Abstract
The development of the pancreas and determination of endocrine cell fate are controlled by a highly complex interplay of signaling events and transcriptional networks. It is now known that an interconnected epigenetic program is also required to drive these processes. Recent studies using genome-wide approaches have implicated epigenetic regulators, such as DNA and histone-modifying enzymes and non-coding RNAs, to play critical roles in pancreas development and the maintenance of cell identity and function. Furthermore, genome-wide analyses have implicated epigenetic changes as a casual factor in the pathogenesis of diabetes. In the future, genomic approaches to further our understanding of the role of epigenetics in endocrine cell development and function will be useful for devising strategies to produce or manipulate β-cells for therapies of diabetes.
Introduction
Epigenetics refers to mechanisms that alter gene expression patterns in the absence of changes in the nucleotide sequence of the DNA. Epigenetic marks, which include DNA modifications (such as methylation) and post-translational modifications of histones (such as acetylation, phosphorylation, ubiquitination and sumoylation), are deposited on chromatin by DNA and histone-modifying enzymes. In addition, long non-coding RNAs (lncRNAs) are emerging as important epigenetic regulators by functioning as molecular scaffolds to initiate and sustain epigenetic changes 1. Epigenetic regulators are now known to contribute to pancreas development as well as the differentiation, maintenance and function of pancreatic endocrine cells, most notably the insulin-producing β-cells. In addition, studies have shown that an altered epigenetic landscape is associated with the pathogenesis of diabetes. In this review, we highlight the growing evidence for the importance of epigenetic regulation in pancreas development, maintenance of β-cell identity and function, and the pathogenesis of diabetes.
Epigenetic programming of pancreatic organ fate commitment
The pancreas originates from the endodermal germ layer, which also gives rise to the esophagus, stomach, intestine and organs lining the gastrointestinal tract, such as the liver, thyroid and lungs. The specification of endodermal lineage intermediates toward these different organ fates occurs stepwise and is initiated by localized signals that induce the expression of lineage-specific transcription factors (TFs). While the last two decades have provided a detailed understanding of the TFs that mediate the differentiation steps toward the different organ fates, less is known about how changes at the level of chromatin influence these developmental decisions.
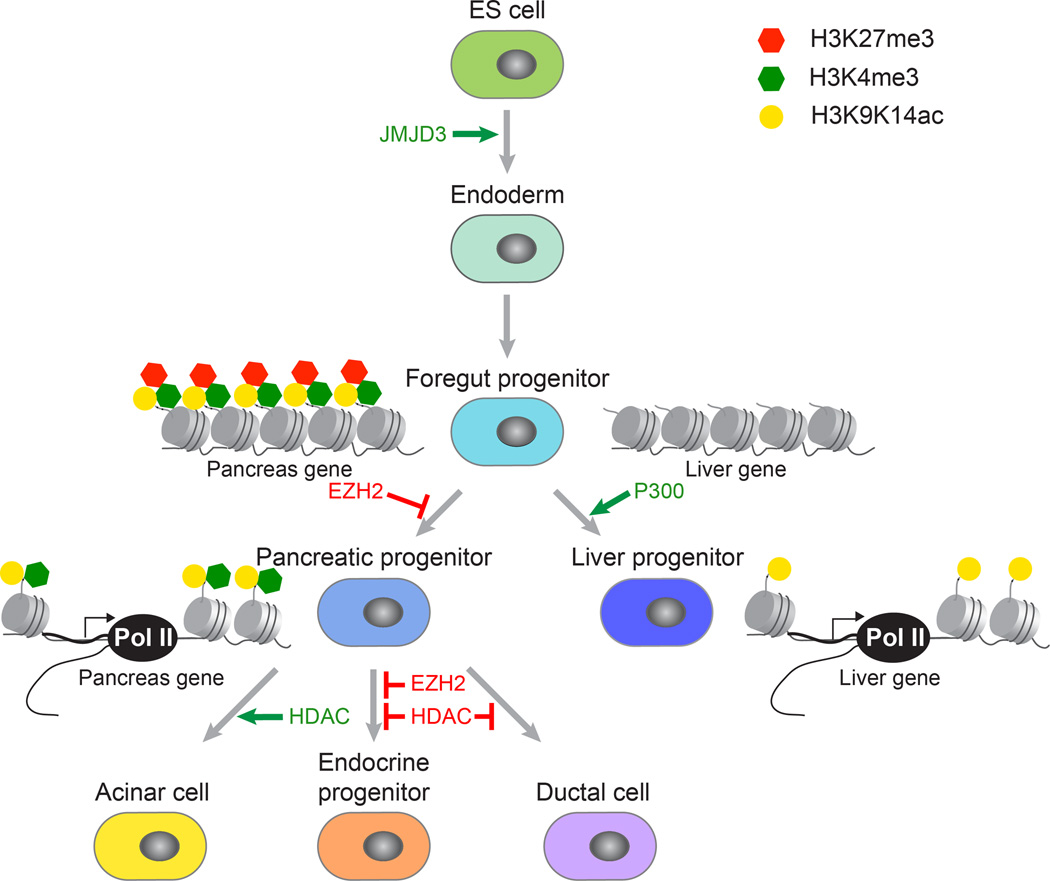
The pancreas and liver arise from a common population of cells in the ventral foregut. Inductive signals for the pancreas and liver must act upon these bipotent progenitors to activate pancreas- or liver-specific genes. Work by Zaret and colleagues explored whether specific chromatin modifications are established at liver- and pancreas-specific regulatory sequences prior to gene activation and whether chromatin “pre-patterns” play a role in cell fate induction 2. Employing analysis of select candidate genes, they found that liver- and pancreas-specific regulatory regions exhibit distinct chromatin patterns in bipotent foregut progenitors. Pancreas regulatory elements are marked by both the active H3K9K14ac and the repressive H3K27me3 marks, reflecting a “poised” state for future gene activation (Figure 1). In contrast, these marks are poorly represented at liver regulatory elements in foregut progenitors, and liver genes acquire H3K9acK14ac active marks de novo when cells differentiate into hepatoblasts. The authors tested whether these histone modifications direct the cell fate choice of foregut progenitors between liver and pancreas. Consistent with the observation that histones at liver-specific loci undergo de novo acetylation during hepatic fate induction, reduced p300 acetyltransferase activity prevents hepatic fate induction and favors the pancreatic fate 2. Inactivation of the histone methyltransferase Ezh2, a key component of the Polycomb Gene (PcG) complex mediating H3K27me3 deposition, similarly favors the pancreatic over the hepatic fate, presumably because “poised” pancreatic genes are activated upon removal of the repressive H3K27me3 mark. These findings demonstrate distinct mechanisms for the activation of pancreas and liver genes during development and illustrate how this mechanism is predetermined by specific chromatin “pre-patterns” in developmental intermediates.
Figure 1. Epigenetic programming of pancreatic lineage specification.
Overview of the key steps in pancreas development and the role of epigenetic regulators in these transitions. As cells transition from an undifferentiated to a differentiated state, the chromatin undergoes cell type-specific alterations that are highly regulated. For example, removal of the repressive mark H3K27me3 from promoters of endodermal regulators by the H3K27 demethylase JMJD3 results in endoderm induction. In foregut progenitor cells, loci for liver- and pancreas-specific genes exhibit different chromatin pre-patterns, and the histone methyltransferase EZH2 and the histone acetyltransferase P300 affect the cell fate choice between liver and pancreas. The differentiation of pancreatic progenitor cells into the different pancreatic cell types is influenced by histone deacetylases (HDACs) and EZH2.
It is clear from these studies that the chromatin undergoes important alterations that are highly regulated as cells transition from an undifferentiated to a differentiated state. In pluripotent embryonic stem (ES) cells, a majority of developmental genes that contain the repressive H3K27me3 mark is also enriched for the active H3K4me3 mark, which has been coined a bivalent state 3. Lineage-specific differentiation of ES cells is associated with a resolution of this bivalent state, with a loss of either H3K27me3 or H3K4me3 leading to the activation or stable repression of lineage-specific genes, respectively. However, bivalent domains are not only resolved but also gained during development and these dynamic changes in H3K27me3 deposition during developmental progression are thought to help facilitate rapid changes in gene expression 4. What has been unclear is when and how the bivalent state is resolved or gained during the progression of ES cells toward a differentiated state. Early studies had been limited in that only select developmental stages were examined, usually only comparing one undifferentiated population with differentiated cells 3, 5. However, recent in vitro differentiation protocols using pluripotent stem cells (PSCs) have enabled the use of genome-wide approaches to identify changes in epigenetic modifications as cells progress through lineage intermediates toward a differentiated state 6, 7. The development of such protocols for the pancreatic endocrine cell lineage 8, 9 has offered an opportunity to study critical epigenetic events occurring during endocrine cell development. Studies using these approaches have shown that at each step during progression toward pancreas, changes in the bivalent state of a promoter coincide with changes in the expression of the associated gene 10. Further studies have shown that these epigenetic changes are directly relevant for the regulation of gene expression, as inactivation of H3K27me3 demethylases in ES cells prevents the induction of endodermal genes during endoderm formation 10–12 (Figure 1). The stage-specific loss of H3K27me3 repression is specifically observed at genes encoding developmental regulators, as for example EOMES at the endoderm stage and PDX1 at induction of the pancreatic fate 10. Similar observations have been reported during the development of other lineages 6, 7, suggesting that the combined analysis of gene expression and bivalent promoter states could be globally employed to identify novel developmental regulators.
It is currently unclear how these ubiquitously expressed histone-modifying enzymes, such as the PcG component EZH2, can have such specific roles in differentiation. One might expect that modulating the activity of these epigenetic regulators would widely change a cell’s chromatin state, resulting in deleterious consequences to the cell. Studies so far suggest that these regulators are acting on select genes and therefore have highly specific context-dependent effects. Indeed, genome-wide studies have shown that PcG-dependent H3K27 trimethylation is not a universal repression mechanism, but actually represses a subset of genes that encode for developmental regulators 13. In addition, the H3K27me3-mediated repressive mechanism is only employed in certain cellular contexts 14. One hypothesis that could explain why histone-modifying enzymes act in such a specific manner is that cell type-specific TFs recruit histone modifiers to select loci during differentiation. It was recently shown that the T-box TF TBX3 directs the H3K27me3 demethylase JMJD3 to the regulatory elements of the endodermal regulator EOMES resulting in derepression of EOMES and subsequent endoderm induction 12. By systematically mapping where epigenetic regulators are recruited and how their recruitment changes during differentiation, we may be able to define the roles for these regulators in the development of the pancreatic as well as other organ lineages.
The above studies provide initial insights at a system-wide level into how pancreatic lineage progression is regulated. There are likely additional layers of regulation, such as differential promoter and enhancer usage, DNA methylation, and non-coding RNAs that orchestrate these changes in gene expression. Genome-wide analysis of how these modifications and regulators change during endodermal differentiation will further elucidate the molecular mechanisms regulating organ specification.
Regulation of pancreatic endocrine cell development by epigenetic mechanisms
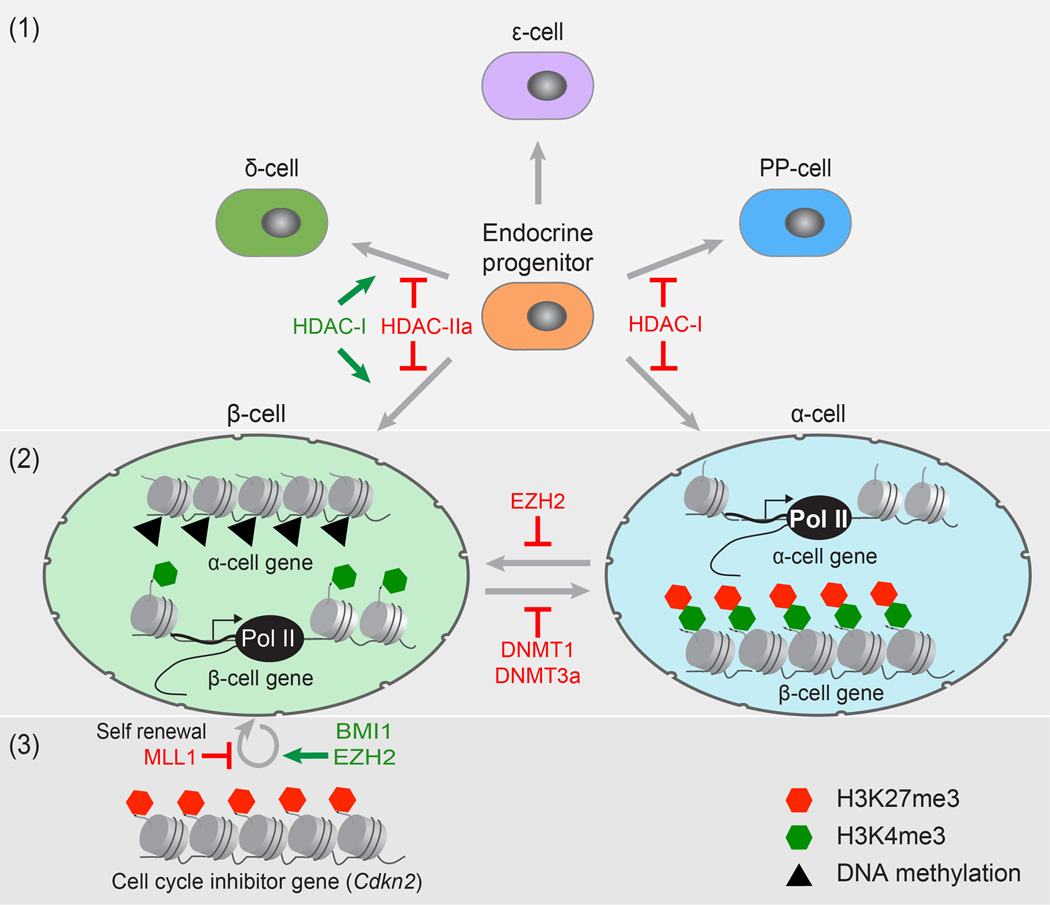
Changes in the epigenome are also important during the differentiation of the different cell types of the pancreas. Studies using inhibitors for histone deacetylases (HDACs) in embryonic pancreas explants have demonstrated that the decision of multipotent pancreatic progenitor cells whether to produce endocrine, acinar or ductal cells relies on modifications of histones (Figure 1). Maintenance of acetylation by HDAC inhibition suppresses acinar cell differentiation and promotes commitment to the ductal and endocrine fates 15. Dynamic changes in the acetylation of histones are also important for terminal endocrine cell differentiation and the specification of the different endocrine cell types, namely alpha (α)-, beta (β)-, delta (δ), pancreatic polypeptide (PP)- and epsilon (ε)-cells. In this context, different classes of HDACs appear to have distinct effects on the development of individual endocrine cell types (Figure 2). For example, HDAC class I (HDAC-I) members specifically restrict the formation of α- and PP-producing cells, while promoting β- and δ-cell differentiation 15. Further supporting the idea that HDACs have distinct functions during endocrine subtype specification, genetic deletion of different HDAC class IIa (HDAC-IIa) members results in either an increase of β-cells or δ-cells 16. Thus, histone-modifying enzymes have highly specific roles during the specification of the different endocrine cell types. This knowledge could be utilized to direct the differentiation of specific endocrine cell types during in vitro differentiation of PSCs.
Figure 2. Epigenetic regulation in pancreatic endocrine differentiation and maintenance of β-cell function.
The diagram depicts the role of epigenetic regulators as endocrine progenitor cells differentiate into the different endocrine subtypes (1), in regulating cell plasticity (2), and β-cell proliferation (3). (1) Histone deacetylase (HDAC) class I (HDAC-I) and HDAC class IIa (HDAC-IIa) family members have distinct effects on the development of individual endocrine cell types. (2) While genes critical for β-cell function are expressed and marked by H3K4me3 in β-cells, these same genes are silent and bivalently modified in α-cells. This bivalency suggests a plastic epigenetic state for key β-cell genes in α-cells. Additionally, to maintain β-cell identity, genes important for α-cell function have to be actively repressed by the DNA methytransferases DNMT1 and DNMT3a in β-cells. (3) Epigenetic modification of the Cdkn2 locus, encoding a cell cycle inhibitor, by BMI1 and EZH2 promotes β-cell proliferation and regeneration, whereas the methyltransferase MLL1 contributes to β-cell senescence.
In the last decade, numerous groups have tried to generate functional β-cells from human PSCs. Early efforts in achieving proper endocrine cell differentiation in vitro had been met with limited success. Insulin+ cells produced with these early protocols expressed multiple hormones, were not glucose responsive and more closely resembled fetal than adult β-cells in regard to gene expression 10, 17, 18. However, developmental precursors produced with these protocols develop into glucose-responsive, mature β-cells upon implantation into mice 19, 20, and based on transcriptome analysis, these in vivo-differentiated endocrine cells are remarkably similar to primary human endocrine cells 10. This strategy of differentiating β-cells in vivo by implanting PSC-derived pancreatic precursors is currently been tested in humans in the first stem cell-based diabetes clinical trial. Furthermore, two independent studies from the Kieffer and Melton labs have recently described in vitro differentiation protocols that can produce functional β-like cells from human PCSs 8, 9. Kieffer and colleagues demonstrated these cells were able to permanently reverse hyperglycemia when transplanted into diabetic mice; however the effects were not immediate, suggesting that some degree of maturation still has to occur within the host.
Earlier studies have shown that the insufficient induction of β-cell genes during terminal differentiation in vitro was associated with aberrant chromatin remodeling. In pancreatic progenitors, the expression of core β-cell genes is largely suppressed by H3K27me3 modifications, which become selectively removed during terminal endocrine differentiation 10, 21. However, in vitro differentiated cells had inadequate changes in H3K27me3 and H3K4me3 modification patterns, especially at key β-cell gene loci 10. Now with the Kieffer and Melton protocols in hand, it would be interesting to determine if these in vitro-produced β-like cells have the same epigenetic makeup as primary β-cells. If there are differences, targeted manipulation of specific epigenetic regulators could help achieve complete β-cell differentiation in vitro. Indeed, Zaret and colleagues have shown that inhibition of EZH2 during pancreatic differentiation of human PSCs leads to an increase in endocrine progenitors and insulin-expressing cells in vitro 22. However, it was not determined if the insulin+ cells generated were functional β-cells.
Roles of the epigenome in cell plasticity and β-cell identity
There have been significant advances in generating insulin-producing cells from a variety of adult cell types. Several reports have achieved transdifferentiation of endoderm-derived cell types, such as liver, intestinal and gall bladder epithelium, into insulin-producing cells by ectopic expression of TFs critical for pancreas and β-cell development 23–26. These reprogramming strategies often lack robustness and tend to be slow and inefficient, which might explain why these strategies have not found widespread applications. However, it has been noted that there is great plasticity among the different cell types of the pancreas, particularly α-cells and δ-cells, towards β-cells. In comparison to pancreatic exocrine cells, which require TF-based reprogramming with multiple endocrine TFs 27, reprogramming of α- or δ- to β-cells can be achieved by manipulating a single TF or simply by ablating the β-cell population 28–31. Why these endocrine cell types can be more easily converted into β-cells and why similar experimental approaches are insufficient to facilitate this conversion in other cell types had been unclear. Recent studies indicate that epigenetic similarities between different but developmentally related cell types could explain some of these differences on a global scale.
Kaestner and colleagues found that the intrinsic plasticity of α-cells associates with specific histone methylation profiles 32. They show that α-cells not only contain more bivalently marked genes than β-cells, but while most genes critical for β-cell development and function are H3K4me3 marked in β-cells, the majority of these genes is bivalently marked in α-cells (Figure 2). This bivalency may enable α-cells to be plastic upon suitable stimuli, as these β-cell-specific genes are poised for activation. This might suggest that the histone methylation profile is an indicator of a cell’s potential for plasticity, with α-cells having greater plasticity toward the β-cell phenotype than, for example, exocrine cells, which carry far less of these bivalent marks and have limited potential for conversion into β-cells. Interestingly, Herrera and colleagues have found that δ-cells, like α-cells, are capable of spontaneously reprogramming into β-cells after β-cell ablation; however, the ability of δ-cells to reprogram does not extend beyond puberty 28. Further studies on the epigenetic state of young and aged endocrine cells could reveal novel mechanisms of how the epigenome affects cellular plasticity.
Recent studies have revealed that once endocrine cells have differentiated, the epigenetic landscape is actively maintained to stabilize cellular identity. For example, to maintain β-cell identity, the α-cell fate regulator Arx needs to be actively repressed. In β-cells, the Arx promoter is highly methylated and this is facilitated by the DNA methyltransferases Dnmt3a and Dnmt1 33, 34(Figure 2). Deletion of either of these enzymes leads to derepression of Arx, and subsequent transcriptional activation of the α-cell program, resulting in β- to α-cell conversion. In β-cells, the Arx promoter is associated with methyl-specific binding proteins that recruit enzymatic complexes capable of locally altering histone modifications. Dnmt3, for example, facilitates transcriptional silencing of Arx by recruiting HDAC1 to the promoter 34. In addition, the methyl-DNA binding protein MeCP2 recruits a complex of proteins to the Arx promoter, including the methyltransferase PRMT6, which can counteract activating H3K4me3 marks by mediating histone arginine methylation 33. These studies illustrate the importance of epigenetic factors in maintaining adult β-cell identity and safeguarding β-cells from transdifferentiation.
It is possible that manipulation of specific epigenetic regulators may loosen these epigenetic constraints and increase plasticity of terminally differentiated cells. It has already been shown that treatment of islets with an inhibitor to PcG proteins results in increased expression of the β-cell TF PDX1 and insulin in α-cells 32 (Figure 2). Therefore, it will be critical in the future to identify epigenetic regulators involved in modulating cell plasticity for improved approaches to direct reprogramming toward the β-cell phenotype. Further studies are needed to systematically identify common epigenetic changes that occur during the transdifferentiation of various cell types into β-cells. Based on these marks, we may be able to predict the epigenetic regulators facilitating this process and confirm their role by genetic deletion. Employing locus-specific editing approaches 35, important epigenetic regulators could then be guided to the regulatory regions of critical lineage determinants to induce reprogramming toward the β-cell phenotype.
Epigenetic control of β-cell function and mass
Epigenetic modifiers are not only important for preventing activation of alternative lineage programs, but also for regulating cell function. For instance, epigenetic factors have been shown to dynamically regulate insulin secretion in β-cells. Mirmira and colleagues have demonstrated that the histone methyltransferase SET7/9 is required for the expression of a subset of genes involved in glucose-stimulated insulin secretion 36. Depletion of SET7/9 results in repression of these genes through loss of activating H3K4me2 marks. SET7/9 catalyzes the transfer of methyl group(s) to histones from the diet-induced co-factor S-adenosyl-L-methionine (SAM). Indeed, the establishment of many epigenetic marks is dependent on the availability of metabolic cofactors such as SAM, nicotinamide adenine dinucleotide (NAD), flavin adenine dinucleotide (FAD) and α-ketoglutarate (α-KG) 37. Because the availability of these cofactors is regulated by nutrient status, diet could alter gene expression in β-cells through epigenetic modifiers. Recently, an impact of a methyl-deficient diet on endocrine pancreas mass and insulin secretion has been reported 38. Prenatal methyl deficiency in rats reduces pancreatic mass and impairs glucose tolerance and insulin secretion. Since dietary methyl donors have a critical role in DNA and histone methylation, the effects mediated by this diet could be attributed to alterations in the epigenome. There is indeed evidence that poor maternal diet or an adverse intrauterine environment has a transgenerational influence on endocrine cell function through epigenetic silencing of developmental regulators, such as Hnf4a and Pdx1 39, 40. It is likely that environmental influences early on in development has long-lasting effects on the epigenome, affecting β-cell function and the risk for diabetes later in life.
The capacity of β-cells to proliferate is critical for their ability to adapt to changing metabolic demands. An example of this is the adaptive expansion of β-cell mass in response to pregnancy, which has been linked to epigenetic changes. During pregnancy β-cell expansion is controlled by the transcriptional regulator/tumor suppressor Men1 41. Men1 associates with MLL1, a H3K4 methyltransferase that trimethylates H3K4 and maintains expression of the cyclin-dependent kinase inhibitors p27Kip1 and p18INK4c to prevent β-cell proliferation 42. Reduction of Men1 during pregnancy facilitates β-cell expansion by reducing active marks on the loci of these cell cycle inhibitors.
The adaptive capacity of β-cells to proliferate declines with age 43 and this has been shown to be regulated by p16INK4a, a cyclin-dependent kinase inhibitor encoded by the Cdkn2a gene. Increased expression of p16INK4a with age mediates an age-associated decline in β-cell proliferation and ability to regenerate 44. Epigenetic modulation of the Cdkn2a locus plays an important role in regulating the capacity of β-cells to proliferate (Figure 2). Studies have shown that an age-dependent decrease in expression of two PcG proteins, Bmi1 and Ezh2, is associated with derepression of the Cdkn2a locus and increase in p16INK4a levels 45, 46. In aged mice, the simultaneous knockdown of MLL1, which prevents deposition of activating H3K4me3 marks, and induction of Ezh2 to remove repressive H3K27me3 marks can repress p16INK4a expression and increase proliferation of β-cells 47. Collectively, these studies illustrate that the epigenome is actively modified in differentiated β-cells and that it may be possible to manipulate regulators of the epigenome to reverse β-cell senescence and promote β-cell regeneration.
The epigenetic basis of diabetes
An abundance of literature has implicated epigenetic changes as a casual factor in the pathogenesis of diabetes 48. Early studies using candidate gene approaches have shown that increased DNA methylation at promoters of genes that regulate β-cell function associates with decreased expression of the respective gene and reduced insulin secretion in islets from patients with type 2 diabetes (T2D) 49–52. More recently, genome-wide DNA methylation profiling has led to the identification of hundreds of genes that in diabetic islets display significant changes in the methylation pattern at promoters when compared to non-diabetic donors 53, 54. These studies indicate that altered DNA methylation at promoters of genes critical for β-cell function affects gene expression and contributes to the pathogenesis of T2D. Islet-specific TFs have been implicated in shaping the proper epigenetic landscape to maintain β-cell gene expression. For example, HNF1-α, a homeodomain-containing TF expressed in islets, interacts with co-activator proteins possessing histone acetyltransferase activity to maintain hyperacetylation of histones at promoters of β-cell-specific genes 55. Loss of Hnf1-α results in hypoacetylation of HNF1-α-bound promoters, loss of H3K4me2 activating marks, and enrichment of repressive H3K27me3 marks. Interestingly, mutations in HNF1-α are associated with maturity onset diabetes of the young (MODY) 56, which suggests that dysregulation of epigenetic modifications caused by HNF1-α mutations might contribute to this subtype of diabetes.
In recent years, genome-wide association studies (GWAS) have identified multiple genetic variants that are associated with T2D 57–59. Most disease-associated variants are non-coding and it is unclear how these variants affect cell function. As the loci of the majority of T2D-associated variants are not in protein-coding regions, this would suggest that these loci have a regulatory function. For instance, some lncRNAs within genomic regions map very closely to small nucleotide polymorphisms associated with β-cell dysfunction and T2D, and it may be that these variants affect the expression or function of these lncRNAs 60. Several other sequence variants associated with T2D map to open chromatin regions containing distal regulatory elements that are occupied by islet-specific TFs 61. Studies have experimentally validated that many of these variants reside in enhancer regions and abolish enhancer activity 61–63. For example, by analyzing the chromatin state and TF occupancy in islets, Ferrer and colleagues demonstrated that several non-coding T2D variants map to enhancers that are bound by islet-specific TFs 61. One of the variants was found to abolish enhancer activity in β-cells, and alter sequence-specific DNA binding of the islet-enriched MODY-associated TF NEUROD1.
There are several examples of mutations in distal regulatory elements causing disease. However, it is still unclear how these distant elements affect gene transcription. Specifically, it has been difficult to define how enhancers communicate with promoters and to identify target genes for enhancers. Just recently, Hattersley, Ferrer and colleagues located an enhancer for the PTF1 gene, encoding a TF essential for the development of the pancreas, in a distal region downstream of PTF1 that harbors mutations associated with pancreas agenesis, a rare condition that occurs when the pancreas fails to develop before birth 64. By using chromosome conformation capture (3C) technology, which measures the frequency of physical interaction or proximity among any pair of genomic loci, they were able to show that this region interacts with the PTF1 promoter. Further, they demonstrated that mutations in this region prevent enhancer activity by abolishing TF binding. Recent advances combining 3C technology with deep sequencing now allows a three-dimensional (3D) view of the genome. This new technology (4C-seq) can be utilized to identify target genes of non-coding T2D genetic variants. This has been nicely demonstrated in recent work by Nobrega and colleagues 65. Here they used 4C-seq to profile genomic interactions of obesity-associated SNPs with gene promoters. They found that sequences residing in the first intron of the FTO gene, which harbors a variant associated with increased risk for obesity and T2D, interact with the promoter of the homeobox gene IRX3, located ~500 kilobases away. Their data directly tie the obesity-associated SNPs within the FTO intron to reduced expression of IRX3. These findings were surprising, because the variant does not influence expression of the closest gene, FTO, but instead expression of a very distal gene.
Concluding remarks and perspectives
In this review we have highlighted recent progress showing the important roles of the epigenome in not only pancreas and endocrine cell development, but also in endocrine cell plasticity and function. With recent advances in stem cell technology and genome-wide approaches, we are now just beginning to understand how epigenetic modifications and modifiers affect these processes. In the future, systems approaches to generate high-resolution epigenome maps from isolated pancreatic cell populations, combined with genetic approaches in model organisms or cultured cells should help identify important epigenetic switches that can be manipulated to evoke changes in cell state. Furthermore, genome-wide studies will provide a more comprehensive and systematic view of the contribution of epigenetics to diabetes by uncovering new target genes that may contribute to dysregulation of β-cell function. GWAS have identified numerous genetic variants associated with disease, yet how T2D-associated variants contribute to the pathogenesis of T2D remains largely unknown. In the future, the effects of these non-coding disease variants can be functionally validated and modeled using locus-specific gene-editing approaches in human PSC-based models. In addition, by analyzing disease variants in the context of the epigenome and creating 3D interaction maps between promoters, enhancers and their regulatory proteins, we can start to define the biological mechanisms underlying their association with T2D and potentially develop novel therapies for the treatment of diabetes.
Acknowledgements
We thank members of the Sander laboratory for constructive comments on the manuscript. We apologize to our colleagues whose references were omitted owing to space constraints. Work in the Sander laboratory is supported by grants from the National Institutes of Health, the Juvenile Diabetes Research Foundation, the Helmsley Charitable Trust, and the California Institute for Regenerative Medicine (CIRM). R.X. was supported by CIRM training grant TG2-01154.
References
- 1.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin "prepattern" and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 4.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of Functional Human Pancreatic beta Cells In Vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O'Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 10.Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, Kelly OG, Wang A, D'Amour KA, Robins AJ, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 2013;23:122–130. doi: 10.1038/cr.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kartikasari AE, Zhou JX, Kanji MS, Chan DN, Sinha A, Grapin-Botton A, Magnuson MA, Lowry WE, Bhushan A. The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 2013;32:1393–1408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haumaitre C, Lenoir O, Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. 2008;28:6373–6383. doi: 10.1128/MCB.00413-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenoir O, Flosseau K, Ma FX, Blondeau B, Mai A, Bassel-Duby R, Ravassard P, Olson EN, Haumaitre C, Scharfmann R. Specific control of pancreatic endocrine beta- and delta-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60:2861–2871. doi: 10.2337/db11-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 18.Hrvatin S, O'Donnell CW, Deng F, Millman JR, Pagliuca FW, DiIorio P, Rezania A, Gifford DK, Melton DA. Differentiated human stem cells resemble fetal, not adult, beta cells. Proc Natl Acad Sci U S A. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 20.Rezania A, Bruin JE, Riedel MJ, Mojibian M, Asadi A, Xu J, Gauvin R, Narayan K, Karanu F, O'Neil JJ, et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61:2016–2029. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Arensbergen J, Garcia-Hurtado J, Moran I, Maestro MA, Xu X, Van de Casteele M, Skoudy AL, Palassini M, Heimberg H, Ferrer J. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res. 2010;20:722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu CR, Li LC, Donahue G, Ying L, Zhang YW, Gadue P, Zaret KS. Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO J. 2014 doi: 10.15252/embj.201488671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YJ, Finkbeiner SR, Weinblatt D, Emmett MJ, Tameire F, Yousefi M, Yang C, Maehr R, Zhou Q, Shemer R, et al. De novo formation of insulin-producing"neo-beta cell islets" from intestinal crypts. Cell Rep. 2014;6:1046–1058. doi: 10.1016/j.celrep.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, Barshack I, Seijffers R, Kopolovic J, Kaiser N, et al. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med. 2000;6:568–572. doi: 10.1038/75050. [DOI] [PubMed] [Google Scholar]
- 25.Hickey RD, Galivo F, Schug J, Brehm MA, Haft A, Wang Y, Benedetti E, Gu G, Magnuson MA, Shultz LD, et al. Generation of islet-like cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. 2013;11:503–515. doi: 10.1016/j.scr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima H, Fujimiya M, Matsumura K, Younan P, Imaeda H, Maeda M, Chan L. NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat Med. 2003;9:596–603. doi: 10.1038/nm867. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chera S, Baronnier D, Ghila L, Cigliola V, Jensen JN, Gu G, Furuyama K, Thorel F, Gribble FM, Reimann F, et al. Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature. 2014 doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell. 2009;138:449–462. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtney M, Gjernes E, Druelle N, Ravaud C, Vieira A, Ben-Othman N, Pfeifer A, Avolio F, Leuckx G, Lacas-Gervais S, et al. The inactivation of Arx in pancreatic alpha-cells triggers their neogenesis and conversion into functional beta-like cells. PLoS Genet. 2013;9:e1003934. doi: 10.1371/journal.pgen.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, Kaestner KH. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. J Clin Invest. 2013;123:1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20:419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, Sussel L. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 2011;25:2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–193. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol. 2012;227:3169–3177. doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith GC, Konycheva G, Dziadek MA, Ravelich SR, Patel S, Reddy S, Breier BH, Vickers MH, Owens JA, Ferguson LR. Pre- and postnatal methyl deficiency in the rat differentially alters glucose homeostasis. J Nutrigenet Nutrigenomics. 2011;4:175–191. doi: 10.1159/000330227. [DOI] [PubMed] [Google Scholar]
- 39.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, Jones RH, Marquez VE, Cairns W, Tadayyon M, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A. 2011;108:5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karnik SK, Chen H, McLean GW, Heit JJ, Gu X, Zhang AY, Fontaine M, Yen MH, Kim SK. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science. 2007;318:806–809. doi: 10.1126/science.1146812. [DOI] [PubMed] [Google Scholar]
- 42.Karnik SK, Hughes CM, Gu X, Rozenblatt-Rosen O, McLean GW, Xiong Y, Meyerson M, Kim SK. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proc Natl Acad Sci U S A. 2005;102:14659–14664. doi: 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou JX, Dhawan S, Fu H, Snyder E, Bottino R, Kundu S, Kim SK, Bhushan A. Combined modulation of polycomb and trithorax genes rejuvenates beta cell replication. J Clin Invest. 2013;123:4849–4858. doi: 10.1172/JCI69468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wren JD, Garner HR. Data-mining analysis suggests an epigenetic pathogenesis for type 2 diabetes. J Biomed Biotechnol. 2005;2005:104–112. doi: 10.1155/JBB.2005.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall E, Dayeh T, Kirkpatrick CL, Wollheim CB, Dekker Nitert M, Ling C. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Med Genet. 2013;14:76. doi: 10.1186/1471-2350-14-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling C, Del Guerra S, Lupi R, Ronn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, Groop L, Wollheim CB, Nitert MD, Ling C. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 2011;54:360–367. doi: 10.1007/s00125-010-1967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, Jing X, Renstrom E, Wollheim CB, Nitert MD, Ling C. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinol. 2012;26:1203–1212. doi: 10.1210/me.2012-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dayeh T, Volkov P, Salo S, Hall E, Nilsson E, Olsson AH, Kirkpatrick CL, Wollheim CB, Eliasson L, Ronn T, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parrizas M, Maestro MA, Boj SF, Paniagua A, Casamitjana R, Gomis R, Rivera F, Ferrer J. Hepatic nuclear factor 1-alpha directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol Cell Biol. 2001;21:3234–3243. doi: 10.1128/MCB.21.9.3234-3243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox RD, Lathrop GM, Boriraj VV, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 57.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Replication DIG Meta-analysis C Asian Genetic Epidemiology Network Type 2 Diabetes C South Asian Type 2 Diabetes C Mexican American Type 2 Diabetes C Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples C. Mahajan A, Go MJ, Zhang W, Below JE, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakic N, Garcia-Hurtado J, Rodriguez-Segui S, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Moran I, Gomez-Marin C, van de Bunt M, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SC, Boyle AP, Scott LJ, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodriguez-Segui SA, Shaw-Smith C, Cho CH, Lango Allen H, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]