Abstract
Purpose
To test the hypothesis that uncultured organisms may be present in cases of culture-negative endophthalmitis, by use of deep DNA sequencing of vitreous biopsies.
Design
Single center consecutive prospective observational study.
Participants and Controls
Aqueous or vitreous biopsies from 21 consecutive patients presenting with presumed infectious endophthalmitis, and seven vitreous samples from patients undergoing surgery for non-infectious retinal disorders.
Methods
Traditional bacterial and fungal culture, 16S quantitative polymerase chain reaction (qPCR) and a representational deep-sequencing method (Biome Representational in Silico Karyotyping [BRiSK]) were applied in parallel to samples to identify DNA sequences corresponding to potential pathogens.
Main Outcome Measures
Presence of potential pathogen DNA in ocular samples.
Results
None of 7 control eyes undergoing routine vitreous surgery yielded positive results for bacteria or virus by culture or 16S PCR. Fourteen of the 21 samples (66.7%) from eyes harboring suspected infectious endophthalmitis were culture-positive, the most common being Staphylococcal and Streptococcal species. There was good agreement among culture, 16S bacterial PCR, and BRiSK methodologies for culture-positive cases (Fleiss’ kappa of 0.621). 16S PCR did not yield a recognizable pathogen sequence in any culture-negative sample, while BRiSK suggested presence of Steptococcus in one culture-negative sample. Surprisingly, using BRiSK, 57.1% of culture-positive and 100% of culture-negative samples demonstrated presence of Torque Teno Virus (TTV) sequences, compared to none in the controls (Fisher exact, p = 0.0005). Presence of TTV viral DNA was confirmed in seven cases by qPCR. No other known viruses or potential pathogens were identified in these samples.
Conclusion
Culture, 16S qPCR, and BRiSK provide complementary information in presumed infectious endophthalmitis. The majority of culture-negative endophthalmitis samples did not contain significant levels of bacterial DNA. ‘culture-negativity’ does not appear to be due to failure of growth of fastidious bacteria. The small DNA virus TTV was unexpectedly found in all culture-negative samples and some culture-positive samples. The current study cannot distinguish whether TTV is a direct intraocular pathogen, an adjuvant for inflammation, a general marker of inflammation, or a commensal virus, but provides a testable hypothesis for a pathogenic mechanism in culture-negative endophthalmitis.
Introduction
Infectious endophthalmitis is among the most serious post-surgical complications of ophthalmic surgery. Although a rare complication of cataract surgery or intravitreal injection, with an incidence of 0.05% to 0.3%,1–7 endophthalmitis often leads to poor visual outcomes. 3, 8, 9 Because of the high volume of current and anticipated cataract surgery worldwide (with VISION2020 goals of 32 million cataract surgeries per year), and the large and increasing number of intravitreal injections performed, endophthalmitis will continue to affect tens of thousands of individuals annually worldwide.
The standard technique for diagnosing endophthalmitis is microbial culture. Surprisingly, despite the unambiguous presentation of most cases of post-operative endophthalmitis, microbial culture has a yield of only ~70%. 9 More recent studies examining endophthalmitis following intravitreal injection have found less than 50% of cases to be culture-positive. 10, 11. In recent years several studies have examined the utility of bacterial ribosomal 16S polymerase chain reaction (PCR) and sequencing in identifying bacterial pathogens in endophthalmitis.12–18 In this technique, a set of DNA primers that recognize the conserved 16S ribosomal gene found in nearly all bacteria are used to detect the presence of bacterial DNA. PCR products can then be sequenced to determine the genus of bacteria present. These studies have shown that 16S PCR is more sensitive and specific than traditional culture techniques. However 16S PCR has significant limitations: its sensitivity is sufficiently high that false-positive and artefactual products may be produced, 19 and determination of the causative organism requires subsequent sequencing or further analysis of PCR products. These limitations can be overcome by employing quantitation via quantitative PCR (qPCR) combined with sequencing of product; however, this approach has rarely employed to date in the study of endophthalmitis. 14 Additionally, 16S amplification is limited to bacteria, and cannot detect fungi (which require separate fungal rDNA ribosomal PCR), parasites, or viruses.
With the advent of massively parallel DNA sequencing platforms, the availability of the complete sequence of the human genome, and with increasing computational capacities, it is becoming possible to sequence all DNA in a biopsy sample and identify all non-human DNA present in order to detect potential occult or novel pathogens. At present, it remains prohibitively labor- and cost-intensive to completely sequence all genomes present in routine biopsy samples. However, it is possible to purify a defined fraction of all DNA present in a sample and sequence this to near saturation. One technique for achieving this is Biome Representational in Silico Karyotyping (BRiSK). 20 This technique is capable of identifying most known bacteria, as well as identifying phage, viruses, and previously unknown organisms. We report here the first application of deep DNA sequencing to vitreous and aqueous biopsies from patients with endophthalmitis, and compare results from this technique to traditional culture and 16S qPCR. We find that BRiSK has sensitivity and specificity comparable to the culture and qPCR techniques. Unexpectedly, we have identified DNA from a known anellovirus (Torque Teno Virus, TTV) in a majority of endophthalmitis cases, including in all culture-negative endophthalmitis cases.
Methods
This prospective study was approved by the Wills Eye Hospital Institutional Review Board and the University of Washington Human Studies Division Institutional Review Board. Research adhered to the tenets of the Declaration of Helsinki and was conducted in accordance with Health Insurance Portability and Accountability Act regulations. Written informed consent was obtained from all subjects for testing of samples. All participants were enrolled from the clinical offices of the Retina Service of the Wills Eye Hospital. In a prospective fashion, 21 consecutive patients diagnosed with infectious endophthalmitis based on clinical history and examination by a retina specialist (SG) at Wills Eye Hospital were enrolled. Sample size for this pilot study was chosen to provide at least 5 culture-negative samples for analysis, and limited by cost of deep DNA sequencing (at ~$1,000/sequencing run). The clinical diagnosis of endophthalmitis was made based on a combination of ocular pain, conjunctival injection, anterior chamber cellular reaction, and posterior vitritis. Vitreous tap or aqueous tap was obtained in standard clinical fashion following povidone-iondine antisepsis. As a control group, 7 consecutive patients with uninflamed eyes undergoing vitrectomy for non-infectious retinal disorders (epiretinal membrane or and macular hole) were consented for vitreous tap done during the procedure. Briefly, the vitreous samples in the control group were obtained as follows: Patients with epiretinal membranes and/or full thickness macular holes were included. After informed consent was obtained, patients were prepped and draped in the usual sterile fashion for intraocular surgery. A 23-gauge vitrectomy system (Constellation, Alcon, Fort Worth, Texas) was used in all cases. The cannulas were placed 3.5–4mm posterior to the limbus in a beveled fashion. The infusion line was affixed to the inferotemporal quadrant but not turned on. The vitrectomy instrument was introduced, and using proportional vitrectomy at 5000 CPM with manual aspiration, approximately 0.25 mL of vitreous was removed and included in the study. In all cases, approximately 0.2 to 0.3 mL of vitreous sample was sent for standard microbiological culture (including blood agar, chocolate agar, Sabaraud’s medium, and thioglycolate broth) and the remaining 0.05 to 0.10 mL were immediately frozen for subsequent analysis by 16S PCR and BRiSK.
DNA purification
Genomic DNA (gDNA) was isolated from 20–50 μL of vitreous or aqueous fluid using the DNeasy Blood & tissue kit (Qiagen, Inc) as per protocol. The DNA was eluted in the kit elution buffer and stored at −20°C. DNA was quantified using the NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). Phi29 amplification of the gDNA was performed by the Repli-g midi kit (Qiagen, Inc) as per instructions. The Phi amplified gDNA was also stored at −20°C.
16S bacterial, TTV, and actin qPCR
Pan bacterial PCR was performed using 16S rRNA universal primers (Integrated DNA Technologies IDT, San Diego, CA). The sequences of the primers were: 5′-GAGGAAGGTGGGGATGACGT-3′ and 5′-AGGCCCGGGAACGTATTCAC-3′. HotStarTaq plus DNA polymerase was used for the PCR reactions. For each reaction 100 ng of genomic DNA was used. The master mix containing 10x Buffer, Taq polymerase, dNTP mix and the primers was treated with 8-methoxypsoralen (25μg/mL) and UV nicked for 5 min (Bio-Rad GS gene linker, UV chamber) to bind any contaminating DNA. PCR amplification was performed in a MasterCycler gradient (Eppendorf, Hamburg, Germany). Cycling conditions were 10-minute denaturation at 94°C, followed by 30 cycles of 45-second denaturation at 94°C, 30 second annealing at 58°C and one minute extension at 72°C. Elongation step was for 10 minutes at 72°C.
The primer pairs for TTV PCR were 5′-AGGTGAGTTTACACACCGCAGTCA-3′ and 5′-AATGAAGACCCTAAGAGCCTTGCC-3′. The primers for β-Actin were 5′-TGCTCCTCCTGAGCGCAAG-3′ and 5′-GCCGGACTCGTCATACTCC-3′. Cycling conditions for the TTV primers were ten-minute denaturation at 95°C, followed by 25 cycles of 30 second denaturation at 95°C, 15 second annealing at 58°C and one minute extension at 72°C. Final elongation was for 10 minutes at 72°C.
qPCR assay was performed on the Applied Biosystems 7500 Fast Real-Time PCR system platform (Applied Biosystems, Foster City, CA). The final PCR mix contained 0.8 μL each of forward and reverse primers (final concentration of each 0.4 μM), 10 μL of the Absolute Blue qPCR SYBR low ROX Mix (Thermo Fisher Scientific) and 1 ul of unamplified gDNA. For 16S DNA PCR, the master mix without the template was treated with 8-methoxypsoralen and UV-treated for 5 minutes. The final reaction volume was 20 μL. For standard curve, a plasmid cDNA of the cloned gene of interest (e.g., the target sequence for 16S, TTV, or actin) was serially diluted ten-fold to obtain copy numbers ranging from 1x101 to 108 copy/mL. qPCR routinely was able to detect 10 copies/mL of each control cDNA. The run consisted of initial holding stages at 50°C for 2 minutes, at 95°C for 10 minutes followed by cycling stage (25–28 cycles) at 95°C for 15 seconds and 60°C for 1 minute.
Copy number of experimental samples were calculated by interpolation of delta CT number against the standard curve derived from the cloned product.
BRiSK
BRiSK is a representational deep DNA sequencing technique. 20 Briefly, total DNA from a biopsy is digested with the Type IIB DNA restriction enzyme BsaXI, which cuts a 33 bp fragment surrounding the DNA sequence ACNNNNNCTCC (which occurs randomly once every 4096 bp on average). These 33-mer fragments are sequenced in multiplex on a massively parallel DNA sequencing platform, typically yielding > 3 x 107 bp per sample or greater than 1 x 106 sequence ‘tags’. Each tag is compared against a database containing all known BsaXI sites in the National Library of Medicine NCBI ‘non-redundant’ dataset. Human sequences are identified, mapped, and quantified. Non-human sequences are then compared against a database containing all known bacterial, fungal, parasitic, and viral sequences.
BRiSK was performed as described. 20 3 μg of extracted DNA samples were digested by BsaX1 to create 33-mer tags, which were ligated to Illumina sequencing primers and purified on a biotin-streptavidin column. The resulting tags were submitted to a massively parallel deep-sequencing platform (Illumina HiSeq 750) and the sequenced tags were matched against a database constructed of 27-mer tags (the double-stranded portion of the 33 bp BsaX1 digestion product) virtually digested by BsaX1 from the NIH Genbank. A one edit-distance Levenshtein permutation matching heuristic was performed to account for sequencing errors and polymorphisms. Only tags that matched to a unique organism were included for further analysis.
Statistical Analysis
Fisher exact testing was performed for categorical variables and Student’s t-test was performed for continuous variables. The Fleiss kappa test was used to test for concordance among the different diagnostic techniques. The Kruskal-Wallis test was performed to compare the distribution of tag recovery rates with adjustment for multiple comparisons by Dunn’s test. All statistical tests were performed with R (http://www.r-project.org).
Results
A total of 21 consecutive infectious endophthalmitis biopsies (18 vitreous and 3 aqueous) and 7 vitreous control samples were collected by aqueous or vitreous tap. Baseline characteristics of enrolled subjects are given in Table I. The most common procedure linked with presumed infectious endophthalmitis was cataract surgery (52.4%) followed by intravitreal injection (19.0%).
Table 1.
Clinical characteristics of Infectious Endophthalmitis Cases and Controls.
| Cases (n=21) | Controls (n=7) | |||
|---|---|---|---|---|
|
|
||||
| Age (mean, SD) | 65.7 | 17.8 | 77.5 | 8.17 |
| Gender (n, %) | ||||
| Male | 9 | 42.9 | 1 | 14.3 |
| Female | 12 | 57.1 | 6 | 85.7 |
| Tap location (n, %) | ||||
| Anterior chamber | 3 | 14.3 | - | - |
| Vitreous | 18 | 85.7 | 7 | 100.0 |
| Etiology (n, %) | ||||
| Cataract | 11 | 52.4 | - | - |
| Intravitreal Injection | 4 | 19.0 | - | - |
| Bleb-associated | 3 | 14.3 | - | - |
| IOL exchange | 2 | 9.5 | - | - |
| Endogenous | 1 | 4.8 | - | - |
| Retained Lens Fragment | - | - | 3 | 42.8 |
| Proliferative Retinopathy | - | - | 2 | 28.6 |
| Vitreomacular Traction | - | - | 2 | 28.6 |
Bacterial culture results
A total of 14 of the 21 samples were positive by microbial culture (Table 2). Seven samples were positive for coagulase-negative Staphylococcus (one was read as ‘light growth’), and four were positive for Streptococcus species (one read as ‘light growth’). The remaining three culture-positive samples grew Moraxella, Bacillus (light growth), and Prevotella. None of the 7 control samples yielded growth.
Table 2.
Bacterial Genus Results of Traditional Culture, Bacterial 16S Ribosomal PCR, and Biome Representational in-Silico Karyotyping (BRiSK)
| Culture | 16S PCR | BRiSK |
|---|---|---|
| Coagulase negative Staphylococcus | Staphylococcus epidermidis | Staphylococcus epidermidis |
| Coagulase negative Staphylococcus | Staphylococcus epidermidis | Staphylococcus epidermidis |
| Coagulase negative Staphylococcus | Staphylococcus epidermidis | Staphylococcus epidermidis |
| Coagulase negative Staphylococcus | Staphylococcus epidermidis | Staphylococcus epidermidis |
| Coagulase negative Staphylococcus | Staphylococcus epidermidis | Staphylococcus epidermidis |
| Coagulase negative Staphylococcus | - | - |
| Coagulase negative Staphylococcus | - | - |
| Streptococcus intermedius | Streptococcus gordonii | Streptococcus gordonii |
| Beta hemolytic Streptococcus | Streptococcus agalactiae | Streptococcus agalactiae |
| Streptococcus viridans | Streptococcus mitis | Streptococcus mitis |
| ‘Light growth Streptococcus’ | - | - |
| Moraxella catarhallis | Moraxella catarhallis | Moraxella catarhallis |
| Prevotella melaninogenica | Streptococcus mitis | Streptococcus species |
| Bacillus (broth only) | - | - |
| - | - | Pseudomonas aeruginosa |
| - | - | Streptococcus parasanguinus |
| - | - | - |
| - | - | - |
| - | - | - |
| - | - | - |
| - | - | - |
16S qPCR
Analysis of DNA revealed high quality amplification for human actin in all samples (Figure 1). 16S PCR yielded strong signals on gel electrophoresis for 10 of the 21 samples. None of the 7 negative control samples yielded visible 16S PCR products. Quantitation of 16S levels revealed a nearly 4-log range of 16S copies/human actin copy, ranging from ~0.005 to 35 bacteria per human genome copy (Figure 1). Of the 10 samples with less than 0.1 16S bacterial DNA copy per human actin copy, all were either read as culture-negative or ‘light growth’. Sequencing results were only analyzed for samples that showed visible band to be cloned on PCR. Sequencing of DNA products revealed five sequences matching Staphylococcus, four matching Streptococcus, and one matching Moraxella. Sequencing of extremely faint PCR bands yielded unusual organisms not associated with human disease including Acidovorax caeni, Variovorax paradoxus, Citrobacter youngae, Geobacter argillaceus, Chryseobacterium hominis, Hyphomicrobium sulfonivorans which were taken to be artifact. None of these organisms were detected by culture or BRiSK in any sample.
Figure 1.
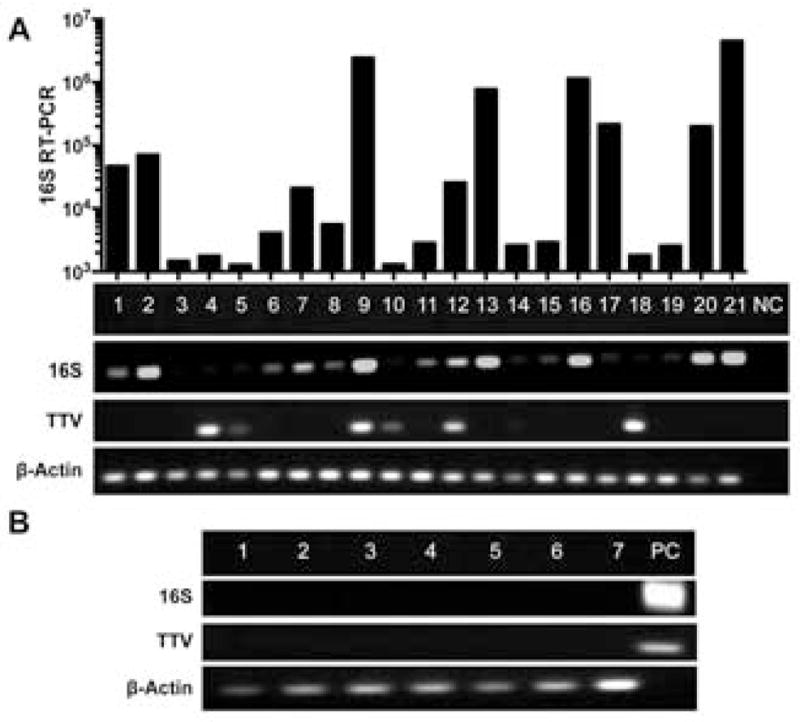
Results of RT-PCR analysis of universal bacterial 16S ribosomal primers (16S), universal set of primers for Torque Teno Virus (TTV), and β-Actin in (A) endophthalmitis samples with negative control (NC) and quantitative results for 16S and (B) normal uninflamed vitreous samples with positive control (PC). Bar graph shows bacterial RT-PCR quantitation 16S copies per mL fluid.
Bacterial BRiSK results
A mean of 2.2 x 106 sequence tags were recovered from each sample. All endophthalmitis samples yielded a majority of high quality human sequences (mean 97% human sequence). Bacterial sequences were detected in 12 of the 21 samples. Five samples were positive for Staphylococcus, five for Streptococcus species, and one each for Moraxella and Pseudomonas.
Concordance between pathogen detection techniques
Because some cultures were read as genera without speciation (for instance, ‘coagulase-negative Staphylococcus’), concordance analysis between methods was performed at the genus level (Table 2). Overall traditional culture, bacterial 16S RT-PCR, and BRiSK showed good agreement among the three different methodologies with complete agreement in 21/28 cases and controls (Fleiss’ kappa = 0.621), including 14/21 endophthalmitis cases and 7/7 negative controls. Bacterial 16S and BRiSK had kappa values of 0.605 and 0.624 respectively when compared to traditional culture, while bacterial 16S qPCR and BRiSK had a higher agreement, with kappa of 0.793.
Discrepancies between methods included four culture-positive samples that were negative for recognizable pathogens by 16S PCR and BRiSK. Two of these grew coagulase-negative Staphylococcus, one was ‘light growth Streptococcus’, and one grew Bacillus in broth only. These samples had four of the five lowest 16S/actin ratios, with levels less than 0.02 bacterial 16S sequences/human actin sequence, suggesting presence of less than one bacterium per 50 human cells. One sample was positive with Prevotella (a bacteriodes bactrerium) by culture, but Streptococcus by 16S and BRiSK, suggesting this bacterium may have been misidentified in culture. Two samples yielded bacterial sequences (for Pseudomonas and Streptococcus) by BRiSK but were culture-negative and did not yield these pathogens by 16S PCR.
Bacterial speciation by BRiSK and 16S were in concordance, and showed all cases of coagulase-negative Staphylococus to be Staphylococcus epidermidis. Speciation of Streptococcocal species was also in agreement between 16S PCR and BRiSK in the four positive Streptococcocal cases, and included two cases of Streptococcus mitis, and one case each of Streptococcus agalactiae and Streptococcus gordonii.
Non-bacterial sequences
BRiSK revealed presence of sequences belonging to TTV in numerous endophthalmitis samples. 8/14 (57.1%) of the culture-positive and 7/7 (100.0%) of culture-negative samples contained sequences specific for TTV. In contrast, none of the negative control samples were positive for TTV (Table 3) (p = 0.0005 by Fisher Exact test). The number of tags recovered ranged from 1 to greater than 3,000 per sample (Figure 2). The median number of tags recovered was higher in culture-negative than culture-positive samples (mean 26 vs. 2 tags per sample) but was not statistically significant. Quantitation of tags in BRiSK suggested multiplicity of infection ranging from less than one copy per human genome to greater than 200 copies. TTV presence was verified by qPCR in seven endophthalmitis samples (five culture-negative and two culture-positive, Figure 1). qPCR copy numbers for TTV ranged from 581 to greater than 400,000 per mL.
Table 3.
Presence of Torque Teno Virus in Culture Positive, Culture Negative, and Control Samples
| Culture + | Culture − | Control | ||||
|---|---|---|---|---|---|---|
| n | % | n | % | N | % | |
|
| ||||||
| Torque Teno + | 8 | 57.1 | 7 | 100 | - | - |
| Torque Teno − | 6 | 42.9 | - | - | 7 | 100 |
Figure 2.
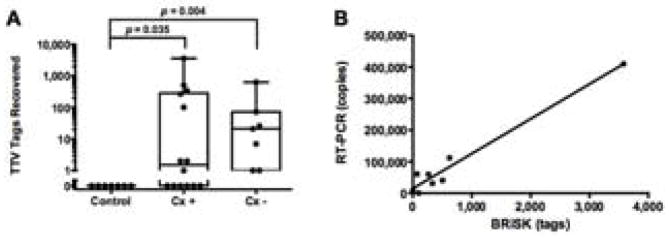
(A) Box-whisker plot of tag recovery of Torque Teno Virus (TTV) by BRiSK in culture positive (Cx +), culture negative (Cx −), and control samples (p values are adjusted for multiple comparisons). (B) Correlation between TTV tag recovery by BRiSK and RT-PCR (r2 = 0.96).
Discussion
To understand the genesis of culture-negative endophthalmitis, and to compare multiple methods for diagnosing infectious endophthalmitis, we applied three techniques (traditional culture, 16S qPCR, and representational deep DNA sequencing [BRiSK]) for detection of potential pathogens in 21 samples of presumed infectious endophthalmitis, and seven control cases without inflammation or sign of infection. Consistent with previous results, approximately one-third of the studied cases were culture-negative. We found perfect concordance amongst the three techniques for the 12 cases that were both culture-positive and showed high levels of bacterial DNA by 16S qPCR. Remarkably, only one culture-negative case yielded evidence of bacterial infection by either molecular technique, and this case (which was positive for BRiSK for Pseudomonas aeruginosa) was negative for bacterial pathogens by 16S. Although the sample size was small, this suggests that culture-negative cases of presumed infectious endophthalmitis are indeed largely devoid of abundant fastidious or novel bacteria.
A number of previous studies have examined the use of 16S PCR in the diagnosis of endophthalmitis. 12–18, 21–25 The majority of these studies have suggested superior sensitivity and specificity for PCR-based techniques. Although we could detect bacteria by high cycle-number PCR in the majority of cases, they were associated with cloned fragments belonging to unusual organisms which, in most cases, have not been associated with human disease, and these results were not corroborated by deep representational DNA sequencing, While it is possible that BRiSK may have detected unannotated sequences associated with these organisms, an alternative explanation is that these sequences represent very low level contaminants present in either the ocular sample or in reagents used for PCR. 26–28
The etiologic basis of culture-negative endophthalmitis thus remains unclear. Possibilities for this condition include: 1) acute infection with non-bacterial pathogens, 2) strong immunological reaction to scant (undetectable) bacteria, or 3) true ‘sterile’ endophthalmitis associated with antigenic response to a non-infectious antigen. 29 The unexpected finding of copious sequence tags associated with TTV is potentially consistent with the first hypothesis.
TTV is a human anellovirus discovered approximately 15 years ago from the serum of a patient with post-transfusion hepatitis30. Exposure to TTV is nearly universal. 31, 32 While TTV has not been definitively linked to any specific human disease, high titers of TTV in the serum have been associated with multiple sclerosis, systemic lupus erythematosis, hepatitis, rheumatic diseases, asthma, and fever of unknown origin (reviewed in 33). In two previous survey studies, TTV was isolated from the tears of 59% 34 and 66% 35 of subjects. Recently Smits et al. 36 described recovery of TTV from the vitreous of children affected seasonal hyperacute panuveitis (SHAPU), a severe form of panuveitis that resembles endophthalmitis, which is endemic to Nepal. 37 The authors reported that TTV was found in~90% of vitreous samples from SHAPU patients, as well as ~50% of endophthalmitis samples, but not in vitreous samples recovered from retinal detachment surgery. SHAPU is associated with the eclosion of the Tussock moth, and it is thought that migration of moth ‘hairs’ into the vitreous leads may track infection into the vitreous cavity. 37 However, SHAPU cases have been bacterial culture-negative to date. Given the presence of TTV on the ocular surface, it is possible that recovery of TTV in these samples may represent a similar inoculation route to that of post-operative endophthalmitis.
The presence of TTV in the vitreous of patients with endophthalmitis could be consistent with any of four hypotheses. First, TTV could be a bystander or marker of generalized inflammation or leukocytic infiltration. TTV may infect leukocytes.38 Thus, any condition causing a severe vitritis could be associated with recovery of TTV. TTV also can be detected in the serum and breakdown of the blood-retinal barrier due to endophthalmitis might allow TTV to enter the eye from the serum. Arguing against this, Smits et al. found ~9 fold higher incidence of TTV recovery from vitreous in SHAPU patients than in patients with other forms of uveitis. 37 Additionally, in the present study, BRiSK tag recovery of TTV was sometimes on the order of 100’s to 1000’s of copies per human genome, suggesting active replication. Second, TTV could be replicating in the eye in a facultative manner, but not be involved in the pathogenesis of disease. While this hypothesis cannot be excluded from the present experiments, it is known that TTV activates the innate immune system via toll-like receptor-9 signaling, 39 as well as generating both serologic and T-cell mediated responses (reviewed in 40). Third, TTV could be a factor influencing pathogenesis of endophthalmitis but not directly causative of disease. The finding of TTV in a majority of cases of culture-positive endophthalmitis could be consistent with this hypothesis. The immune response to TTV might be additive in this model to the response to bacterial infection. TTV is known to encode a microRNA that renders infected lymphocytes resistant to immunomodulation by endogenous interferon. 41 Finally, in the fourth model, TTV could itself by a pathogen causing endophthalmitis. These models are not mutually exclusive; for instance, TTV could be sufficient to cause endophthalmitis, but could also be an adjunct to pathogenesis for bacterial endophthalmitis.
Experimental discrimination between these models is hampered by lack of a robust in vitro system for viral replication 42 and the absence of small animal models of TTV. There is a TTV species endemic to pigs 43, 44 but it has not yet been established whether this virus is found in eye or in eye disease in these animals. Additionally, the very high sequence variation in TTV also creates challenges for understanding viral pathogenesis. At least 50 molecular variants have been documented. 33 No single set of PCR primers can detect all viral strains; this may account for the lower number of samples positive for PCR than by BRiSK in the present experiment. Whether different strains of virus (which can vary substantially in sequence) display differential pathologies is presently unknown. 33 Nonetheless, correlative data on the prevalence, strain types, and dynamic behavior TTV on the ocular surface, in larger series of endophthalmitis, and in other forms of ocular inflammation would provide data that may constrain these models.
In conclusion, we find no single technique sufficient for complete analysis of endophthalmitis for infectious pathogens. Bacterial culture and quantitative 16S PCR are comparable and complementary; but PCR is subject to potential false positive results, while speciation may be inexact using traditional culture techniques. Both molecular methods suggest that, at least in this relatively small series, culture-negative cases truly have limited bacterial loads. Deep DNA sequencing offers an additional method for analysis of ocular fluids. In addition to good corroboration with both other methods, this technique has allowed identification of significant levels of viral DNA in culture-negative and culture-positive cases. The pathologic significance of this finding remains to be elucidated.
Acknowledgments
The authors wish to thank Adam Gersenblith, MD with his help in managing specimens. This work was supported by NIH R01EY022038 (RVG), Burroughs-Wellcome Clinical Scientist Award in Translational Science (RVG), the Danforth Foundation (RVG) and the J. Arch McNamara Research Fund (SJG). This work was supported in part by NIH CORE Grant P30EY001730 and an unrestricted grant from Research to Prevent Blindness.
Footnotes
Conflict of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson MW, Doft BH, Kelsey SF, et al. The Endophthalmitis Vitrectomy Study. Relationship between clinical presentation and microbiologic spectrum. Ophthalmology. 1997;104(2):261–72. doi: 10.1016/s0161-6420(97)30326-1. [DOI] [PubMed] [Google Scholar]
- 2.Nentwich MM, Yactayo-Miranda Y, Schwarzbach F, et al. Endophthalmitis after intravitreal injection: decreasing incidence and clinical outcome-8-year results from a tertiary ophthalmic referral center. Retina. 2014;34(5):943–50. doi: 10.1097/IAE.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 3.Mithal K, Mathai A, Pathengay A, et al. Endophthalmitis following intravitreal anti-VEGF injections in ambulatory surgical centre facility: incidence, management and outcome. Br J Ophthalmol. 2013;97(12):1609–12. doi: 10.1136/bjophthalmol-2013-303222. [DOI] [PubMed] [Google Scholar]
- 4.Friling E, Lundstrom M, Stenevi U, Montan P. Six-year incidence of endophthalmitis after cataract surgery: Swedish national study. J Cataract Refract Surg. 2013;39(1):15–21. doi: 10.1016/j.jcrs.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 5.Khanna RC, Garudadri C. Incidence of post-cataract endophthalmitis at Aravind Eye Hospital. Indian J Ophthalmol. 2010;58(6):562. doi: 10.4103/0301-4738.71704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gower EW. Estimating the incidence of endophthalmitis using a national medical database. Ophthalmic Epidemiol. 2008;15(6):357–8. doi: 10.1080/09286580802624459. [DOI] [PubMed] [Google Scholar]
- 7.Mollan SP, Gao A, Lockwood A, et al. Postcataract endophthalmitis: incidence and microbial isolates in a United Kingdom region from 1996 through 2004. J Cataract Refract Surg. 2007;33(2):265–8. doi: 10.1016/j.jcrs.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Krause L, Bechrakis NE, Heimann H, et al. Incidence and outcome of endophthalmitis over a 13-year period. Can J Ophthalmol. 2009;44(1):88–94. doi: 10.3129/i08-160. [DOI] [PubMed] [Google Scholar]
- 9.Group EVS. Microbiologic factors and visual outcome in the endophthalmitis vitrectomy study. Am J Ophthalmol. 1996;122(6):830–46. doi: 10.1016/s0002-9394(14)70380-0. [DOI] [PubMed] [Google Scholar]
- 10.Shah CP, Garg SJ, Vander JF, et al. Outcomes and risk factors associated with endophthalmitis after intravitreal injection of anti-vascular endothelial growth factor agents. Ophthalmology. 2011;118(10):2028–34. doi: 10.1016/j.ophtha.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 11.Storey P, Dollin M, Pitcher J, et al. The role of topical antibiotic prophylaxis to prevent endophthalmitis after intravitreal injection. Ophthalmology. 2014;121(1):283–9. doi: 10.1016/j.ophtha.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 12.Bharathi MJ, Rameshkumar G, Ramakrishnan R, et al. Comparative evaluation of uniplex, nested, semi-nested, multiplex and nested multiplex PCR methods in the identification of microbial etiology of clinically suspected infectious endophthalmitis. Curr Eye Res. 2013;38(5):550–62. doi: 10.3109/02713683.2013.772205. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa M, Sugita S, Shimizu N, et al. Broad-range real-time PCR assay for detection of bacterial DNA in ocular samples from infectious endophthalmitis. Jpn J Ophthalmol. 2012;56(6):529–35. doi: 10.1007/s10384-012-0174-z. [DOI] [PubMed] [Google Scholar]
- 14.Joseph CR, Lalitha P, Sivaraman KR, et al. Real-time polymerase chain reaction in the diagnosis of acute postoperative endophthalmitis. Am J Ophthalmol. 2012;153(6):1031–7. e2. doi: 10.1016/j.ajo.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Sugita S, Shimizu N, Watanabe K, et al. Diagnosis of bacterial endophthalmitis by broad-range quantitative PCR. Br J Ophthalmol. 2011;95(3):345–9. doi: 10.1136/bjo.2009.171504. [DOI] [PubMed] [Google Scholar]
- 16.Chiquet C, Cornut PL, Benito Y, et al. Eubacterial PCR for bacterial detection and identification in 100 acute postcataract surgery endophthalmitis. Invest Ophthalmol Vis Sci. 2008;49(5):1971–8. doi: 10.1167/iovs.07-1377. [DOI] [PubMed] [Google Scholar]
- 17.Okhravi N, Adamson P, Matheson MM, et al. PCR-RFLP-mediated detection and speciation of bacterial species causing endophthalmitis. Invest Ophthalmol Vis Sci. 2000;41(6):1438–47. [PubMed] [Google Scholar]
- 18.Knox CM, Cevallos V, Margolis TP, Dean D. Identification of bacterial pathogens in patients with endophthalmitis by 16S ribosomal DNA typing. Am J Ophthalmol. 1999;128(4):511–2. doi: 10.1016/s0002-9394(99)00181-6. [DOI] [PubMed] [Google Scholar]
- 19.Osborne CA, Galic M, Sangwan P, Janssen PH. PCR-generated artefact from 16S rRNA gene-specific primers. FEMS Microbiol Lett. 2005;248(2):183–7. doi: 10.1016/j.femsle.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 20.Muthappan V, Lee AY, Lamprecht TL, et al. Biome representational in silico karyotyping. Genome Res. 2011;21(4):626–33. doi: 10.1101/gr.115758.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Therese KL, Anand AR, Madhavan HN. Polymerase chain reaction in the diagnosis of bacterial endophthalmitis. Br J Ophthalmol. 1998;82(9):1078–82. doi: 10.1136/bjo.82.9.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll NM, Jaeger EE, Choudhury S, et al. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol. 2000;38(5):1753–7. doi: 10.1128/jcm.38.5.1753-1757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varghese B, Rodrigues C, Deshmukh M, et al. Broad-range bacterial and fungal DNA amplification on vitreous humor from suspected endophthalmitis patients. Mol Diagn Ther. 2006;10(5):319–26. doi: 10.1007/BF03256207. [DOI] [PubMed] [Google Scholar]
- 24.Chiquet C, Lina G, Benito Y, et al. Polymerase chain reaction identification in aqueous humor of patients with postoperative endophthalmitis. J Cataract Refract Surg. 2007;33(4):635–41. doi: 10.1016/j.jcrs.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Navarro-Noya Y, Hernandez-Rodriguez C, Zenteno JC, et al. 16S rRNA gene-based identification of bacteria in postoperative endophthalmitis by PCR-Denaturing Gradient Gel Electrophoresis (PCR-DGGE) fingerprinting. Braz J Microbiol. 2012;43(1):283–7. doi: 10.1590/S1517-838220120001000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philipp S, Huemer HP, Irschick EU, Gassner C. Obstacles of Multiplex Real-Time PCR for Bacterial 16S rDNA: Primer Specifity and DNA Decontamination of Taq Polymerase. Transfus Med Hemother. 2010;37(1):21–8. doi: 10.1159/000265571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugita S, Ogawa M, Shimizu N, et al. Use of a comprehensive polymerase chain reaction system for diagnosis of ocular infectious diseases. Ophthalmology. 2013;120(9):1761–8. doi: 10.1016/j.ophtha.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 28.Cherkaoui A, Emonet S, Ceroni D, et al. Development and validation of a modified broad-range 16S rDNA PCR for diagnostic purposes in clinical microbiology. J Microbiol Methods. 2009;79(2):227–31. doi: 10.1016/j.mimet.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Otsuka H, Kawano H, Sonoda S, et al. Particle-induced endophthalmitis: possible mechanisms of sterile endophthalmitis after intravitreal triamcinolone. Invest Ophthalmol Vis Sci. 2013;54(3):1758–66. doi: 10.1167/iovs.12-11247. [DOI] [PubMed] [Google Scholar]
- 30.Naoumov NV, Petrova EP, Thomas MG, Williams R. Presence of a newly described human DNA virus (TTV) in patients with liver disease. Lancet. 1998;352(9123):195–7. doi: 10.1016/S0140-6736(98)04069-0. [DOI] [PubMed] [Google Scholar]
- 31.Handa A, Dickstein B, Young NS, Brown KE. Prevalence of the newly described human circovirus, TTV, in United States blood donors. Transfusion. 2000;40(2):245–51. doi: 10.1046/j.1537-2995.2000.40020245.x. [DOI] [PubMed] [Google Scholar]
- 32.Vasilyev EV, Trofimov DY, Tonevitsky AG, et al. Torque Teno Virus (TTV) distribution in healthy Russian population. Virol J. 2009;6:134. doi: 10.1186/1743-422X-6-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hino S, Miyata H. Torque teno virus (TTV): current status. Rev Med Virol. 2007;17(1):45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- 34.Matsubara H, Michitaka K, Horiike N, et al. Existence of TT virus DNA in extracellular body fluids from normal healthy Japanese subjects. Intervirology. 2000;43(1):16–9. doi: 10.1159/000025018. [DOI] [PubMed] [Google Scholar]
- 35.Emre S, Otlu B, Cankaya C, et al. Transfusion-transmitted virus DNA in serum, tear and aqueous humour of patients undergoing cataract operation. Clin Experiment Ophthalmol. 2007;35(8):759–62. doi: 10.1111/j.1442-9071.2007.01575.x. [DOI] [PubMed] [Google Scholar]
- 36.Smits SL, Manandhar A, van Loenen FB, et al. High prevalence of anelloviruses in vitreous fluid of children with seasonal hyperacute panuveitis. J Infect Dis. 2012;205(12):1877–84. doi: 10.1093/infdis/jis284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manandhar A. Seasonal hyperacute panuveitis: an update. Curr Opin Ophthalmol. 2011;22(6):496–501. doi: 10.1097/ICU.0b013e32834bcbf4. [DOI] [PubMed] [Google Scholar]
- 38.de Oliveira JC, Nasser TF, Oda JM, et al. Detection of TTV in peripheral blood cells from patients with altered ALT and AST levels. New Microbiol. 2008;31(2):195–201. [PubMed] [Google Scholar]
- 39.Rocchi J, Ricci V, Albani M, et al. Torquetenovirus DNA drives proinflammatory cytokines production and secretion by immune cells via toll-like receptor 9. Virology. 2009;394(2):235–42. doi: 10.1016/j.virol.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 40.Gergely P, Jr, Perl A, Poor G. Possible pathogenic nature of the recently discovered TT virus: does it play a role in autoimmune rheumatic diseases? Autoimmun Rev. 2006;6(1):5–9. doi: 10.1016/j.autrev.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Kincaid RP, Burke JM, Cox JC, et al. A human torque teno virus encodes a microRNA that inhibits interferon signaling. PLoS Pathog. 2013;9(12):e1003818. doi: 10.1371/journal.ppat.1003818. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Desai M, Pal R, Deshmukh R, Banker D. Replication of TT virus in hepatocyte and leucocyte cell lines. J Med Virol. 2005;77(1):136–43. doi: 10.1002/jmv.20426. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto H. TT viruses in animals. Curr Top Microbiol Immunol. 2009;331:35–52. doi: 10.1007/978-3-540-70972-5_3. [DOI] [PubMed] [Google Scholar]
- 44.Kekarainen T, Segales J. Torque teno virus infection in the pig and its potential role as a model of human infection. Vet J. 2009;180(2):163–8. doi: 10.1016/j.tvjl.2007.12.005. [DOI] [PubMed] [Google Scholar]


