Abstract
Yersinia pestis, the causative agent of plague, can be transmitted by fleas by two different mechanisms: by early-phase transmission (EPT), which occurs shortly after flea infection, or by blocked fleas following long-term infection. Efficient flea-borne transmission is predicated upon the ability of Y. pestis to be maintained within the flea. Signature-tagged mutagenesis (STM) was used to identify genes required for Y. pestis maintenance in a genuine plague vector, Xenopsylla cheopis. The STM screen identified seven mutants that displayed markedly reduced fitness in fleas after 4 days, the time during which EPT occurs. Two of the mutants contained insertions in genes encoding glucose 1-phosphate uridylyltransferase (galU) and UDP-4-amino-4-deoxy-l-arabinose-oxoglutarate aminotransferase (arnB), which are involved in the modification of lipid A with 4-amino-4-deoxy-l-arabinose (Ara4N) and resistance to cationic antimicrobial peptides (CAMPs). These Y. pestis mutants were more susceptible to the CAMPs cecropin A and polymyxin B, and produced lipid A lacking Ara4N modifications. Surprisingly, an in-frame deletion of arnB retained modest levels of CAMP resistance and Ara4N modification, indicating the presence of compensatory factors. It was determined that WecE, an aminotransferase involved in biosynthesis of enterobacterial common antigen, plays a novel role in Y. pestis Ara4N modification by partially offsetting the loss of arnB. These results indicated that mechanisms of Ara4N modification of lipid A are more complex than previously thought, and these modifications, as well as several factors yet to be elucidated, play an important role in early survival and transmission of Y. pestis in the flea vector.
Introduction
Yersinia pestis can undergo transmission from fleas to mammalian hosts by at least two different mechanisms: bacteria may be transmitted shortly after infection of a flea vector [early-phase transmission (EPT), 1–4 days post-infection] in a biofilm-independent manner (Eisen et al., 2006; Kartman et al., 1958; Vetter et al., 2010) or after long-term infection, during which time they form a biofilm and block the feeding ability of the flea (Bacot & Martin, 1914; Burroughs, 1947; Chouikha & Hinnebusch, 2012). Currently, there is little known about the mechanisms by which EPT occurs, but many flea species are able to efficiently transmit Y. pestis in this manner (Eisen & Gage, 2012; Eisen et al., 2007), which may play a significant role in the spread of plague.
Plague transmission via blocked fleas was described 100 years ago (Bacot & Martin, 1914) and, although there is still much that is unknown about the mechanisms involved, a small number of transmission factors required for survival and biofilm formation in fleas have been identified. These factors include the ymt (Yersinia murine toxin) gene, which encodes a product with phospholipase D activity. Ymt is important for maintaining Y. pestis infections in the flea vector, ostensibly by protecting the bacteria from unknown toxic byproducts generated during the digestion of the flea’s blood meal (Hinnebusch et al., 2000, 2002; Rudolph et al., 1999). Additionally, the ability of Y. pestis to form stable biofilms is important in its ability to block fleas and thus be transmitted by the classic route. The hms, gmhA and phoPQ genes are necessary for flea blockage through their involvement in biofilm formation (Darby et al., 2005; Hinnebusch et al., 1996; Jarrett et al., 2004; Rebeil et al., 2013). Although these genes play important roles in the association of Y. pestis with fleas, they do not fully explain the organism’s ability to survive in arthropods and undergo successful transmission (Eisen et al., 2006; Hinnebusch et al., 1996; Houhamdi & Raoult, 2008). Therefore, it is likely that there are other transmission factors yet to be identified that enable Y. pestis to utilize a flea vector and spread to vertebrate hosts.
Here, we describe the construction and screening of a Y. pestis signature-tagged mutagenesis (STM) library (Hensel et al., 1995; Mazurkiewicz et al., 2006) to identify genes that may play a role in survival within a well-recognized flea vector of plague, Xenopsylla cheopis. This approach identified seven mutants with significant survival or colonization defects in the flea vector, including at least two involved in lipid A modification, which is predicted to allow Y. pestis to resist the action of cationic antimicrobial peptides (CAMPs) produced by the insect. Furthermore, we identified a novel role for WecE, an enzyme normally involved in enterobacterial common antigen (ECA) biosynthesis, in modification of lipid A.
Methods
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Y. pestis strains were routinely grown in heart infusion (HI) broth (Difco; BD) or on HI agar at 28 or 37 °C, as indicated. Escherichia coli strains were grown in Luria–Bertani medium at 37 °C. Unless otherwise indicated, antibiotics were used at the following concentrations: 50 µg kanamycin ml−1, 100 µg ampicillin ml−1, 2 µg irgasan ml−1, 15 µg tetracycline ml−1 and 12.5 µg chloramphenicol ml−1. The pUT-miniTn5Km2 (Apr, Kmr) STM plasmid library (a gift from D. Holden, Imperial College, London, UK) was used as the source of tagged transposons (Hensel et al., 1995).
Table 1. Bacterial strains and plasmids.
| Strain or plasmid | Relevant characteristics/usage | Reference/source |
| Y. pestis strains | ||
| KIM6+ | Parent strain; pCD−, pMT+, pPCP+ | Perry et al. (1990) |
| 3C1 | KIM6+ STM mutant with transposon insertion in galU | This study |
| 3C1-C | 3C1 complemented with pGEN-MCS plus galU | This study |
| 3C1-EV | 3C1 harbouring pGEN-MCS empty vector | This study |
| 9H9 | KIM6+ STM mutant with transposon insertion in arnB | This study |
| 9H9-C | 9H9 complemented with pCR2.1-TOPO plus arnBCADTEF operon | This study |
| 9H9-EV | 9H9 harbouring pCR2.1-TOPO empty vector | This study |
| KIM6+ΔarnB | In-frame deletion of arnB | This study |
| KIM6+ΔarnB-C | KIM6+ΔarnB complemented with pCR2.1-TOPO plus arnB | This study |
| KIM6+ΔarnB-EV | KIM6+ΔarnB harbouring pCR2.1-TOPO empty vector | This study |
| KIM6+ ΔwecE | In-frame deletion of wecE | This study |
| KIM6+ ΔwecE-C | KIM6+ΔwecE complemented with pCR2.1-TOPO plus wecE | This study |
| KIM6+ ΔwecE-EV | KIM6+ΔwecE harbouring pCR2.1-TOPO empty vector | This study |
| KIM6+ ΔarnBΔwecE | In-frame deletions of both arnB and wecE | This study |
| KIM6+ ΔarnBΔwecE-Ca | KIM6+ΔarnBΔwecE complemented with pCR2.1-TOPO plus arnB | This study |
| KIM6+ ΔarnBΔwecE-Cw | KIM6+ΔarnBΔwecE complemented with pCR2.1-TOPO plus wecE | This study |
| KIM6+ ΔarnBΔwecE-Caw | KIM6+ΔarnBΔwecE complemented with pCR2.1-TOPO plus arnB, wecE | This study |
| KIM6+ ΔarnBΔwecE-EV | KIM6+ΔarnBΔwecE harbouring pCR2.1-TOPO empty vector | This study |
| E. coli strains | ||
| DH5α | Cloning strain for complementation constructs | Invitrogen |
| S17-1λpir | Cloning strain for STM library generation | Simon et al. (1983) |
| Plasmids | ||
| pKD46 | λRed recombinase expression plasmid; Apr | Datsenko & Wanner (2000) |
| pCR2.1-TOPO | Complementation cloning plasmid; Apr, Kmr | Invitrogen |
| pFLP2 | Source of Flp recombinase; Apr | Hoang et al. (1998) |
| pFKM1 | Kanamycin resistance cassette with flanking FRT sites; Kmr | Choi et al. (2005) |
| pGEN-MCS | Complementation cloning plasmid; Apr | Lane et al. (2007) |
| pUTmini-Tn5Km2 | Pool of plasmids with uniquely tagged transposons; Kmr | Hensel et al. (1995) |
STM library generation.
Tagged pUT-miniTn5Km2 plasmids were transformed into E. coli S17-1λpir and isolated colonies were picked into three 96-well plates, and screened for amplification and hybridization efficiency using a modified colony blot assay. Briefly, 50 µl overnight culture of each clone was transferred to a positively charged Nytran nylon membrane (Whatman) by vacuum filtration with a 96-well dot-blot apparatus (Bio-Rad). Bacterial cells were lysed and DNA was denatured with 1 N NaOH and washed with 2× SSC (saline sodium citrate) prior to hybridization to labelled probes. Clones with the strongest hybridization signals were assembled into pools and screened for cross-hybridization as described by Hensel et al. (1995). Cross-reactive clones were removed prior to assembly of the final optimized set of 96 STM vectors.
Separate matings were performed between Y. pestis KIM6+ and each of the 96 E. coli S17-1λpir clones carrying uniquely tagged pUT-miniTn5Km2 using a 96-well broth mating strategy. Approximately equal numbers of cells from overnight cultures of Y. pestis and each E. coli clone were mixed, pelleted (10 000 g, 1 min) and allowed to mate for 3 h at 28 °C. Cells were resuspended in 1 ml 0.9 % saline and aliquots were plated onto HI agar containing kanamycin (300 µg ml−1), ampicillin and irgasan. Individual colonies from each mating were picked to a series of 39 separate 96-well plates such that each sequence tag resided in the same position of each plate. Based on the Poisson distribution, we estimated that ~53 % of Y. pestis genes were mutated in this library (Sambrook et al., 1989). Genuine transposition events were confirmed by screening each clone for kanamycin resistance and ampicillin sensitivity.
In vitro infection of X. cheopis fleas.
Adult X. cheopis fleas that had been starved for 7–10 days were fed through freshly prepared mouse skins attached to glass feeders containing defibrinated Sprague–Dawley rat blood (Bioreclamation) as described previously (Eisen et al., 2006). Briefly, for STM screening, each pool of 96 clones was grown overnight and harvested by centrifugation. Cells were washed in 0.1 vol. 0.85 % NaCl and resuspended in rat blood at a concentration of 1–4×109 c.f.u. ml−1. Mouse skins were recovered after euthanasia of animals used for other purposes under protocols approved by the Centers for Disease Control and Prevention, Division of Vector-Borne Diseases Animal Care and Use Committee (06-014, 08-022, 11-015). For in vivo competition assays, individual STM mutant and WT bacteria were mixed in rat blood and plated as described below to determine initial infection ratios. Up to 150 fleas per feeder were allowed to feed for 1 h and then scored by light microscopy for the presence or absence of a fresh blood meal. Unfed fleas were discarded. The remaining fed fleas were maintained for 4 days at 23 °C and 80–85 % relative humidity, and then live fleas were transferred to vials and maintained at −80 °C until processed. To determine the concentration of bacteria in each feeder, the infected blood was serially diluted and plated in triplicate onto blood agar base containing 6 % sheep blood supplemented with kanamycin where appropriate. Plates were incubated at 28 °C for 48 h prior to enumeration of colonies.
Identification of STM mutants with possible survival defects in the flea.
Viable Y. pestis mutants were recovered by culturing disrupted fleas (18 per pool) in HI supplemented with kanamycin (Eisen et al., 2006). Genomic DNA was isolated from recovered mutants (output DNA) as well as viable Y. pestis isolated from infected blood meals (input DNA) using a Wizard Genomic DNA Purification kit (Promega). Probes for hybridization were generated essentially as described previously (Hensel et al., 1995; Mei et al., 1997). Briefly, transposon tags were PCR-amplified from genomic DNA samples using primers P2 and P4 (Table S1, available in the online Supplementary Material), and the resulting ~80 bp product was gel-purified. This pool of amplified tags was used as template for generation of DIG-dUTP-labelled probes (PCR DIG Probe Synthesis kit; Roche) using primers P2 and P4 (Mei et al., 1997). Probes were digested with HindIII and the labelled tag sequences were excised from 4 % agarose gels (Hensel et al., 1995). The gel slices were melted and denatured in hybridization buffer (DIG Easy Hyb; Roche) and hybridized at 42 °C overnight to pairs of dot-blot membranes (Mei et al., 1997) containing arrays of the 96 unique sequence-tagged plasmids that had been vacuum blotted, alkali denatured and UV cross-linked to Nytran nylon (Sambrook et al., 1989). Following hybridization, membranes were washed under high stringency, incubated in DIG-blocking solution and blocking solution containing anti-DIG-alkaline phosphatase antibody conjugate for 30 min each, then washed three times with wash buffer according to the manufacturer’s recommendations (DIG Wash and Block Buffer set; Roche). Membranes were incubated 5 min with detection buffer containing CSPD luminescent substrate (DIG Luminescent Detection kit; Roche), excess buffer was decanted and the membranes were incubated for an additional 10 min at 35 °C prior to exposing to autoradiography film. Mutants that displayed decreased hybridization signals in the output compared with the input DNA pools were rescreened by flea infection in a different background of clones to confirm the loss of signal.
Insertion sites of STM transposons in mutants with possible survival defects were determined using the single-primer PCR (spPCR) technique as described by Karlyshev et al. (2000). Briefly, spPCR was performed on each STM mutant using the transposon-anchored, outward-facing primers, P6U and P7U (Table S1), which bind to sequences on opposite ends of the STM transposon. The resulting spPCR products were purified using ExoSAP-IT (USB), and sequenced on an ABI PRISM 3730 genetic analyser using the nested primers P6M and P7M (Table S1). Insertion sites were identified by performing sequence similarity searches using blastn, and verified by gene-specific PCR and sequencing.
Competition assays.
STM mutants that showed repeated loss of signal in the pool-based screening were analysed for survival relative to WT bacteria both in vivo and in vitro. Mixtures of a single STM mutant and WT Y. pestis were used to infect fleas, and were also inoculated into HI broth. After 4 days, fed fleas were collected and stored at −80 °C until use. Fleas (n = 17–39) were triturated in 250 µl HI broth plus 10 % glycerol, and then serial dilutions were plated on HI agar with and without kanamycin to calculate the output ratio of the total numbers of mutant (number of kanamycin-resistant colonies) to WT (total number of colonies minus number of kanamycin-resistant colonies) Y. pestis. Input blood meals and both time 0- and 4-day HI broth competition cultures were similarly diluted and plated to calculate input and output ratios. Competitive indices were calculated as the output ratio/input ratio for each competition.
RNA extraction and reverse transcription (RT)-PCR.
RNA was extracted from Y. pestis strains using RiboZol RNA Extraction Reagent (Amresco) as directed. RT was performed using Moloney murine leukemia virus reverse transcriptase (Thermo Scientific) as directed, using 100 ng RNA and random hexamers for cDNA synthesis. Control reactions without the addition of reverse transcriptase were also performed. First-strand cDNA was amplified using Taq polymerase (Roche) and primers for each gene in the arn operon (y1917–y1923) as well as a reference gene, rpoD (Table S1). Products were detected by electrophoresis on a 4 % agarose gel containing ethidium bromide.
Generation and complementation of mutants.
Unmarked, in-frame deletions of the arnB and wecE genes were generated in KIM6+ using the λRed recombineering approach (Datsenko & Wanner, 2000; Derbise et al., 2003). Briefly, a kanamycin resistance gene flanked by FRT (flippase recognition target) sites was amplified from pFKM1 using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and primers with 75 bp oligonucleotide tails homologous to the sequences up and downstream of the target gene (Table S1). The linear PCR product was transformed into KIM6+ electrocompetent cells harbouring the pKD46 plasmid (Datsenko & Wanner, 2000) that had been induced with 0.2 % arabinose for 4–6 h. Recombinants were selected on HI plates with kanamycin and subsequently screened for loss of pKD46. Ampicillin-sensitive recombinants were transformed with pFLP2 (Hoang et al., 1998) to remove the kanamycin cassette, leaving a 126 bp in-frame scar site that contained the start codon as well as the last 12 codons of the target gene. The STM and λRed mutant strains were complemented by amplifying the target gene, including its native promoter region, from KIM6+ using the Expand High Fidelity PCR System (Roche) and primers indicated in Table S1. The resulting amplicons were then cloned into pGEN-MCS (a gift from H. Mobley, University of Michigan, Ann Arbor, MI, USA) or pCR2.1-TOPO (Invitrogen) as indicated in Table 1, and mutant strains were transformed with complementation vectors by electroporation. Recombinants and complementation vectors were sequenced for verification prior to further experimentation.
CAMP susceptibility assays.
CAMP susceptibility assays were performed as described by Felek et al. (2010), except as otherwise noted. Briefly, twofold serial dilutions of the CAMPs polymyxin B and cecropin A were prepared in HI broth in 96-well round-bottom plates. Bacterial strains were grown overnight at 28 or 37 °C and 105 cells were added to each well in triplicate. Plates were incubated overnight at 28 or 37 °C and the MIC for each strain was recorded as the lowest CAMP concentration at which bacterial growth was not observed. E. coli ATCC 25922 was used for quality control.
Lipid A extraction.
LPS was extracted from overnight bacterial cultures grown at 28 °C using an LPS extraction kit (iNtRON Biotechnology) as directed, except 20 ml culture was initially harvested for use. The extracted LPS was subjected to mild acid hydrolysis as described by Kawasaki (2009). Briefly, the dried LPS pellet was suspended in 150 µl 100 mM sodium acetate buffer (pH 4.5) and incubated for 3 h at 95 °C. Then, 600 µl chloroform/methanol mixture (1 : 2), 200 µl chloroform and 100 µl PBS were added sequentially followed by centrifugation at 2000 r.p.m. for 5 min. The aqueous phase was discarded and 600 µl chloroform/methanol/0.1 M KCl mixture (3 : 47 : 48, by vol.) was added to the chloroform phase and mixed, followed by centrifugation at 2000 r.p.m. for 5 min. Finally, the aqueous phase was discarded and the remaining lipid A-containing chloroform phase was transferred to a new tube and dried under a stream of nitrogen. Lipid A samples were resuspended in 50 µl chloroform/methanol/water mixture (4 : 4 : 1, by vol.).
MS of lipid A.
Lipid A preparations were analysed by MALDI-TOF MS in the negative-ion mode using a Bruker microflex LRF instrument (Bruker Daltonics). Lipid A samples were mixed with an equal volume of 5-chloro-2-mercaptobenzothiazole [20 mg ml−1 in chloroform/methanol/water (4 : 4 : 1, by vol.)] supplemented with 20 mM EDTA/ammonium hydroxide (Zhou et al., 2010) and 0.5 µl aliquots were spotted onto the target plate. Spectra were acquired in the reflectron mode and analysed using flexAnalysis (Bruker Daltonics). External calibration was performed using diphosphoryl lipid A from E. coli F583 (Sigma-Aldrich). To identify relative differences in levels of 4-amino-4-deoxy-l-arabinose (Ara4N) modification, Ara4N-modified lipid A peaks were compared to their cognate, unmodified lipid A peaks [(sum of Ara4N-modified peaks at 1455, 1535, 1717 and 1953 m/z)/(sum of unmodified peaks at 1324, 1404, 1586 and 1822 m/z)] for the following Y. pestis strains: KIM6+ (n = 10 biological replicates), 9H9 (n = 8), 9H9-C (n = 5), ΔarnB (n = 12), ΔwecE (n = 10), ΔarnBΔwecE (n = 5), ΔarnBΔwecE-Ca (n = 5), ΔarnBΔwecE-Cw (n = 5) and ΔarnBΔwecE-Caw (n = 5). Mean ratios (derived from 500 shots per biological replicate)±sd were plotted for each strain. Mann–Whitney tests were performed in GraphPad Prism to determine whether the mean proportions of Ara4N differed significantly between the mutant strains and WT.
Results
STM mutant generation and negative selection
To identify genes involved in the survival or maintenance of Y. pestis in its flea vector, STM was used to randomly generate ~3800 mutants with 96 sequence-tagged transposons. Pools of 96 mutants were used to infect fleas and were evaluated for loss of individual mutants after 4 days of infection. Mutants lost in the STM screen were re-pooled and reintroduced into fleas for verification. Those with reproducible STM results were subjected to in vivo and in vitro competition with WT Y. pestis KIM6+. Seven mutants showed dramatically reduced ability to compete with WT in the flea. These defects were not due to general growth deficiencies, as all mutants were able to compete effectively with WT in vitro over the same time period (Table 2).
Table 2. STM mutants with decreased competitive index in fleas.
| STM mutant | Y. pestis KIM10+ locus tag | Gene | Tn5 insertion site* | Product | Competitive index† | |
| In vivo | In vitro | |||||
| 3C1 | y2631 | galU | 2 895 235 | UTP-glucose 1-phosphate uridylyltransferase | 0.0037 | 5.23 |
| 3D9 | y2283 | 2 519 947 | Hypothetical | 0.0383 | 2.07 | |
| 5F7 | y1748 | 1 929 492 | Putative transposase | 0.0024 | 1.67 | |
| 9F7 | y2422 | 2 689 958 | Hypothetical | 0.0306 | 1.20 | |
| 9H9 | y1917 | arnB | 2 110 322 | UDP-4-amino-4-deoxy-l-arabinose-oxoglutarate aminotransferase | 0.0222 | 1.10 |
| 10A3 | y3386 | 3 734 381 | Carbohydrate symporter permease | 0.0259 | 1.32 | |
| 17B4 | y4035‡ | 4 474 082‡ | Hypothetical | 0.0331 | 3.28 | |
Base pair location of transposon insertion site according to KIM10+ genome.
Competitive index of STM mutant to KIM6+ at 4 days post-infection/inoculation.
Transposon inserted into promoter region.
Identification of genes disrupted in STM mutants
STM transposon insertion sites were identified using spPCR (Karlyshev et al., 2000). Insertion sites were within ORFs for six of the seven STM mutants, whilst one was located in a promoter region (Table 2). Three mutants had insertions in genes encoding hypothetical proteins: one in a predicted permease and one in a predicted transposase. Two mutants had insertions in galU and arnB, which were predicted to encode proteins involved in LPS modification with Ara4N and resistance to CAMPs (Breazeale et al., 2003; Fernández et al., 2012; Klein et al., 2012).
CAMP susceptibility
Modification of bacterial LPS with Ara4N confers resistance to CAMPs, some of which are produced by the insect host immune system (Bulet & Stöcklin, 2005; Ravi et al., 2011; Vadyvaloo et al., 2010). Both the 3C1 (galU) and 9H9 (arnB) mutants showed increased sensitivity to the CAMPs polymyxin B and cecropin A (Table 3). Complementation of the mutants with the galU gene or the complete arn operon in trans restored WT levels of CAMP resistance (Table 3). Complementation of 9H9 with only arnB did not restore resistance to CAMPs, indicating that the transposon insertion had polar effects on the arn operon. RT-PCR confirmed that the expression of the arn operon genes was essentially eliminated in the 9H9 mutant. To define the contribution of arnB to CAMP resistance, an in-frame deletion was generated (KIM6+ΔarnB). Interestingly, this mutant displayed intermediate levels of resistance to CAMPs (Table 3), indicating that Y. pestis may have partial redundancy in the pathway for Ara4N modification of lipid A.
Table 3. Resistance of Y. pestis strains to cationic antimicrobial peptides.
| Strain | MIC (μg ml−1) | |||
| Polymyxin B | Cecropin A | |||
| 28 °C | 37 °C | 28 °C | 37 °C | |
| KIM6+ | ≥256 | 2 | ≥80 | 20 |
| 3C1 | 0.25 | 0.125 | 5 | 5 |
| 3C1-C (pGEN-galU) | ≥256 | 0.25 | ≥80 | 20 |
| 3C1-EV (pGEN-MCS) | 0.125 | 0.125 | 5 | 2.5 |
| 9H9 | 0.25 | 0.125 | 5 | 5 |
| 9H9-C (pCR2.1-arnBCADTEF) | ≥256 | 1 | ≥80 | 20 |
| 9H9-EV (pCR2.1-TOPO) | 0.25 | 0.125 | 5 | 5 |
| KIM6+ΔarnB | 32 | 0.25 | ≥80 | 10 |
| KIM6+ΔarnB-C (pCR2.1-arnB) | 128 | 1 | ≥80 | 20 |
| KIM6+ΔarnB-EV (pCR2.1-TOPO) | 32 | 0.25 | ≥80 | 10 |
| KIM6+ΔwecE | 0.25 | 0.25 | ≥80 | 10 |
| KIM6+ΔwecE-C (pCR2.1-wecE) | ≥256 | 2 | ≥80 | 20 |
| KIM6+ΔwecE-EV (pCR2.1-TOPO) | 0.25 | 0.25 | ≥80 | 10 |
| KIM6+ΔarnBΔwecE | 0.25 | 0.125 | 10 | 10 |
| KIM6+ΔarnBΔwecE-Ca (pCR2.1-arnB) | 1 | 0.25 | ≥80 | 10 |
| KIM6+ΔarnBΔwecE-Cw (pCR2.1-wecE) | 16 | 0.25 | ≥80 | 10 |
| KIM6+ΔarnBΔwecE-Caw (pCR2.1-arnB, wecE) | ≥256 | 1 | ≥80 | 10 |
To further explore this pathway, in-frame deletions of a related sugar aminotransferase gene, wecE (KIM6+ΔwecE), as well as a double mutant (KIM6+ΔarnBΔwecE) were generated. The double mutant was more susceptible to CAMPs than the arnB mutant, with MICs similar to the whole-operon mutant (9H9). Surprisingly, the wecE mutant was nearly as susceptible as the double mutant, indicating a significant role in CAMP resistance (Table 3). Complementation of the double mutant with both the arnB and wecE genes restored WT levels of CAMP resistance, whilst complementation with either arnB or wecE alone restored moderate levels of resistance, similar to those of the respective single mutants (Table 3).
MALDI-TOF MS of lipid A
As galU and arnB were previously implicated in CAMP resistance via Ara4N modification of lipid A (Gunn et al., 1998; Jiang et al., 2010; Klein et al., 2012), the LPS of these mutants was examined. Y. pestis has rough LPS, lacking O-antigen (Skurnik et al., 2000), and its structure varies depending on growth temperature, with Ara4N modifications less prevalent at 37 °C than at lower temperatures (Anisimov et al., 2005; Rebeil et al., 2004). After growth at 28 °C, SDS-PAGE revealed no gross differences in LPS structure, so MALDI-TOF MS was performed on lipid A to identify more subtle alterations. The WT strain displayed peaks corresponding to tetra (1HPO3)-, tetra (2HPO3)-, penta- and hexa-acylated lipid A species at m/z 1324, 1404, 1586 and 1822, respectively, indicating unmodified lipid A, plus additional peaks corresponding to the Ara4N modification of each of these lipid A species (+131 m/z) (Fig. 1a). The lipid A spectrum of the 9H9 mutant displayed only the four unmodified lipid A peaks, with an absence of the peaks representing Ara4N modification (Fig. 1). Consistent with data from Klein et al. (2012), the 3C1 (galU) mutant showed a lipid A pattern nearly identical to the 9H9 mutant, indicating loss of Ara4N modifications (Fig. 2). Complementation of the 9H9 mutant with the full arn operon, but not arnB alone, resulted in a WT spectral pattern indicating restoration of Ara4N modification, which is consistent with the increased CAMP MICs (Fig. 1). As expected, the complemented 3C1 mutant displayed a WT lipid A profile, and the spectra for the 9H9-EV and 3C1-EV (empty vector control) strains appeared identical to those of their respective mutant strains. Taken together, these data confirmed that galU and the arn operon were involved in Ara4N modification of lipid A in Y. pestis.
Fig. 1.
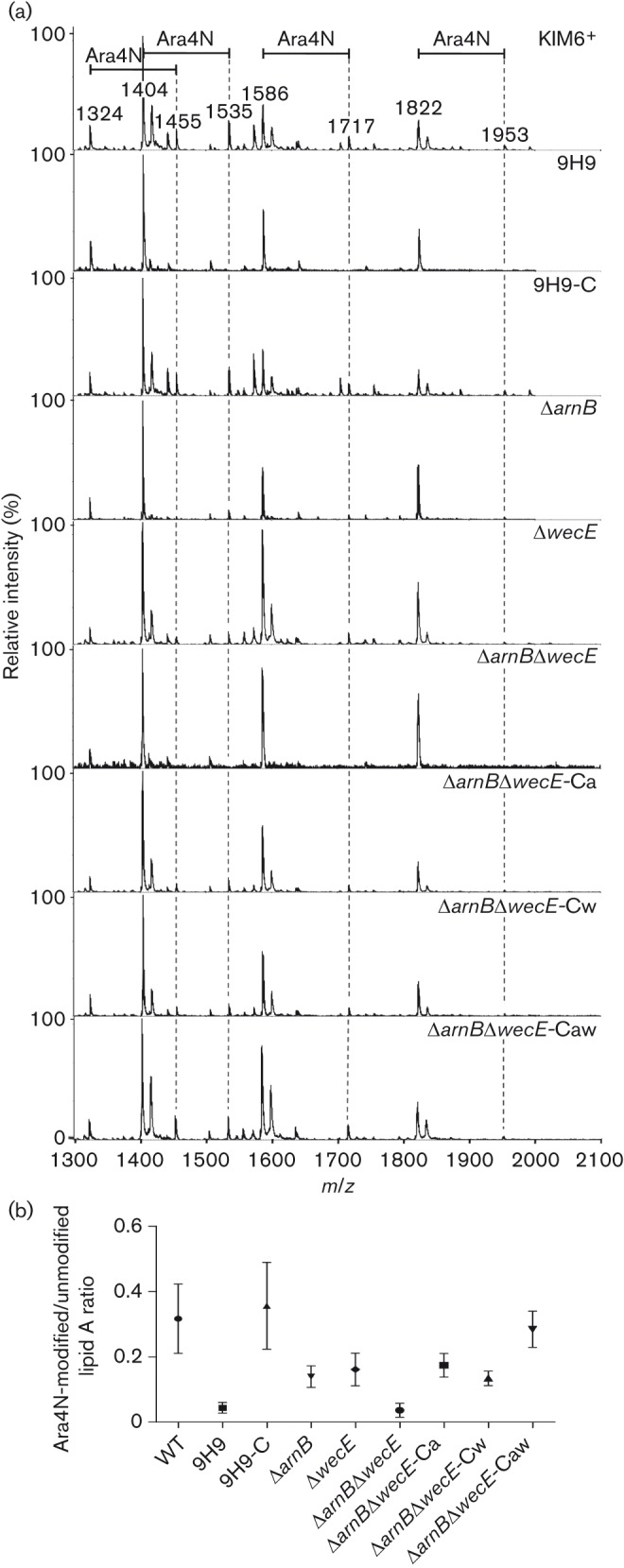
MALDI-TOF MS analysis of lipid A from Y. pestis. (a) Representative spectra of Y. pestis strains. Four major lipid A species consisting of tetra (1HPO3)-, tetra (2HPO3)-, penta- and hexa-acylated forms are present in all strains (1324, 1404, 1586 and 1822 m/z, respectively). Peaks indicating the addition of an Ara4N group (+131 m/z) to each of these species are indicated by the dotted lines. (b) Mean ratios of Ara4N-modified (sum of peak intensities at 1455, 1535, 1717 and 1953 m/z) to unmodified (sum of peak intensities at 1324, 1404, 1586 and 1822 m/z) lipid A in Y. pestis strains. Bar, sd.
Fig. 2.
MALDI-TOF MS analysis of lipid A from WT and 3C1 mutants. (a) Representative WT KIM6+ Y. pestis spectrum. Four major lipid A species consisting of tetra (1HPO3)-, tetra (2HPO3)-, penta- and hexa-acylated forms are present (1324, 1404, 1586 and 1822 m/z, respectively). Peaks indicating the addition of an Ara4N group (+131 m/z) to each of these species are indicated by the dotted lines. (b) Representative STM mutant 3C1 spectrum. Peaks representing Ara4N modifications are absent. (c) Representative STM mutant 3C1 complemented with galU (3C1-C) spectrum. Ara4N peaks are restored.
In contrast to the 9H9 insertion mutant, which effectively lacked expression of the entire arn operon, the KIM6+ΔarnB mutant displayed some Ara4N-modified lipid A, albeit at significantly lower levels than WT (Fig. 1b, P = 0.0001), which was consistent with the intermediate CAMP susceptibility observed for this strain. Complementation with the arnB gene restored WT levels of Ara4N modification, whilst the empty vector alone did not alter the mutant’s spectra. KIM6+ΔwecE also showed significantly lower levels of Ara4N-modified lipid A than WT (Fig. 1b, P = 0.0040), consistent with its susceptibility to CAMPs and complementation with wecE yielded spectra similar to WT. The KIM6+ΔarnBΔwecE lipid A profile was similar to that of the 9H9 mutant, with no evidence of Ara4N modification (Fig. 1b). Complementation with both genes restored WT modification of lipid A (P = 0.8371), whilst complementation with arnB or wecE individually restored partial Ara4N modification (P = 0.0159 and P = 0.0028, respectively, compared with double mutant), similar to that seen in the single mutants (Fig. 1b).
Discussion
In this study, STM was used to generate an insertional mutant library to identify genes that contribute to the survival or maintenance of Y. pestis in its flea vector through the first 4 days of infection – the period of time during which Y. pestis can efficiently undergo EPT. STM is a powerful negative selection technique that allows for pool-based mutant analysis, enabling the identification of mutants unable to be effectively maintained in the flea (Hensel et al., 1995; Mazurkiewicz et al., 2006). A small number of other mutagenesis studies have aimed to dissect other parts of the Y. pestis infection cycle (Flashner et al., 2004; Klein et al., 2012) or used alternative model systems to approximate transmission dynamics (Darby et al., 2005). In contrast, this study utilized a proven Y. pestis vector, X. cheopis, to identify genes that likely play a direct role in plague transmission. Few of the putative transmission factors identified in this screen have been extensively characterized in Y. pestis. However, the galU gene and the arnBCADTEF operon were shown in other organisms to be involved in Ara4N modification of lipid A (Gunn et al., 1998, 2000). This modification plays an important role in other bacterial species by increasing their resistance to CAMPs produced by eukaryotic hosts (Gunn et al., 1998; Raetz et al., 2007). Recently, galU and the arn (or pmr) operon were shown to play an important role in allowing Y. pestis to survive within murine macrophages (Klein et al., 2012). Therefore, the galU and arnB mutants identified from this STM screen were investigated to further examine the role of these genes in the flea vector. The 3C1 (galU) and 9H9 (arn) mutants both had dramatic survival or colonization defects in fleas at 4 days post-infection and showed decreased resistance to representatives of two major classes of CAMPs, cecropin A and polymyxin B. Cecropin A belongs to a well-studied class of linear, α-helical peptides often produced by insects, including X. cheopis (Vadyvaloo et al., 2010), whilst polymyxin B belongs to a family of cyclic polypeptides and many insect-associated bacteria show resistance to this CAMP (Anisimov et al., 2005; McCoy et al., 2001). The production of CAMPs is a major defence mechanism against micro-organisms for many insects and may play an important role in protecting fleas from infectious agents, particularly within the midgut, as is seen in other blood-feeding insects (Lehane et al., 1997; Vadyvaloo et al., 2010). MS of these mutants confirmed that they were unable to modify their lipid A with Ara4N, rendering them susceptible to CAMPs. Together, these data confirm the role of galU and the arn operon in the modification of lipid A with Ara4N, as described in other organisms, and indicate that this modification of lipid A allows Y. pestis to be maintained early in the infection of fleas, facilitating transmission to other hosts.
The CAMP susceptibility of the 9H9 STM mutant, which contained a transposon insertion in the arnB gene, was not complemented with arnB alone. This indicated that the transposon had a polar effect on other genes in the operon, which was confirmed by RT-PCR. Consistent with this observation, complementation with the full arn operon restored WT levels of CAMP resistance and Ara4N-modified lipid A. Interestingly, an in-frame deletion of the arnB gene alone exhibited an intermediate phenotype. ArnB is an aminotransferase involved in the conversion of UDP-4-ketopentose to UDP-l-Ara4N during biosynthesis of Ara4N-modified lipid A (Raetz et al., 2007). The decreased levels of Ara4N-modified lipid A in KIM6+ΔarnB, together with its intermediate level of CAMP resistance, suggested that at least one alternative transamination mechanism exists in Y. pestis in the absence of ArnB. This is in contrast to previously studied organisms that appear dependent on arnB for Ara4N modification and associated CAMP resistance (Gunn et al., 2000). ArnB is a member of the DegT/DnrJ/EryC1/StrS family of aminotransferases and orthologues of this gene have been found in bacteria that modify their lipid A with Ara4N (Breazeale et al., 2003; Hwang et al., 2004). The closest homologue to arnB in the Y. pestis genome is the sugar aminotransferase wecE (y0369; 30 % amino acid identity, 48 % similarity), which is part of an operon for ECA synthesis (Danese et al., 1998). In Y. pestis and other organisms, these orthologues maintain similar active-site residues and form a similar overall structure (Hwang et al., 2004; Noland et al., 2002). The wecE/arnB double mutant showed greater CAMP sensitivity than KIM6+ΔarnB and completely lacked Ara4N-modified lipid A, indicating that WecE may contribute to the Ara4N modification of lipid A. The wecE mutant was also more sensitive to polymyxin B and showed significantly less Ara4N-modified lipid A than WT (P = 0.0040), yet significantly more than the wecE/arnB double mutant (P = 0.0159, Fig. 1c). This is particularly interesting as the polymyxin B MIC for the wecE mutant is equivalent to that of mutants completely lacking Ara4N, even though this mutant maintains some Ara4N modification capability. The double mutant complemented with either the arnB or wecE gene alone restored low levels, and complementation with both genes restored WT levels of Ara4N modification. It is also interesting to note that complementation of this mutant with wecE alone yielded a 16-fold increase in polymyxin B resistance relative to complementation with arnB. Together, these data suggest that Y. pestis WecE plays not only a previously undescribed role in Ara4N modification of lipid A, but perhaps additional Ara4N-independent roles in CAMP resistance.
ECA is a cell-surface carbohydrate found among members of the Enterobacteriaceae that is composed of ‘→3)-α- d-Fucp4NAc-(1→4)-β-d-ManpNAcA-(1→4)-α-d-GlcpNAc-(1→’ trisaccharide repeats of varying chain lengths that can exist in multiple forms (Mäkelä & Mayer, 1976; Rick & Silver, 1996). The predominant form of ECA, ECAPG, is shared by all members of the Enterobacteriaceae and is linked via phosphodiester bonds to diacylglycerol in the outer membrane (Rick et al., 1998). In some species, ECA can also be covalently linked to the LPS core (Gozdziewicz et al., 2014; Muszyński et al., 2013) or can exist in a free cyclic form in the periplasmic space (Rick & Silver, 1996; Vinogradov et al., 1994). Cyclic ECA has been described from Y. pestis (Vinogradov et al., 1994), but there is limited information on other forms it may produce. The role of ECA is not well understood, but it may be involved in virulence (Gilbreath et al., 2012) or provide a protective function against environmental stress (Barua et al., 2002). In E. coli, wecE mutants accumulate the lipid II intermediate of ECA, which results in structural disturbances in the outer membrane (Danese et al., 1998). Although the E. coli wecE mutants do not show increased CAMP sensitivity (Nichols et al., 2011), it is conceivable that similar membrane disturbances in the Y. pestis wecE mutant may affect CAMP sensitivity. Additionally, LPS-anchored ECA may alter CAMP affinity and susceptibility in Y. pestis. Taken together, our data point to a novel role for WecE in polymyxin B resistance and suggest that Y. pestis has evolved additional mechanisms of CAMP resistance that are distinct from those of other currently described systems.
Rebeil et al. (2013) recently demonstrated that Y. pestis mutants deficient in their ability to modify LPS with Ara4N were able to maintain long-term infections in fleas (4 weeks) and develop proventricular blockages. This initially appears to contrast with our data, which show that Ara4N modification is important for survival of Y. pestis in the early stages of flea infection. However, the methods used in these studies differ in important ways, which may explain this apparent difference. Rebeil et al. (2013) infected fleas using heparinized mouse blood rather than defibrinated rat blood as used in this study. Heparin is highly negatively charged, and is known to bind and inhibit the antibacterial activity of CAMPs (Barańska-Rybak et al., 2006; Park et al., 2001; Schmidtchen et al., 2001). In addition, heparin was recently shown to be protective for Y. pestis mutants in blood by an unknown mechanism (Minato et al., 2013). Furthermore, the difference in rat versus mouse blood could lead to alterations in host response to Y. pestis, infection prevalence and bacterial loads within the flea (Eisen et al., 2008). Finally, the difference in time post-infection when the fleas were evaluated (4 days versus 4 weeks) could have a significant impact on the results. Early killing followed by recovery at later time points has been seen with the well-described Y. pestis transmission factor ymt (Hinnebusch et al., 2002). As arn mutants are still able to form biofilms (Rebeil et al., 2013), cells that are not initially eliminated may be afforded an additional level of protection by the biofilm, allowing their numbers to increase over a 4-week infection such as that used by Rebeil et al. (2013) (Otto, 2006). Overall, these two studies imply that Ara4N modification, and CAMP resistance in general, is important for survival or colonization during the EPT period, but may not be essential for long-term colonization.
The ymt gene is important for survival in the flea midgut, and bacteria lacking this gene are deficient in flea colonization (Hinnebusch et al., 2002); however, ymt mutants were not identified in this STM screen. This is not surprising as the STM mutants were pooled and any ymt mutants would be surrounded by bacteria producing functional Ymt, providing protection against the cytotoxic agents this protein is presumed to inactivate. In fact, Hinnebusch et al. (2002) showed that 81 % of fleas maintained an infection with a ymt mutant strain (ymtH188N) in the proventriculus, the midgut or both when fed ymtH188N mixed with WT KIM6+. In the midgut, the mutant clustered within masses of WT bacteria, indicating it may not survive on its own, but could replicate when protected by Ymt-producing bacteria. Given the data, it is likely that in the pools of mutants, other strains were able to inactivate or serve as a sink for toxic compounds, minimizing their effects on ymt mutants. By extension, there may be additional genes required for survival in fleas that were not identified by this STM screening strategy due to protection the other pooled mutants may have provided. Other previously described transmission factors, hms and gmhA, were also not observed in the STM screen. This was expected as both are involved in biofilm formation, which is only required for late-phase transmission, and neither are required for survival in the flea (Darby et al., 2005; Hinnebusch et al., 1996). As this screen was designed to look for mutants that were unable to survive during the period required for EPT, mutants such as these would not be expected to be identified.
Investigation into the remaining STM mutants shows that the 3D9 and 9F7 mutants had transposon insertions in genes annotated as hypothetical in the KIM10+ genome (y2422 and y2283). There is little homology to known genes for y2422, whilst y2283 shows some similarity to a glycerophosphoryl diester phosphodiesterase (ugpQ). In E. coli, this enzyme is upregulated under limiting phosphate conditions and liberates phosphate from glycerophosphodiesters (Ohshima et al., 2008). The 10A3 mutation is in y3386, which is annotated in Yersinia spp. as a sodium : galactoside symporter family protein. Inactivation of this gene may result in a metabolic defect, leading to decreased survival in the flea or perhaps it is involved in uptake of a precursor of Ara4N that may be required for lipid modification. Further investigation may elucidate the substrate specificity of this transporter as well as the molecules it uses as co-transporters, providing insight into the possible role of this gene. The 17B4 mutation is located between y4035 (hypothetical) and y4036 (proY), and may affect the promoter of y4035, interfering with gene expression. The role of y4035 is currently unknown, but it is interesting to note that immediately downstream (y4034) is a phosphoethanolamine transferase gene. It is possible that expression of this transferase is also affected, which could interfere with LPS modification by phosphoethanolamine (Reynolds et al., 2005). In other species, phosphoethanolamine modifications to the LPS core have been shown to be involved in maintaining outer membrane integrity and may increase resistance to some CAMPs (Cullen et al., 2013; Reynolds et al., 2005; Viau et al., 2011). Lastly, the 5F7 mutant contains an insertion in a predicted transposase gene (y1748). It is unclear how this might affect bacterial survival in the flea; however, adjacent to this transposase is a small, unannotated ORF with similarity to bssS, which in E. coli regulates a wide variety of genes involved in biofilm formation, catabolite repression and stress response (Domka et al., 2006). Altered expression of this gene may therefore disrupt survival in the flea. Further study is needed to understand exactly how these other mutants affect the ability of Y. pestis to colonize or survive in the flea vector.
There are likely other factors required by Y. pestis for flea survival that remain to be discovered, given that the mutant library was not fully saturated. However, even without reaching saturation, many studies in other systems have identified important survival or virulence factors, increasing our understanding of host–pathogen interactions (Flashner et al., 2004; Merrell et al., 2002). The approach of using STM to investigate Y. pestis–flea interactions revealed hitherto unidentified mechanisms by which this significant human pathogen can move between hosts to cause its devastating disease. Some of the mutants with maintenance defects in fleas have disruptions in hypothetical genes, and further investigation of these mutants may provide insight into novel factors that are important for arthropod infection and transmission.
Acknowledgements
We thank Phillip Nielson, Matt Cady and Patricia Getna for technical assistance, and Dr Robert Perry (University of Kentucky, Lexington, KY, USA) for providing Y. pestis KIM6+. This work was supported in part by a NIAID-RMRCE Career Development Award (U54 AI-065357 to M.A.F.).
Abbreviations:
- Ara4N
4-amino-4-deoxy-l-arabinose
- CAMP
cationic antimicrobial peptide
- ECA
enterobacterial common antigen
- EPT
early-phase transmission
- FRT
flippase recognition target
- RT
reverse transcription
- spPCR
single-primer PCR
- STM
signature-tagged mutagenesis
Footnotes
One supplementary table is available with the online Supplementary Material.
References
- Anisimov A. P., Dentovskaya S. V., Titareva G. M., Bakhteeva I. V., Shaikhutdinova R. Z., Balakhonov S. V., Lindner B., Kocharova N. A., Senchenkova S. N. & other authors (2005). Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect Immun 73, 7324–7331. 10.1128/IAI.73.11.7324-7331.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacot A. W., Martin C. J. (1914). LXVII. Observations on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 13 (Suppl), 423–439. [PMC free article] [PubMed] [Google Scholar]
- Barańska-Rybak W., Sonesson A., Nowicki R., Schmidtchen A. (2006). Glycosaminoglycans inhibit the antibacterial activity of LL-37 in biological fluids. J Antimicrob Chemother 57, 260–265. 10.1093/jac/dki460 [DOI] [PubMed] [Google Scholar]
- Barua S., Yamashino T., Hasegawa T., Yokoyama K., Torii K., Ohta M. (2002). Involvement of surface polysaccharides in the organic acid resistance of Shiga Toxin-producing Escherichia coli O157 : H7. Mol Microbiol 43, 629–640. 10.1046/j.1365-2958.2002.02768.x [DOI] [PubMed] [Google Scholar]
- Breazeale S. D., Ribeiro A. A., Raetz C. R. (2003). Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose in polymyxin-resistant mutants of Escherichia coli. An aminotransferase (ArnB) that generates UDP-4-deoxyl-l-arabinose. J Biol Chem 278, 24731–24739. 10.1074/jbc.M304043200 [DOI] [PubMed] [Google Scholar]
- Bulet P., Stöcklin R. (2005). Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett 12, 3–11. 10.2174/0929866053406011 [DOI] [PubMed] [Google Scholar]
- Burroughs A. L. (1947). Sylvatic plague studies: the vector efficiency of nine species of fleas compared with Xenopsylla cheopis. J Hyg (Lond) 45, 371–396. 10.1017/S0022172400014042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H., Gaynor J. B., White K. G., Lopez C., Bosio C. M., Karkhoff-Schweizer R. R., Schweizer H. P. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2, 443–448. 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- Chouikha I., Hinnebusch B. J. (2012). Yersinia–flea interactions and the evolution of the arthropod-borne transmission route of plague. Curr Opin Microbiol 15, 239–246. 10.1016/j.mib.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen T. W., O’Brien J. P., Hendrixson D. R., Giles D. K., Hobb R. I., Thompson S. A., Brodbelt J. S., Trent M. S. (2013). EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect Immun 81, 430–440. 10.1128/IAI.01046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese P. N., Oliver G. R., Barr K., Bowman G. D., Rick P. D., Silhavy T. J. (1998). Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J Bacteriol 180, 5875–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C., Ananth S. L., Tan L., Hinnebusch B. J. (2005). Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect Immun 73, 7236–7242. 10.1128/IAI.73.11.7236-7242.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbise A., Lesic B., Dacheux D., Ghigo J. M., Carniel E. (2003). A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol 38, 113–116. 10.1016/S0928-8244(03)00181-0 [DOI] [PubMed] [Google Scholar]
- Domka J., Lee J., Wood T. K. (2006). YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72, 2449–2459. 10.1128/AEM.72.4.2449-2459.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen R. J., Gage K. L. (2012). Transmission of flea-borne zoonotic agents. Annu Rev Entomol 57, 61–82. 10.1146/annurev-ento-120710-100717 [DOI] [PubMed] [Google Scholar]
- Eisen R. J., Bearden S. W., Wilder A. P., Montenieri J. A., Antolin M. F., Gage K. L. (2006). Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci U S A 103, 15380–15385. 10.1073/pnas.0606831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen R. J., Lowell J. L., Montenieri J. A., Bearden S. W., Gage K. L. (2007). Temporal dynamics of early-phase transmission of Yersinia pestis by unblocked fleas: secondary infectious feeds prolong efficient transmission by Oropsylla montana (Siphonaptera: Ceratophyllidae). J Med Entomol 44, 672–677. 10.1603/0022-2585(2007)44[672:TDOETO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Eisen R. J., Vetter S. M., Holmes J. L., Bearden S. W., Montenieri J. A., Gage K. L. (2008). Source of host blood affects prevalence of infection and bacterial loads of Yersinia pestis in fleas. J Med Entomol 45, 933–938. 10.1603/0022-2585(2008)45[933:SOHBAP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Felek S., Muszyński A., Carlson R. W., Tsang T. M., Hinnebusch B. J., Krukonis E. S. (2010). Phosphoglucomutase of Yersinia pestis is required for autoaggregation and polymyxin B resistance. Infect Immun 78, 1163–1175. 10.1128/IAI.00997-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L., Alvarez-Ortega C., Wiegand I., Olivares J., Kocíncová D., Lam J. S., Martínez J. L., Hancock R. E. (2013). Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57, 110–119. 10.1128/AAC.01583-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner Y., Mamroud E., Tidhar A., Ber R., Aftalion M., Gur D., Lazar S., Zvi A., Bino T. & other authors (2004). Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect Immun 72, 908–915. 10.1128/IAI.72.2.908-915.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbreath J. J., Colvocoresses Dodds J., Rick P. D., Soloski M. J., Merrell D. S., Metcalf E. S. (2012). Enterobacterial common antigen mutants of Salmonella enterica serovar Typhimurium establish a persistent infection and provide protection against subsequent lethal challenge. Infect Immun 80, 441–450. 10.1128/IAI.05559-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozdziewicz T. K., Lugowski C., Lukasiewicz J. (2014). First evidence for a covalent linkage between enterobacterial common antigen and lipopolysaccharide in Shigella sonnei phase II ECALPS. J Biol Chem 289, 2745–2754. 10.1074/jbc.M113.512749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. S., Lim K. B., Krueger J., Kim K., Guo L., Hackett M., Miller S. I. (1998). PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27, 1171–1182. 10.1046/j.1365-2958.1998.00757.x [DOI] [PubMed] [Google Scholar]
- Gunn J. S., Ryan S. S., Van Velkinburgh J. C., Ernst R. K., Miller S. I. (2000). Genetic and functional analysis of a PmrA–PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect Immun 68, 6139–6146. 10.1128/IAI.68.11.6139-6146.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M., Shea J. E., Gleeson C., Jones M. D., Dalton E., Holden D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403. 10.1126/science.7618105 [DOI] [PubMed] [Google Scholar]
- Hinnebusch B. J., Perry R. D., Schwan T. G. (1996). Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273, 367–370. 10.1126/science.273.5273.367 [DOI] [PubMed] [Google Scholar]
- Hinnebusch J., Cherepanov P., Du Y., Rudolph A., Dixon J. D., Schwan T., Forsberg A. (2000). Murine toxin of Yersinia pestis shows phospholipase D activity but is not required for virulence in mice. Int J Med Microbiol 290, 483–487. 10.1016/S1438-4221(00)80070-3 [DOI] [PubMed] [Google Scholar]
- Hinnebusch B. J., Rudolph A. E., Cherepanov P., Dixon J. E., Schwan T. G., Forsberg A. (2002). Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296, 733–735. 10.1126/science.1069972 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Houhamdi L., Raoult D. (2008). Different genes govern Yersinia pestis pathogenicity in Caenorhabditis elegans and human lice. Microb Pathog 44, 435–437. 10.1016/j.micpath.2007.11.007 [DOI] [PubMed] [Google Scholar]
- Hwang B. Y., Lee H. J., Yang Y. H., Joo H. S., Kim B. G. (2004). Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem Biol 11, 915–925. 10.1016/j.chembiol.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Jarrett C. O., Deak E., Isherwood K. E., Oyston P. C., Fischer E. R., Whitney A. R., Kobayashi S. D., DeLeo F. R., Hinnebusch B. J. (2004). Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis 190, 783–792. 10.1086/422695 [DOI] [PubMed] [Google Scholar]
- Jiang S. S., Lin T. Y., Wang W. B., Liu M. C., Hsueh P. R., Liaw S. J. (2010). Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob Agents Chemother 54, 2000–2009. 10.1128/AAC.01384-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev A. V., Pallen M. J., Wren B. W. (2000). Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28, 1078–1082. [DOI] [PubMed] [Google Scholar]
- Kartman L., Prince F. M., Quan S. F., Stark H. E. (1958). New knowledge on the ecology of sylvatic plague. Ann N Y Acad Sci 70, 668–711. 10.1111/j.1749-6632.1958.tb35421.x [DOI] [PubMed] [Google Scholar]
- Kawasaki K. (2009). Alternative procedures for analysis of lipid A modification with phosphoethanolamine or aminoarabinose. J Microbiol Methods 76, 313–315. 10.1016/j.mimet.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Klein K. A., Fukuto H. S., Pelletier M., Romanov G., Grabenstein J. P., Palmer L. E., Ernst R., Bliska J. B. (2012). A transposon site hybridization screen identifies galU and wecBC as important for survival of Yersinia pestis in murine macrophages. J Bacteriol 194, 653–662. 10.1128/JB.06237-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. C., Alteri C. J., Smith S. N., Mobley H. L. (2007). Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104, 16669–16674. 10.1073/pnas.0607898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane M. J., Wu D., Lehane S. M. (1997). Midgut-specific immune molecules are produced by the blood-sucking insect Stomoxys calcitrans. Proc Natl Acad Sci U S A 94, 11502–11507. 10.1073/pnas.94.21.11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä P. H., Mayer H. (1976). Enterobacterial common antigen. Bacteriol Rev 40, 591–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurkiewicz P., Tang C. M., Boone C., Holden D. W. (2006). Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat Rev Genet 7, 929–939. 10.1038/nrg1984 [DOI] [PubMed] [Google Scholar]
- McCoy A. J., Liu H., Falla T. J., Gunn J. S. (2001). Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother 45, 2030–2037. 10.1128/AAC.45.7.2030-2037.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J. M., Nourbakhsh F., Ford C. W., Holden D. W. (1997). Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol 26, 399–407. 10.1046/j.1365-2958.1997.5911966.x [DOI] [PubMed] [Google Scholar]
- Merrell D. S., Hava D. L., Camilli A. (2002). Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43, 1471–1491. 10.1046/j.1365-2958.2002.02857.x [DOI] [PubMed] [Google Scholar]
- Minato Y., Ghosh A., Faulkner W. J., Lind E. J., Schesser Bartra S., Plano G. V., Jarrett C. O., Hinnebusch B. J., Winogrodzki J. & other authors (2013). Na+/H+ antiport is essential for Yersinia pestis virulence. Infect Immun 81, 3163–3172. 10.1128/IAI.00071-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszyński A., Rabsztyn K., Knapska K., Duda K. A., Duda-Grychtoł K., Kasperkiewicz K., Radziejewska-Lebrecht J., Holst O., Skurnik M. (2013). Enterobacterial common antigen and O-specific polysaccharide coexist in the lipopolysaccharide of Yersinia enterocolitica serotype O : 3. Microbiology 159, 1782–1793. 10.1099/mic.0.066662-0 [DOI] [PubMed] [Google Scholar]
- Nichols R. J., Sen S., Choo Y. J., Beltrao P., Zietek M., Chaba R., Lee S., Kazmierczak K. M., Lee K. J. & other authors (2011). Phenotypic landscape of a bacterial cell. Cell 144, 143–156. 10.1016/j.cell.2010.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland B. W., Newman J. M., Hendle J., Badger J., Christopher J. A., Tresser J., Buchanan M. D., Wright T. A., Rutter M. E. & other authors (2002). Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: a 4-amino-4-deoxy-l-arabinose lipopolysaccharide-modifying enzyme. Structure 10, 1569–1580. 10.1016/S0969-2126(02)00879-1 [DOI] [PubMed] [Google Scholar]
- Ohshima N., Yamashita S., Takahashi N., Kuroishi C., Shiro Y., Takio K. (2008). Escherichia coli cytosolic glycerophosphodiester phosphodiesterase (UgpQ) requires Mg2+, Co2+, or Mn2+ for its enzyme activity. J Bacteriol 190, 1219–1223. 10.1128/JB.01223-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2006). Bacterial evasion of antimicrobial peptides by biofilm formation. In Antimicrobial Peptides and Human Disease, pp. 251–258. Edited by Shafer W. M. Berlin: Springer; 10.1007/3-540-29916-5_10 [DOI] [PubMed] [Google Scholar]
- Park P. W., Pier G. B., Hinkes M. T., Bernfield M. (2001). Exploitation of syndecan-1 shedding by Pseudomonas aeruginosa enhances virulence. Nature 411, 98–102. 10.1038/35075100 [DOI] [PubMed] [Google Scholar]
- Perry R. D., Pendrak M. L., Schuetze P. (1990). Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol 172, 5929–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007). Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 76, 295–329. 10.1146/annurev.biochem.76.010307.145803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi C., Jeyashree A., Renuka Devi K. (2011). Antimicrobial peptides from insects: an overview. Res Biotech 2, 1–7. [Google Scholar]
- Rebeil R., Ernst R. K., Gowen B. B., Miller S. I., Hinnebusch B. J. (2004). Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol 52, 1363–1373. 10.1111/j.1365-2958.2004.04059.x [DOI] [PubMed] [Google Scholar]
- Rebeil R., Jarrett C. O., Driver J. D., Ernst R. K., Oyston P. C., Hinnebusch B. J. (2013). Induction of the Yersinia pestis PhoP–PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol 195, 1920–1930. 10.1128/JB.02000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C. M., Kalb S. R., Cotter R. J., Raetz C. R. (2005). A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem 280, 21202–21211. 10.1074/jbc.M500964200 [DOI] [PubMed] [Google Scholar]
- Rick P. D., Silver R. P. (1996). Enterobacterial common antigen and capsular polysaccharides. In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd edn, pp. 104–122. Edited by Neidhardt F. C. Washington, DC: American Society for Microbiology. [Google Scholar]
- Rick P. D., Hubbard G. L., Kitaoka M., Nagaki H., Kinoshita T., Dowd S., Simplaceanu V., Ho C. (1998). Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology 8, 557–567. 10.1093/glycob/8.6.557 [DOI] [PubMed] [Google Scholar]
- Rudolph A. E., Stuckey J. A., Zhao Y., Matthews H. R., Patton W. A., Moss J., Dixon J. E. (1999). Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J Biol Chem 274, 11824–11831. 10.1074/jbc.274.17.11824 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schmidtchen A., Frick I. M., Björck L. (2001). Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial alpha-defensin. Mol Microbiol 39, 708–713. 10.1046/j.1365-2958.2001.02251.x [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Puhler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechol 1, 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Skurnik M., Peippo A., Ervelä E. (2000). Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O : 1b. Mol Microbiol 37, 316–330. 10.1046/j.1365-2958.2000.01993.x [DOI] [PubMed] [Google Scholar]
- Vadyvaloo V., Jarrett C., Sturdevant D. E., Sebbane F., Hinnebusch B. J. (2010). Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6, e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter S. M., Eisen R. J., Schotthoefer A. M., Montenieri J. A., Holmes J. L., Bobrov A. G., Bearden S. W., Perry R. D., Gage K. L. (2010). Biofilm formation is not required for early-phase transmission of Yersinia pestis. Microbiology 156, 2216–2225. 10.1099/mic.0.037952-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viau C., Le Sage V., Ting D. K., Gross J., Le Moual H. (2011). Absence of PmrAB-mediated phosphoethanolamine modifications of Citrobacter rodentium lipopolysaccharide affects outer membrane integrity. J Bacteriol 193, 2168–2176. 10.1128/JB.01449-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov E. V., Knirel Y. A., Thomas-Oates J. E., Shashkov A. S., L’vov V. L. (1994). The structure of the cyclic enterobacterial common antigen (ECA) from Yersinia pestis. Carbohydr Res 258, 223–232. 10.1016/0008-6215(94)84088-1 [DOI] [PubMed] [Google Scholar]
- Zhou P., Altman E., Perry M. B., Li J. (2010). Study of matrix additives for sensitive analysis of lipid A by matrix-assisted laser desorption ionization mass spectrometry. Appl Environ Microbiol 76, 3437–3443. 10.1128/AEM.03082-09 [DOI] [PMC free article] [PubMed] [Google Scholar]


