Abstract
Signaling proteins are flexible in both form and function. They can bind to multiple molecular partners and integrate diverse types of cellular information. When imaged by time-lapse microscopy, many signaling proteins show complex patterns of activity or localization that vary from cell to cell. This heterogeneity is so prevalent that it has spurred the development of new computational strategies to analyze single-cell signaling patterns. A collective observation from these analyses is that cells appear less heterogeneous when their responses are normalized to, or synchronized with, other single-cell measurements. In many cases, these transformed signaling patterns show distinct dynamical trends that correspond with predictable phenotypic outcomes. When signaling mechanisms are unclear, computational models can suggest putative molecular interactions that are experimentally testable. Thus, computational analysis of single-cell signaling has not only provided new ways to quantify the responses of individual cells, but has helped resolve longstanding questions surrounding many well-studied human signaling proteins including NF-κB, p53, ERK1/2, and CDK2. A number of specific challenges lie ahead for single-cell analysis such as quantifying the contribution of non-cell autonomous signaling as well as the characterization of protein signaling dynamics in vivo.
Keywords: cell signaling, single-cell analysis, computational modeling, time-lapse microscopy, NF-κB, p53, ERK1/2, CDK2
1. INTRODUCTION
The ability to visualize signaling proteins in real time and at single-cell resolution has revealed a staggering picture of complexity in cellular signal transduction. Genetically identical cells can show vastly different signaling patterns—even under basal conditions or in response to the same stimulus. In fact, if there has been one lesson learned from single-cell dynamics, it is that variability from cell to cell is the rule rather than the exception. Cells in the same culture dish can show patterns of gene or protein expression that vary over several orders of magnitude [1–3], and signaling patterns measured in real time are noisy and asynchronous [4, 5]. These observations present major challenges for understanding single-cell signaling: How much of the observed heterogeneity from cell to cell is meaningful? Are the observed patterns variations of a single signaling response or are there multiple responses? If there are multiple responses, how can we distinguish among different signaling patterns in individual cells?
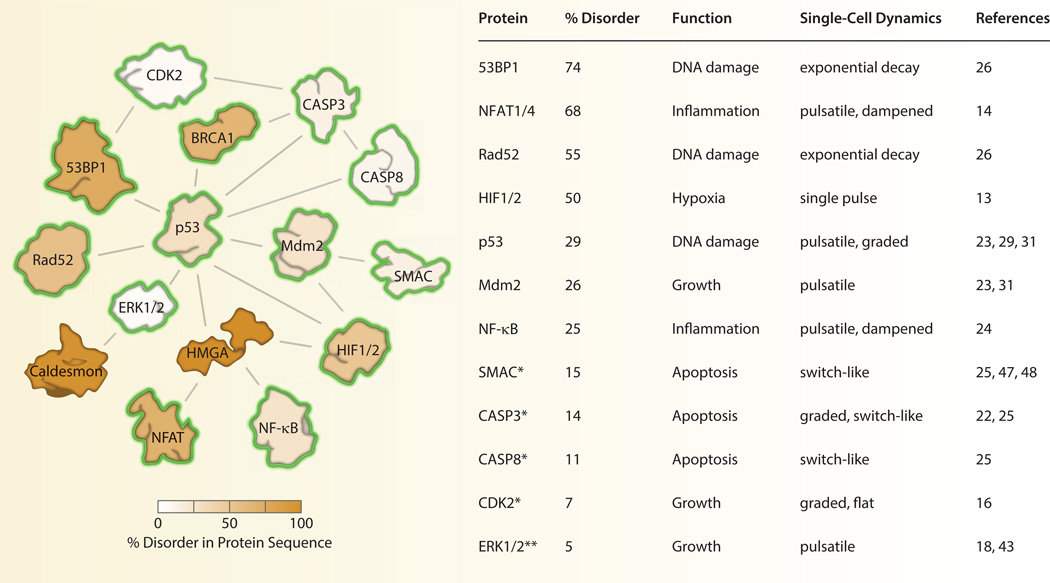
The idea that a single signaling protein can display multiple behaviors is already well appreciated in the field of intrinsically disordered proteins (IDPs), the focus of this special issue. It has long been observed that certain proteins associated with signal transduction have unusually high levels of disordered regions in their peptide sequence [6–8]. This trend may reflect the tendency for signaling proteins to have multiple binding partners as well as multiple functionalities in the cell [6, 8, 9]. It is our observation that many IDPs (or those involved in signaling complexes with disordered proteins) have been examined by fluorescence time-lapse microscopy. Often, these proteins display rich and complex signaling dynamics in single cells (Fig. 1). This is yet another indication that signaling proteins are structurally pliable molecules capable of sophisticated information processing [10]. As such, the variability of single-cell protein dynamics of IDPs adds additional challenges and opportunities for understanding the role of these proteins in cell signaling and disease.
Fig. 1. Dynamics and disorder among human signaling proteins.
Interior protein color indicates the level of structural disorder calculated using the meta approach [75]. A yellow halo indicates that the dynamics of the protein have been characterized in live cells. *The dynamics of SMAC, CASP3, CASP8, and CDK2 were characterized by enzymatic activity rather than protein expression levels. Protein-protein interactions were obtained from the Biomolecular Interaction Network Database [76].
In this review, we examine how signaling patterns among individual cells have been resolved through the use of computational analyses. We first introduce some of the different patterns of signaling observed in individual cells across different cellular pathways. Since this topic has been reviewed elsewhere in depth [11, 12], we provide a brief overview of the variety of signaling patterns observed among mammalian proteins and highlight the insight gained from an exciting set of recent studies [1, 13–18]. These studies are drawn together by a common set of computational approaches used to analyze signaling patterns in individual cells. We find that these approaches are essentially variations of existing methods for normalizing and comparing biological data. However, they are specifically tailored toward the heterogeneous temporal data gathered from single cells. In many cases these approaches not only resolve, but also make use of, cell-to-cell heterogeneity by relating variation in signaling to differences in downstream behaviors. We further show how single-cell signaling has been modeled computationally to predict cellular behaviors and suggest new mechanistic interactions. Finally, we discuss specific challenges for understanding single-cell signaling responses that must build on existing work in the field.
2. SINGLE-CELL DYNAMICS OF HUMAN SIGNALING PROTEINS
An increasing number of human signaling proteins have been characterized in living cells including several proteins with significantly disordered protein structures (Fig. 1). These studies typically quantify protein abundance over time using a fluorescent reporter protein that is covalently linked to the coding region of the protein of interest [19, 20]. In cases where the enzymatic activity of the protein is more biologically relevant than its expression level, it is necessary to use a genetically encoded biosensor that exhibits conformational changes that reflect changes in the enzymatic activity of interest [21]. These biosensors include, for example, fluorescent substrates that mimic endogenous cleavage sites used to measure protease activity [22]. Once the fluorescent reporter is stably expressed, cells are cultured directly on the microscope, relevant perturbations are performed, and images are acquired periodically at a time scale that is appropriate for the biological process under investigation [11]. Segmentation borders are then drawn to separate neighboring cells. The resulting data set is a time series of fluorescence intensity values that reveal how the protein, or enzyme activity, changes over time in individual cells.
Human signaling proteins show a wide range of dynamical behaviors including pulses [13, 18, 23], bursts [14], oscillations [24], switches [25], and decays [26]. One of the best-studied proteins in single cells is the stress response factor NF-κB. NF-κB is a transcription factor that responds to cytokines, inflammation, and other cellular stresses. Upon activation with inflammatory stimuli such as tumor necrosis factor α (TNFα), NF-κB localizes to the nucleus and promotes transcription of IκBα, an inhibitor that binds to NF-κB and triggers export of NF-κB to the cytoplasm. Activation of NF-κB and subsequent expression of IκBα leads to multiples cycles of NF-κB nucleo-cytoplasmic shuttling. Live-cell imaging of NF-κB localization has revealed that, after stimulation with TNFα, NF-κB shows a prominent first pulse of activity followed by a series of long-term pulses [24]. At low doses of TNFα, activation of NF-κB is highly heterogeneous with the majority of cells showing all-or-nothing activity [27].
Additional signaling proteins in the immune response have been characterized in individual cells. Notably, two isoforms of the nuclear factor of activated T-cells (NFAT1 and NFAT4) show dramatically different nuclear localization dynamics in response to calcium stimulation [14]. NFAT1 responds slowly to stimulation, showing prolonged occupation of the nucleus over several hours. In contrast, NFAT4 shows rapid and repeated bursts of nuclear localization that last between 5 and 10 minutes. Although it is premature to make any firm conclusions, it appears that a large proportion of signaling proteins that have been measured in live cells show some form of pulsatile signaling. Whether “frequency modulated” signaling is a pervasive theme in biology remains to be determined [11, 12, 28]. If so, it would suggest that the temporal pattern of protein signaling may be as relevant as its absolute abundance. Such a finding expands our notion of good indicators of functional relevance to include both expression levels and dynamical patterns of activity.
Another well-characterized protein in live cells is the tumor suppressor p53 [23, 29]. Following DNA damage, p53 undergoes posttranslational modification that frees it from Mdm2, an E3 ubiquitin ligase that promotes rapid degradation of the p53 protein. However, because Mdm2 is also a target gene product of p53, the induced elevation of p53 eventually promotes its own degradation, leading to periodic accumulation of nuclear p53. p53 dynamics were originally predicted to be damped oscillations based on population measurements of p53 and Mdm2 by Western blot [30]. When imaged in single cells, however, p53 signaling was shown to occur in a series of pulses with uniform width and height [23, 31]. Rather than increasing the absolute levels of p53, larger doses of DNA damage increases the number of consecutive pulses.
As a well-recognized intrinsically disordered protein, p53 has also been examined at the single-molecule level to understand its binding and oligomerization properties [32]. Following initial work to characterize the binding affinity of p53 to DNA using ensemble methods such as analytical ultracentrifugation [33], fluorescence correlation spectroscopy was used to determine the precise kinetics of p53 oligomerization [34]. More recently, analysis of p53 oligomerization dynamics in single cells confirmed that dimers are the predominant form under basal conditions. Interestingly, after DNA damage, formation of p53 tetramers precedes increases in p53 protein levels suggesting that p53 oligomerization is dynamically regulated in response to genotoxic stress [35]. These examples show how the structural disorder of a signaling molecule may affect its cellular function and regulation.
Additional components of the DNA damage response were recently characterized in single human cells. Two members of the hypoxia-inducible factor family of transcription factors (HIF-1α and -2α), which can bind both Mdm2 [36] and p53 [37] to alter cellular stress responses, show a single 3 h pulse that is rapidly terminated under continuous hypoxic conditions [13]. The p53 binding protein 1 (53BP1), which localizes to double-strand DNA breaks, shows exponential decay kinetics that reflect the rate of DNA repair in individual cells. When combined with a reporter for yeast homology Rad52, a marker for DNA repair by homologous recombination, the relative rates of repair by two alternative mechanisms—non-homologous end joining and homologous recombination—were determined in individual cells [26]. While the average rates of repair were consistent with rates previously estimated through bulk cell measurements, the simultaneous imaging of 53BP1 and Rad52 dynamics in single cells allowed precise calculation of these rates during each phase of the cell cycle. This led to the discovery that repair processes change continually over the course of the cell cycle and may depend on the extent of active DNA replication. As this study demonstrates, simultaneous examination of two or more signaling proteins in the same cell provides a powerful look at the kinetic and functional relationship between multiple signaling pathways [38].
Gaining a clear picture of protein signaling patterns in individual cells is critically important for forming hypotheses about their functional role. For example, the discovery that p53 undergoes repeated pulses of expression in response to DNA damage, rather than damped oscillations, shifted attention from the amplitude of the p53 response to its duration. This “dose-to-duration” encoding mechanism [39] suggested that the functional role of p53 pulses may be related to the persistence of the signaling response rather than its absolute levels. To test this idea, the duration of p53 pulses was altered using an Mdm2 inhibitor to maintain a constant concentration of total p53. This perturbation led to changes in target gene expression and cellular fate [40]. A similar result was observed for the extracellular signal related kinase (ERK), which relays information about extracellular growth factors to cell cycle decisions. While the absolute amplitude of ERK was relatively constant across all cells, the decision for a cell to enter S phase correlated strongly with the fraction of time ERK spent in an active signaling state [18]. Thus, real-time measurements of protein signaling in single cells have refined our understanding of how cellular information is transmitted. An exciting direction for the IDP field will be to examine how protein disorder is related to signaling dynamics in single cells. For example, sustained expression of an IDP over time may allow it the disordered structure to interact with a new set of molecular partners.
In summary, human proteins show a wide array of dynamical patterns. What is the function of these signaling dynamics? How do they interact with other components of signaling pathways to control downstream behaviors? In the following section, we discuss how functional information has been obtained from live-cell signaling studies through a set of elegant, but conceptually simple, computational approaches. We find that, when normalized to other single-cell measurements, some of the observed heterogeneity in protein signaling—while perhaps functionally relevant—vanishes and dominant patterns of activity emerge. Remaining differences in normalized signaling patterns are then used to classify cells into distinct functional groups. In these cases, differences among cells are most helpful if they can be linked to some measureable difference in phenotypic outcome.
3. COMPUTATIONAL ANALYSIS OF SINGLE-CELL SIGNALING PATTERNS
A single live-cell experiment can easily produce images for thousands of individual cells. The resulting data set is a collection of individual time series traces that often show considerable heterogeneity. One of the exciting aspects of gathering such data is that, very often, novel behaviors are observed using relatively simple experimental designs. Instead of applying complicated perturbations, new biology can be discovered by simply reorganizing the data in a systematic way. To this end, three core approaches have emerged for analyzing single-cell dynamics: dynamic feature normalization, in silico synchronization, and sister-cell analysis.
3.1 DYNAMIC FEATURE NORMALIZATION
Many bulk measurement techniques in molecular biology employ some form of normalization to improve biological interpretation. The basic principle behind these methods is to identify a measurement that is expected to be constant across independent experiments. For example, quantification of mRNA levels by real-time quantitative PCR is often performed by dividing the abundance of the gene of interest to a “housekeeping” gene, such as GAPDH or ACTB, that is expected to show small variation across experimental conditions [41]. If the abundance of the housekeeping gene does change, it is usually not interpreted as biologically meaningful. Instead, the change is interpreted as technical error that should be equally adjusted for both the housekeeping gene and the gene of interest. Thus, normalization is both biologically informed (e.g., selecting a gene that is expected to be constant) and serves to reduce technical variation across experiments, improving the interpretation of experimental results.
As we have seen, individual cells show large variability in gene expression and protein dynamics during signal transduction (Fig. 2A) [31, 42, 43]. When measuring signaling patterns across individual cells, therefore, each cell may be thought of as a separate experiment that requires an internally constant quantity to allow comparison among cells. This task is unique for single cells, however, because the variability observed among cells is generally not due to technical error but is biologically grounded. Thus, careful consideration must be used to determine what is expected to be constant among every cell. While the instantaneous level of a protein can vary substantially from cell to cell, normalizing cellular responses to certain features of their dynamic response curve can show strong consistency between cells (Fig. 2B).
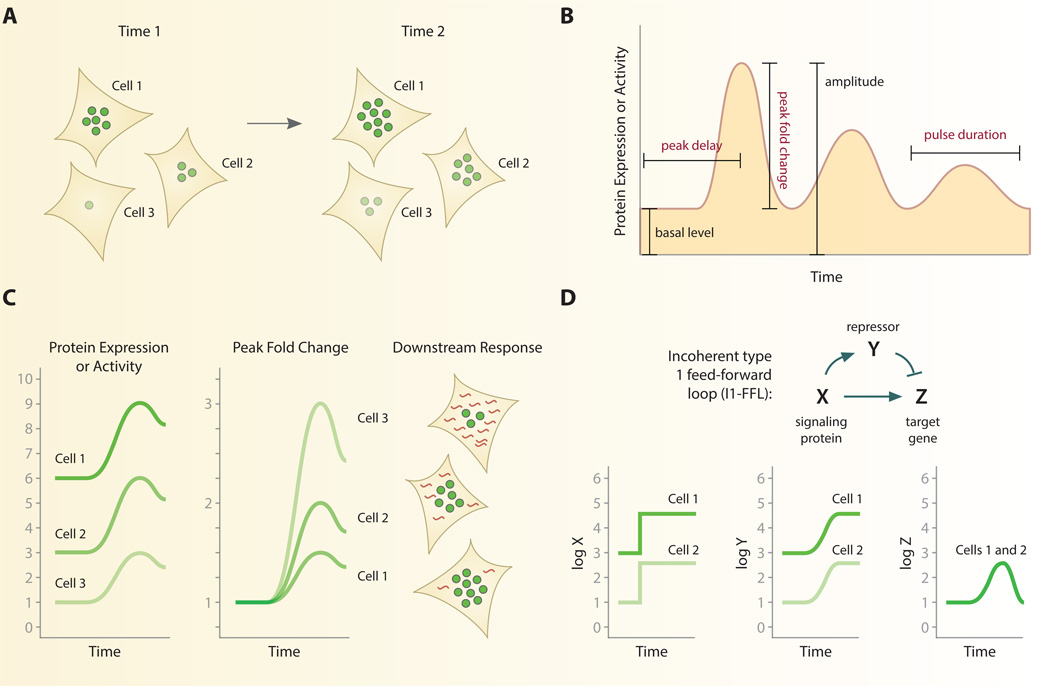
Fig. 2. Normalization of single-cell signaling patterns.
A. Individual cells may vary considerably in the instantaneous level or activity of a signaling protein. B. High-resolution temporal data gathered from live-cell imaging generates a dynamic response curve for individual cells. Specific features of the response curve, such as peak fold change, have proven useful for normalization. Other quantities such as peak delay and pulse duration are also found to be relatively constant among individual cells across multiple signaling pathways. Features in red type depend on time for calculation. C. Normalizing each cell’s response to dynamical features can reverse trends and predict functional outcomes. In response to inflammatory cytokines the peak fold change of NF-kB, but not its absolute abundance, correlates with singlecell gene expression. D. An incoherent feedforward loop can detect fold changes in individual cells. By activating a repressor Y, individual cells with the same fold change response but different absolute changes in a signaling protein, X, can produce identical responses in a target gene Z. See ref. [45].
For example, the fold change between peak fluorescence and baseline fluorescence yields a value that is often consistent among individual cells. This approach was used to analyze the dynamics of ERK2 in response to the epidermal growth factor (EGF), a strong mitogen that leads to cell proliferation [43]. Upon stimulation with EGF, ERK2 enters the nucleus where it can be readily quantified through fluorescent time-lapse imaging. Single-cell analysis demonstrated that basal levels of nuclear ERK2 varied significantly between cells. The increase observed in ERK2 nuclear fluorescence after EGF stimulation was equally variable when measuring both the absolute and mean fluorescence intensity. However, normalizing the maximum fluorescence intensity to the initial intensity, or peak fold change, unmasked a response whose magnitude is relatively consistent between cells. It should be noted that although fold change is a common metric in many measurement techniques, peak fold change differs in that is requires the precise location of a peak response in time and thus depends directly on the timing of the response. Interestingly, other time-dependent features of the ERK response showed small variation between individual cells. These results echoes the findings that p53 [31] and HIF [13] dynamics also show high consistency in the duration of their response curves.
Why might peak fold change be more tightly controlled than the absolute level of a protein? One possibility is that the peak fold change of a protein is a relevant quantity for controlling functional responses. Single cells may detect the relative change over time to control downstream processes [44]. In support of this hypothesis, fold change of NF-κB was recently shown to correlate with the expression of the target genes IL8, TNFAPI3, and NFKBIA in the same cells [1]. Peak fold change showed a stronger correlation with target gene expression than absolute abundance of NF-κB in the nucleus. These functional studies provide strong support that cells adapt to absolute basal levels by detecting fold change, and that the magnitude of the change in signaling regulates downstream responses such as gene expression in single cells [44–46] (Fig. 2C). Goentoro and colleagues have offered a theoretical explanation for how cells may interpret fold changes mechanistically [45]. The incoherent type 1 feed-forward loop (I1-FFL) provides a simplified mechanistic interpretation of the fold-change response (Fig. 2D). In the I1-FFL, an activator X controls both the target gene Z and the repressor gene Y. Simultaneous activation of target and repressor help cells to “remember” past levels of gene Z. Therefore, when X activates the target gene Z, the repressor Y will ensure that levels of Z return to baseline. This explains why many fold change responses in individual cells, such as ERK2 and NF-κB, return to baseline levels after stimulation—a property known as perfect adaptation. The kinetics of repressor gene Y shapes the response of target gene Z to activator X: a slow response of repressor Y to activation by X controls the amplitude of Z; a delayed response of Y to activation by X controls duration of Z. Together, these dynamics define a mechanistic network that generates responses that are proportional to the peak fold change of the stimulus. In the case NF-κB, it is likely that negative feedback from inhibitors IκΒ and A20 mediate adaptation of target gene expression at the transcriptional level, although definitive evidence for this mechanism would require a way to maintain stable elevated levels of active nuclear NF-kB.
Besides peak fold change, other features of the dynamic response curve have been shown to be consistent among individual cells (Fig. 2A). Studies of both ERK2 and NF-kB show that the most tightly controlled quantities are peak fold change, final fold change, and the peak delay [1, 43]. In these studies, quantities that depend on time show the lowest variability from cell to cell [11, 12, 31]. A notable exception to this trend is found in the response to drug-induced apoptosis. The TNFα-related apoptosis-inducing ligand (TRAIL) is a “death ligand” that activates apoptotic pathways leading to cell death. The delay between the addition of TRAIL and the cellular commitment to apoptosis varies widely across a population of cells [22, 25, 47, 48]. In this case, the variable delay in commitment to apoptosis is thought to be advantageous for a population of cells because it prevents the simultaneous death of an entire tissue. By staggering the apoptotic responses of individual cells, certain cells can outlast the death stimulus long enough to engage a survival response. Whatever the functional role may be, these findings indicate that the timing of single-cell signaling responses is a primary controller of downstream behavior.
3.2 IN SILICO SYNCHRONIZATION
Normalization typically involves adjusting measurements by calculating the relative abundance of two or more measured values. However, as we have already seen, many biological processes are characterized not only by abundance but also the timing of their responses. Numerous examples of cellular processes that depend on precise timing include the sequential phases of the cell cycle [49], segmentation of vertebrate development [50], and delayed sporulation of bacteria [51]. While bulk measurements of these biological events can often reveal dominant patterns of activity, these approaches can also mask temporal patterns because of asynchrony among individual cells. As such, bulk measurements are limited in their sensitivity of detection and potentially miss additional patterns of activation that may be biologically relevant to the system [11].
A common way to address asynchrony is through experimental interventions that synchronize cells. For example, chemically inducing cell cycle synchrony using nocodazole, a drug that interferes with microtubule polymerization, can arrest cells just before mitosis [52, 53]. However, this perturbation can also alter the biological event under study and introduce artifacts into the experiment. To alleviate this problem, several studies have employed computational approaches to synchronize cells in silico after the experiment is over. This strategy has facilitated the detection of diverse patterns of activation with better temporal resolution and more physiologically relevant conditions.
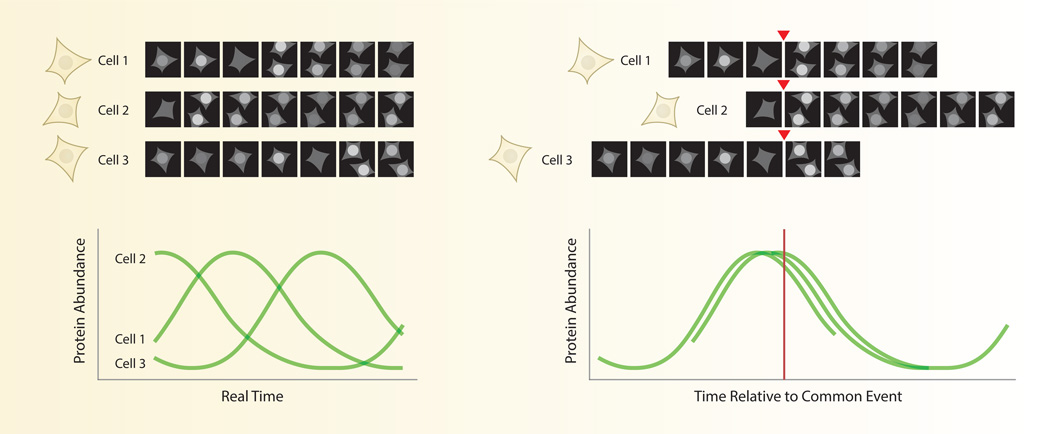
In silico synchronization is a form of normalization in which individual cells are compared on the basis of a common event (Fig. 3). Since the dynamics of each cell are obtained through a sequence of images, these sequences can be easily shifted forward or backward in time to align each cell at a different moment in real time. In many cases, synchronizing single cells computationally has identified dominant patterns of a response that are masked in an asynchronous population. For example, fluorescent reporters for cell cycle phase such as geminin show rapid degradation after mitosis. This pattern is invisible by bulk measurements such as Western blot, which would show constant geminin levels in an unsynchronized population. In addition, in silico synchronization can help separate distinct subsets of cells with different temporal patterns. For example, in silico synchronization of human mammary epithelial cells to the time of mitosis uncovered two populations with differing dynamics of cyclin-dependent kinase 2 (CDK2), a regulator of entry into S phase [16]. One subpopulation (CDK2low) exhibited low CDK2 activity of after mitosis, which corresponded to extended time in a G0-like state. All other cells (CDK2inc) showed a gradual increase in CDK2 activity after mitosis and reduced time in G1. Comparison between different mammalian cell lines showed that CDK2low and CDK2inc cells were present in different proportions, providing a novel and intuitive explanation for well-known differences in the population doubling times of these cell lines.
Fig. 3. In silico synchronization of signaling patterns in single cells.
Individual cells often show heterogeneity in the timing of their responses. Aligning response curves to some common cellular event, such as mitosis, can often reduce the heterogeneity of the individual responses. This strategy has helped identify distinct subsets of cells and suggested functional relationships between the measured protein and the aligning event.
The identification of two dynamical patterns of CDK2 activity also helped resolve a longstanding controversy regarding the location of the cell cycle “restriction point”—a point in time at which the cell commits to division regardless of stimulation by growth factors. Previous work dating back to 1974 had suggested that the restriction point occurred in late G1 in the cell cycle [54], while other studies placed the restriction point just before mitosis of the previous cell cycle [55, 56]. Experiments using cells synchronized by serum starvation were only able to detect the late G1 restriction point due to the fact that the cells had been synchronized in G0. However, when freely cycling cells bearing a live-cell CDK2 activity reporter are monitored and synchronized in silico by the time of mitosis, both a G2/M and a G0/G1 restriction point are observed. Thus, studying proliferating cells in the absence of experimental synchronization helped resolve contradictory evidence in the cell cycle field about the location of a key regulatory event.
Synchronization to mitosis also points to a potential function of pulsatile p53 dynamics. Under basal conditions, with no acute stress, p53 shows spontaneous and asynchronous pulses of activity. Aligning these pulses computationally to the time of mitosis revealed that p53 pulses consistently occur ~5 h after mitosis, a time associated with intrinsic DNA damage [57]. These pulses may be related to double strand DNA breaks that occur during normal cell cycle progression. In view of the bifurcation of CDK2 activity after mitosis, this finding suggests an attractive hypothesis that would cell cycle progression, DNA damage and p53 dynamics: low levels of DNA damage during DNA replication triggers pulses of p53 that lead to the quiescent (CDK2low) state. Such a mechanism would extend the length of time a cell spends in G0/G1, allowing any DNA damage to be adequately repaired before committing to another round of DNA synthesis and cell division. Supporting this idea, a significant correlation was found between the level of damage and the probability of observing a p53 pulse [17]. Future studies that incorporate the precise point in the cell cycle at which the damage occurs could determine whether there is a “restriction threshold” of DNA damage for generating a p53 response.
Another use of in silico synchronization is to define the precise temporal relationship between two closely associated events. During apoptosis, for example, mitochondrial outer membrane permeabilization (MOMP) is a key event that signifies an irreversible commitment to apoptosis [58]. When cytokines bind to death receptors, initiator caspases are activated and MOMP occurs, releasing cytochrome c into the cytoplasm and activating effector caspases that ultimately dismantle the cell. Previous studies in cell populations suggested that activation of initiator and effector caspases may occur at the same time. However, synchronization of live-cell reporters for both initiator and effector caspase activity with respect to a MOMP sensor determined that initiator caspases are activated first following exposure to TRAIL [25]. Steady increases in initiator caspase activity reach a threshold level that induces MOMP followed by rapid activation of effector caspases. In addition, high time-resolution imaging of cells undergoing apoptosis identified an intermediate pre-MOMP state where effector caspases are cleaved but their activity is temporarily inhibited. The key to detecting these patterns was synchronization with respect to MOMP, a discrete biological event that is easily identifiable in single cells [59].
3.3 SISTER-CELL ANALYSIS
Another approach to understand signaling in single cells is to make systematic comparisons between specific pairs of cells. While individual responses may be highly variable across a population because of variation in extracellular signals or cell cycle position, “sister cells” that were generated from the same mitotic event are expected to show less variability in these factors. Conceptually, sister-cell analysis is akin to the use of twin studies in behavioral genetics [60]. Since monozygotic twins share nearly identical genomes, any observed differences between the twins can be attributed to environmental influences. In a similar way, sister cells are likely to share a more common epigenetic traits, such as protein levels, inherited from the mother cell [61]. For example, sister cells show 98% correspondence in choice of CDK2 activity states after mitosis, suggesting a heritable factor that regulates proliferation state after mitosis [16]. Using this logic, sister-cell analysis has helped resolve subtle differences in signaling dynamics between pairs of cells and, in some cases, has provided profound insight into why individual signaling responses vary across cell populations, a fact that has confounded single-cell biologists for decades [62].
Sister-cell analysis was used to understand why some cells in a population respond to chemotherapeutic drugs while others do not. Isogenic cells show widely varying apoptotic responses to TRAIL including early MOMP, delayed MOMP, or complete resistance to the ligand [48]. For cells undergoing apoptosis, a relevant quantity is the time that elapses between addition of TRAIL and the instant of MOMP. While this delay time is highly correlated for sister cells, it quickly diverges as a function of time since the last mitosis. In fact, cells show a complete loss of correlation in MOMP delay time in less than one cell cycle. What causes such quick divergence in cell fate among sister cells? This question was answered in part by inhibiting protein synthesis, which extended the correlation of MOMP for two generations [61]. Thus, for resistance to death ligands, de novo protein synthesis constitutes a major source of heterogeneity among individual cell fate decisions.
The functional consequences of protein signaling patterns can also be discerned by comparing sister cells. An analysis of ERK activity in over 200 pairs of sister cells revealed a subtle but highly significant “bump” in ERK activity approximately 12 hours before entry into S phase. Though modest, this difference was sufficient to discriminate whether or not a cell was destined to commit to DNA replication. Interestingly, the most significant metric that discriminated between sister cells with divergent behaviors was the duration of the ERK activity (Fig. 2B) [18], again underscoring the potential functional importance of the temporal features of the signaling response curve.
4. MODELING SINGLE-CELL SIGNALING MECHANISMS
Computational models serve to synthesize our understanding of cell signaling within a quantitative framework [63]. A good model allows researchers to simulate signaling responses and make predictions about how the system works. Most often, computational models are based on existing knowledge of a molecular pathway. Introducing slight modifications to an existing model can be used to test the feasibility of hypothetical mechanisms. In this section, we provide examples of computational models that have advanced our quantitative understanding of single-cell signaling.
Perhaps the most widely used framework for modeling cellular signaling networks is a system of ordinary or delayed differential equations (ODEs and DDEs). Differential equation models are desirable because they are mechanistically interpretable, capable of simulating the system under conditions that were not directly measured, and allow one to make forward predictions about the signaling responses that can be tested experimentally. For example, a simple DDE model was used to describe the dynamics of p53 in response to DNA damage in single cells [64]. Although the negative feedback between p53 and Mdm2 provided the oscillatory dynamics of p53, it was unable to explain the sudden cessation of pulses observed in single cells. By proposing a putative negative feedback between p53 and upstream DNA damage signal, the DDE model showed strong agreement with single-cell data. Additional experimental studies identified the phosphatase Wip1 as a key factor in terminating a series of p53 pulses.
Introduction of a hypothetical interaction into a model of NF-κB dynamics was used to reconcile the dynamics of NF-κB with target gene expression in single cells [1]. TNFα stimulation triggers changes in several genes whose expression levels correlate with the peak fold change in NF-κB activation. However, when the data were fit to a previously published computational model of NF-κB signaling [65], no suitable set of parameter values were obtained that could accurately predict target gene expression. Drawing on earlier work suggesting that I1-FFLs can detect fold changes [45], Lee et al. proposed the existence of a competitive inhibitor of transcription that is activated by NF-κB. This revised model produced an accurate prediction of the number of transcripts per cell based on NF-κB dynamics, guiding further molecular studies aimed at identifying the putative inhibitor of transcription.
Computational models have also been used to guide experimental perturbations. To understand the function of p53 dynamics in response to DNA damage, a computational model was used to predict the effect of the p53 stabilizing drug, Nutlin-3 [66]. Nutlin-3 binds to Mdm2 and prevents ubiquitination of p53. This leads to stabilization of p53 but also the concomitant increase in Mdm2 as a downstream target of p53. Using a derivation of a previous model of p53 dynamics [64], the highly nonlinear effect of Nutlin-3 was modeled to suggest a specific dosing scheme that would maintain constant p53 levels over time. When put to the test, p53 levels were experimentally modified as predicted by the computational model. These altered p53 dynamics were found to affect target gene expression and drive cells into senescence [40].
To understand how ERK dynamics are decoded to make decisions about entering S phase, Albeck and colleagues proposed an ODE model capable of discriminating transient from persistent ERK pulses [18]. In short, rapidly fluctuating pulses of ERK activity are decoded by slowly changing levels of Fra-1, which is a strong indicator of the decision to enter S phase. A key factor in the model is the slow decay rate of Fra-1, which requires persistent upstream signals to produce a robust downstream response. This same mechanism was identified previously in cell populations as a set of “immediate early genes”, which must accumulate to sufficient levels to detect persistent ERK signaling [67]. A similar mechanism may be at work in the DNA damage response to decode persistent pulses of p53 [40, 68].
A conclusion from these examples is that each single-cell measurement represents a unique perturbation that often can be linked to a specific cellular outcome. A vivid example of this approach, coined as “noise genetics”, was recently reported in which protein levels were correlated with cell movement—a phenotype that is easily quantifiable with high temporal resolution [69]. Without any explicit perturbation to the cells, correlations between natural fluctuations in cell motility and protein levels were used to identify hundreds of candidate genes that regulate cell movement. In this way, heterogeneity observed at single-cell resolution has been repurposed as a powerful means of experimental perturbation.
5. OUTLOOK AND CHALLENGES FOR SINGLE-CELL SIGNALING
Time-lapse fluorescence microscopy provides an exciting glimpse at the signaling activity in individual cells. Here, we have examined the heterogeneity of protein dynamics that is observed at the single-cell level. We have discussed both experimental approaches to measure single-cell responses as well as computational approaches to model, perturb, and understand the role of protein dynamics. Just as genome sequences have spurred the development of bioinformatics methods to analyze sequencing data, the immense imaging data collected from live-cell experiments has spurred the development of unique computational strategies to analyze and model biological phenomenon in single cells over time.
Despite rapid progress in the field, there are specific challenges for single-cell analysis that must be met to bring the field to a state of maturity. To date, most live-cell studies of human proteins have been performed in transformed or immortalized cell lines such as HeLa and MCF7. This is likely due to the practical challenges of culturing cells in microscope enclosure systems and repeatedly exposing them to fluorescent light [19]. A critical step forward therefore is to characterize signaling in primary cells, stem cells, explants, and other tissue-like systems. It is known that populations of hematopoetic stem cells and retinal progenitor cells show extensive intrinsic molecular heterogeneity [70]. Furthermore, there is increasing evidence that single cell dynamics are relevant to normal growth and development in vivo [71]. Live-cell time-lapse microscopy of single molecule dynamics in cells transitioning from multi-potency to lineage commitment could resolve the kinetics of molecular events involved in cell fate specification [72]. For example, recent studies demonstrate that the oscillatory expression of transcription factors in neural progenitor cells regulates cell fate choice in the developing mammalian brain [73].
Another unexplored direction in single-cell dynamics is the role of non-cell autonomous signaling. Technological advances in single-cell measurements have thus far emphasized the characteristics of single cells as independent entities. In contrast, relatively little attention has been given to cell-to-cell signaling, although some work in this area is beginning to emerge [74]. In preparation for these data, computational approaches are needed to account for cell-to-cell communication in order to quantify how the signaling of one cell influences another. One predicted outcome of such analyses would be to resolve additional types of single-cell heterogeneity. For example, two cells that share a similar combination of neighboring cells may show similar signaling patterns. If paracrine signals are at work, it is likely that signaling dynamics for certain proteins are correlated between neighboring cells, even if those two cells do not share a recent ancestor. To detect such patterns, new computational tools are needed to analyze single-cell signaling patterns within the context of large cell populations.
An ultimate goal for analyzing single-cell dynamics, in both individual and coordinated populations of cells, will be to understand how signaling patterns change in the context of disease. With adequate computational tools to simplify and classify single-cell signaling patterns, perturbations of signaling caused by disease can be more readily identified. Such methods could also indicate how unwanted molecular interactions shape and respond signaling patterns in diseased cells. This a natural way forward for the IDP field. By integrating single-cell approaches with existing tools for protein structure analysis, the field will further its ultimate goal of understanding how disordered proteins, both natural and diseased, may function as sources of cellular heterogeneity.
ACKNOWLEDGEMENTS
We thank Jean Cook, Sabrina Spencer, John Albeck, Nan Hao, and all members of our laboratory for helpful discussions, comments and reference suggestions. This research was supported by the National Institutes of Health grant R00-GM102372 (J.E.P.) and Diversity Training Award R00-GM102372-S (D.M.D. and J.E.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lee RE, Walker SR, Savery K, Frank DA, Gaudet S. Fold change of nuclear NF-kappaB determines TNF-induced transcription in single cells. Molecular cell. 2014;53:867–879. doi: 10.1016/j.molcel.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder B, Sacher R, Ramo P, Damm EM, Liberali P, Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 3.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, et al. A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Sci Signal. 2009;2:ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heemskerk JW, Willems GM, Rook MB, Sage SO. Ragged spiking of free calcium in ADP-stimulated human platelets: regulation of puff-like calcium signals in vitro and ex vivo. J Physiol. 2001;535:625–635. doi: 10.1111/j.1469-7793.2001.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 7.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, et al. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. Journal of proteome research. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cumberworth A, Lamour G, Babu MM, Gsponer J. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem J. 2013;454:361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- 9.Hsu WL, Oldfield CJ, Xue B, Meng J, Huang F, Romero P, et al. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22:258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically Disordered Proteins in Human Diseases: Introducing the D2 Concept. Annual Reviews Biophysics. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 11.Purvis JE, Lahav G. Encoding and Decoding Cellular Information through Signaling Dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagnall J, Leedale J, Taylor SE, Spiller DG, White MR, Sharkey KJ, et al. Tight control of hypoxia-inducible factor-alpha transient dynamics is essential for cell survival in hypoxia. J Biol Chem. 2014;289:5549–5564. doi: 10.1074/jbc.M113.500405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yissachar N, Sharar Fischler T, Cohen AA, Reich-Zeliger S, Russ D, Shifrut E, et al. Dynamic response diversity of NFAT isoforms in individual living cells. Molecular cell. 2013;49:322–330. doi: 10.1016/j.molcel.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewer A, Karanam K, Mock C, Lahav G. The p53 response in single cells is linearly correlated to the number of DNA breaks without a distinct threshold. BMC biology. 2013;11:114. doi: 10.1186/1741-7007-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albeck JG, Mills GB, Brugge JS. Frequency-Modulated Pulses of ERK Activity Transmit Quantitative Proliferation Signals. Molecular cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters JC. Live-cell fluorescence imaging. Methods in cell biology. 2013;114:125–150. doi: 10.1016/B978-0-12-407761-4.00006-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Herbst-Robinson KJ, Zhang J. Visualizing Dynamic Activities of Signaling Enzymes Using Genetically Encodable FRET-Based Biosensors: From Designs to Applications. Methods in Enzymology. 2012;504 doi: 10.1016/B978-0-12-391857-4.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgson L, Pertz O, Hahn KM. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods in cell biology. 2008;85:63–81. doi: 10.1016/S0091-679X(08)85004-2. [DOI] [PubMed] [Google Scholar]
- 22.Rehm M, Düβmann H, Jänicke RU, Tavare JM, Kögel D, Prehn JHM. Single-cell Fluorescence Resonance Energy Transfer Analysis Demonstrates that Caspase Activation during Apoptoisis is a Rapid Process: ROLE OF CASPASE-3. Journal of Biological Chemistry. 2002;277:24506–24514. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- 23.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 24.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 25.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Molecular cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Molecular cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yosef N, Regev A. Impulse control: temporal dynamics in gene transcription. Cell. 2011;144:886–896. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batchelor E, Loewer A, Mock C, Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol Syst Biol. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100068. 2006 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, et al. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative Binding of Tetrameric p53 to DNA. Journal of Molecular Biology. 2004;341 doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 34.Rajagopalan S, Huang F, Fersht AR. Single-Molecule Characterization of Oligomerization kinetics and Equilibria of the Tumor Suppressor p53. Nucleic Acids Research. 2011;39 doi: 10.1093/nar/gkq800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaglia G, Guan Y, Shah JV, Lahav G. Activation and Control of p53 Tetramerization in Individual Living Cells. Proceedings of the National Academies of Sciences. 2013;110:15497–15501. doi: 10.1073/pnas.1311126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem. 2003;278:13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki H, Tomida A, Tsuruo T. Dephosphorylated hypoxia-inducible factor 1alpha as a mediator of p53-dependent apoptosis during hypoxia. Oncogene. 2001;20:5779–5788. doi: 10.1038/sj.onc.1204742. [DOI] [PubMed] [Google Scholar]
- 38.Welch CM, Elliott H, Danuser G, Hahn KM. Imaging the coordination of multiple signalling activities in living cells. Nature reviews. 12:749–756. doi: 10.1038/nrm3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behar M, Hao N, Dohlman HG, Elston TC. Dose-to-duration encoding and signaling beyond saturation in intracellular signaling networks. PLoS Comput Biol. 2008;4:e1000197. doi: 10.1371/journal.pcbi.1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 43.Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U. Dynamics and variability of ERK2 response to EGF in individual living cells. Molecular cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Shoval O, Goentoro L, Hart Y, Mayo A, Sontag E, Alon U. Fold-change detection and scalar symmetry of sensory input fields. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15995–16000. doi: 10.1073/pnas.1002352107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Molecular cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goentoro L, Kirschner MW. Evidence that fold-change, and not absolute level, of betacatenin dictates Wnt signaling. Molecular cell. 2009;36:872–884. doi: 10.1016/j.molcel.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flusberg DA, Roux J, Spencer SL, Sorger PK. Cells surviving fractional killing by TRAIL exhibit transient but sustainable resistance and inflammatory phenotypes. Molecular biology of the cell. 2013;24:2186–2200. doi: 10.1091/mbc.E12-10-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH. Distinct Molecular Mechanisms Regulate Cell Cycle Timing at Successive Stages of Drosophila Embryogenesis. Genes & Development. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Resende TP, Andrade RP, Palmeirim I. Timing Embryo Segmentation: Dynamics and Regulatory Mechanisms of the Vertebrate Segmentation Clock. BioMed Research International. 2014;2014:1–12. doi: 10.1155/2014/718683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine JH, Fontes ME, Dworkin J, Elowitz MB. Pulsed Feedback Defers Cellular Differentiation. Plos Biology. 2012;10:e1001252-e. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Molecular and cellular biology. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pardee AB. A restriction point for control of normal animal cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chassot AA, Lossaint G, Turchi L, Meneguzzi G, Fisher D, Ponzio G, et al. Confluence-induced cell cycle exit involves pre-mitotic CDK inhibition by p27(Kip1) and cyclin D1 downregulation. Cell Cycle. 2008;7:2038–2046. doi: 10.4161/cc.7.13.6233. [DOI] [PubMed] [Google Scholar]
- 56.Hitomi M, Stacey DW. Cellular ras and cyclin D1 are required during different cell cycle periods in cycling NIH 3T3 cells. Molecular and cellular biology. 1999;19:4623–4632. doi: 10.1128/mcb.19.7.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 59.Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell. 2011;144:926–939. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouchard TJ, Propping P. Twins as a Tool of Behavioral Genetics. Wiley: 1993. [Google Scholar]
- 61.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 62.Schroeder T. Heterogeneity of sister cell fates. Nature reviews. 2013;14:327. doi: 10.1038/nrm3594. [DOI] [PubMed] [Google Scholar]
- 63.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 64.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Molecular cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 67.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 68.Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat Rev Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farkash-Amar S, Zimmer A, Eden E, Cohen A, Geva-Zatorsky N, Cohen L, et al. Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells. Plos Genetics. 2014;10:1–10. doi: 10.1371/journal.pgen.1004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nature reviews. 2013 doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahrends R, Ota A, Kovary KM, Kudo T, Park BO, Teruel MN. Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science. 2014;344:1384–1389. doi: 10.1126/science.1252079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, et al. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- 74.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014 doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 76.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic acids research. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]