F-box protein in the ubiquitin-mediated protein degradation pathway.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Primers list.
Supplementary Fig. S2. A/C i curves generated from gas-exchange.
Supplementary Fig. S3. Analysis of dark-adapted F v/F m.
Supplementary Fig. S4. Analysis of stomatal conductance (g s) in response to an alteration in cuvette humidity.
Supplementary Fig. S5. Rice leaf surface impression.
Supplementary Fig. S6. Correlation between stomatal parameters and stomatal conductance.
Supplementary Fig. S7. Rice leaf section showing the measurements of anatomical structure.
Supplementary Fig. S8. Single mesophyll cells from leaves of ep3 mutants and their wild-type controls.
Supplementary Fig. S9. SDS-PAGE gel running of Rubisco larger subunit and standard sample.
Supplementary Fig. S10. Gene expression analysis of EP3 in rice tissues.
Acknowledgements
This work is supported by the PhD programme of the Chinese Scholarship Council. We are grateful to Dr Hee-Jong Koh (Seoul National University) and Dr G An (Pohang University of Science and Technology; Republic of Korea) who provided the ep3 NMU mutant and ep3 T-DNA insertion seeds and their wild type controls, and to Dr Hongying Li, Dr Caiyun Yang, Dr Zhefeng Lin, Dr Chungui Lu, Dr Stella Edwards, Liang Zhao, Dr Ajigboye Olubukola, Dr Mohamed Ahamadeen Nagoor, and Dr Ian Smillie for their technical support. We also thank Mark Meacham and Sue Flint for the growth room preparation and thank Dr Poh Lim for advice on data analysis.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with Image. J Biophotonics International 11, 36–43. [Google Scholar]
- Adachi S, Nakae T, Uchida M, Soda K, Takai T, Oi T, Yamamoto T, Ookawa T, Miyake H, Yano M. 2013. The mesophyll anatomy enhancing CO2 diffusion is a key trait for improving rice photosynthesis. Journal of Experimental Botany 64, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. 2007. Stomatal development. Annual Review of Plant Biology 58, 163–181. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Doheny-Adams T, Hunt L, Franks PJ, Beerling DJ, Gray JE. 2012. Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier G, Livingston N. 2004. On the need to incorporate sensitivity to CO2 transfer conductance into the Farquhar–von Caemmerer–Berry leaf photosynthesis model. Plant, Cell and Environment 27, 137–153. [Google Scholar]
- Farquhar GD, Sharkey TD. 1982. Stomatal conductance and photosynthesis. Annual Review of Plant Physiology 33, 317–345. [Google Scholar]
- Feldman AB, Murchie EH, Leung H, Baraoidan M, Coe R, Yu SM, Lo SF, Quick WP. 2014. Increasing leaf vein density by mutagenesis: laying the foundations for C4 rice. PLoS One 9, e94947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexas J, Barbour MM, Brendel O, Cabrera HM, Carriquí M, Díaz-Espejo A, Douthe C, Dreyer E, Ferrio JP, Gago J. 2012. Mesophyll diffusion conductance to CO2: an unappreciated central player in photosynthesis. Plant Science 193, 70–84. [DOI] [PubMed] [Google Scholar]
- Flexas J, Ribas Carbó M, Bota J, Galmés J, Henkle M, Martínez Cañellas S, Medrano H. 2006. Decreased Rubisco activity during water stress is not induced by decreased relative water content but related to conditions of low stomatal conductance and chloroplast CO2 concentration. New Phytologist 172, 73–82. [DOI] [PubMed] [Google Scholar]
- Franks P, Beerling D. 2009. CO2-forced evolution of plant gas exchange capacity and water-use efficiency over the Phanerozoic. Geobiology 7, 227–236. [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA. 2007. HAWAIIAN SKIRT: an F-box gene that regulates organ fusion and growth in Arabidopsis . Plant Physiology 144, 1370–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, van der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM. 2000. The HIC signalling pathway links CO2 perception to stomatal development. Nature 408, 713–716. [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI. 2003. The role of stomata in sensing and driving environmental change. Nature 424, 901–908. [DOI] [PubMed] [Google Scholar]
- Hubbart S, Bird S, Lake JA, Murchie EH. 2013. Does growth under elevated CO2 moderate photoacclimation in rice? Physiologia Plantarum 148, 297–306. [DOI] [PubMed] [Google Scholar]
- Kang C-Y, Lian H-L, Wang FF, Huang JR, Yang HQ. 2009. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. The Plant Cell 21, 2624–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. 2010. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology 61, 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Hirotsuka S, Kumamaru T, Iba K. 2012. Increased leaf photosynthesis caused by elevated stomatal conductance in a rice mutant deficient in SLAC1, a guard cell anion channel protein. Journal of Experimental Botany 63, 5635–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Blatt MR. 2014. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiology 164, 1556–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Kramer DM, Raines CA. 2012. Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Current Opinion in Biotechnology 23, 215–220. [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. 2006. F-box proteins everywhere. Current Opinion in Plant Biology 9, 631–638. [DOI] [PubMed] [Google Scholar]
- Leuning R. 1995. A critical appraisal of a combined stomatal-photosynthesis model for C3 plants. Plant, Cell and Environment 18, 339–355. [Google Scholar]
- Long S, Bernacchi C. 2003. Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. Journal of Experimental Botany 54, 2393–2401. [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC. 2006. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540. [DOI] [PubMed] [Google Scholar]
- Makino A. 2011. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiology 155, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell J, Osterman JC, Mitchell JL. 1995. Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynthesis Research 46, 467–472. [DOI] [PubMed] [Google Scholar]
- McKown AD, Dengler NG. 2007. Key innovations in the evolution of Kranz anatomy and C4 vein pattern in Flaveria (Asteraceae). American Journal of Botany 94, 382–399. [DOI] [PubMed] [Google Scholar]
- Murchie E, Hubbart S, Peng S, Horton P. 2005. Acclimation of photosynthesis to high irradiance in rice: gene expression and interactions with leaf development. Journal of Experimental Botany 56, 449–460. [DOI] [PubMed] [Google Scholar]
- Murchie EH, Yang J, Hubbart S, Horton P, Peng S. 2002. Are there associations between grain filling rate and photosynthesis in the flag leaves of field grown rice? Journal of Experimental Botany 53, 2217–2224. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. 2006. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. The Plant Cell 18, 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi A, Kanemura T, Homma K, Horie T, Shiraiwa T. 2007. Genotypic variation of stomatal conductance in relation to stomatal density and length in rice (Oryza sativa L.). Plant Production Science 10, 322–328. [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM. 2013. Rubisco activity and regulation as targets for crop improvement. Journal of Experimental Botany 64, 717–730. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nature Reviews Molecular Cell Biology 6, 9–20. [DOI] [PubMed] [Google Scholar]
- Piao R, Jiang W, Ham TH, Choi MS, Qiao Y, Chu SH, Park JH, Woo MO, Jin Z, An G. 2009. Map-based cloning of the ERECT PANICLE 3 gene in rice. Theoretical and Applied Genetics 119, 1497–1506. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. 2006. Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. 2012. Mechanisms of stomatal development. Annual Review of Plant Biology 63, 591–614. [DOI] [PubMed] [Google Scholar]
- Pyke K, Leech R. 1987. The control of chloroplast number in wheat mesophyll cells. Planta 170, 416–420. [DOI] [PubMed] [Google Scholar]
- Raven JA. 2002. Selection pressures on stomatal evolution. New Phytologist 153, 371–386. [DOI] [PubMed] [Google Scholar]
- Raven JA. 2014. Speedy small stomata? Journal of Experimental Botany 65, 1415–1424. [DOI] [PubMed] [Google Scholar]
- Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC. 2013. recent advances in dissecting stress-regulatory crosstalk in rice. Molecular Plant 6, 250–260. [DOI] [PubMed] [Google Scholar]
- Smillie I, Pyke K, Murchie E. 2012. Variation in vein density and mesophyll cell architecture in a rice deletion mutant population. Journal of Experimental Botany 63, 4563–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vico G, Manzoni S, Palmroth S, Katul G. 2011. Effects of stomatal delays on the economics of leaf gas exchange under intermittent light regimes. New Phytologist 192, 640–652. [DOI] [PubMed] [Google Scholar]
- Wong S, Cowan I, Farquhar G. 1979. Stomatal conductance correlates with photosynthetic capacity. Nature 282, 424–426. [Google Scholar]
- Xing Y, Zhang Q. 2010. Genetic and molecular bases of rice yield. Annual Review of Plant Biology 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Zhang Q. 2007. Strategies for developing green super rice. Proceedings of the National Academy of Sciences, USA 104, 16402–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Ye, Xu W, Li Z, Deng XW, Wu W, Xue Y. 2008. F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis . Plant Physiology 148, 2121–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR. 2010. Improving photosynthetic efficiency for greater yield. Annual Review of Plant Biology 61, 235–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


