Abstract
This study aimed to evaluate the genes that were expressed in the healing bones around SLA-treated titanium orthodontic mini-implants in a beagle at early (1-week) and late (4-week) stages with RNA-sequencing (RNA-Seq). Samples from sites of surgical defects were used as controls. Total RNA was extracted from the tissue around the implants, and an RNA-Seq analysis was performed with Illumina TruSeq. In the 1-week group, genes in the gene ontology (GO) categories of cell growth and the extracellular matrix (ECM) were upregulated, while genes in the categories of the oxidation-reduction process, intermediate filaments, and structural molecule activity were downregulated. In the 4-week group, the genes upregulated included ECM binding, stem cell fate specification, and intramembranous ossification, while genes in the oxidation-reduction process category were downregulated. GO analysis revealed an upregulation of genes that were related to significant mechanisms, including those with roles in cell proliferation, the ECM, growth factors, and osteogenic-related pathways, which are associated with bone formation. From these results, implant-induced bone formation progressed considerably during the times examined in this study. The upregulation or downregulation of selected genes was confirmed with real-time reverse transcription polymerase chain reaction. The RNA-Seq strategy was useful for defining the biological responses to orthodontic mini-implants and identifying the specific genetic networks for targeted evaluations of successful peri-implant bone remodeling.
1. Introduction
The development of temporary skeletal anchorage devices (TSADs) in orthodontics has made the mechanics of treatment simpler and more effective than conventional techniques [1–3]. Orthodontic mini-implants, which are generally made of a titanium alloy, are cylindrically shaped with a diameter of 1.5–2.0 mm and a length of 6.0–9.0 mm [4]. Orthodontic force is loaded on the day of mini-implant insertion because primary retention is obtained by mechanically locking the implant onto the cortical plate of the alveolar bone. Thus, cortical bone thickness is the most important factor in determining the initial stability of mini-implants [5].
Previous survival analyses of orthodontic mini-implants showed that the failure rate is greater than 10% [6–8]. The known risk factors for failure are root proximity, insertion site, and inflammation of the peri-implant soft tissue. Many attempts have been made to increase the survival rate of mini-implants [1–3, 9]. Studies have shown that the healing period between mini-implant insertion and orthodontic force application is not critical. Deguchi et al. [10] applied orthodontic force after different healing times (3 weeks, 6 weeks, or 12 weeks) in a dog model, and they found no significant differences in the survival rates. Lee et al. [11] reported that a mini-implant placement angle of less than 60° reduces stability when orthopedic forces are applied in various directions. The design and surface treatment of the mini-implants have been modified similar to those of dental implants, and Kim et al. [12] introduced a two-component mini-implant with a sand blasted with large grit and acid-etched- (SLA-) treated surface.
The key to the success of dental prosthetic implants is osseointegration, and several efforts have been made to increase the area of direct contact between bone and the implant surface [13]. The introduction of a proteoglycan and glycosaminoglycan complex at the interface between the titanium implant and the mineralized tissue was reported to increase the mechanical interlocking and biological interfacial adhesion in vitro [14]. Various tools and techniques have been used to investigate the mechanisms of osseointegration. Numerous histologic and histomorphometric analyses and microscopic studies have been performed to examine the bone-implant contact ratio. Transmission electron microscopy, scanning electron microscopy, electron energy loss spectroscopy, and scanning transmission electron microscopy techniques have been introduced to evaluate the titanium-bone interface [15–17].
At the molecular level, only a few transcriptional profiling studies have been performed to characterize osseointegration or alveolar bone remodeling after dental implant placement, and most of these studies have used microarrays. Kojima et al. [18] evaluated gene expression in bone healing around titanium implants in rats with microarray. Ivanovski et al. [19] reported that the transcriptional profile of osseointegration after implant insertion in humans predominantly involved genes that are related to the immune response and extracellular matrix (ECM) formation.
In the field of orthodontics, no studies have examined mini-implant-induced gene expression patterns. The bones healing around orthodontic mini-implants are thought to have unique osteogenic characteristics. Ogawa and Nishimura [20] evaluated the gene expression patterns around implants that have two different surfaces with real-time reverse transcription-polymerase chain reaction (RT-PCR). In the bone surrounding the dual-acid etched surfaced implant, the levels of gene expression of osteonectin and osteocalcin were upregulated compared to their expression in the bone surrounding the turned implant, and bone sialoprotein II, collagen III, and integrin-related genes were upregulated 1 week after implantation [20].
Recently, a technique that combines an RNA sequencing (RNA-Seq) analysis with functional gene classification by whole transcriptome screening has been introduced as an alternative to microarray analysis. Detailed and deep profiling of the transcriptome is possible with this tool [21, 22]. The RNA-Seq analysis can detect a very small amount of RNA compared to that detected by microarray, and the RNA-Seq results show a high level of reproducibility [23]. In addition, fusion gene candidates and insertion and deletion candidates can be identified with RNA-Seq [24]. Moreover, novel genes that have not been annotated in the gene database can be detected [25]. Xu et al. [26] compared transcriptome profiles that were generated by Illumina RNA-Seq and Affymetrix microarray platforms, and their results suggested that RNA-Seq is more advantageous for detecting genes with low-level expression compared with microarrays.
The aim of this study was to use an RNA-Seq analysis to evaluate gene expression in healing bones around titanium orthodontic mini-implants at early and late stages. The molecular mechanisms of bone remodeling around mini-implants were compared with those of bone healing around surgical defects.
2. Materials and Methods
2.1. Animal Subjects and Implantation
C-implants (1.8 mm × 8.5 mm; Dentium Co., Ltd., Suwon, Korea) were used as SLA-treated titanium implants. Six C-implants were inserted into the basal bone of the mandible in a 12-month-old male beagle dog (10 kg body weight). One single animal was optimal for this study in order to exclude individual genetic differences between samples. A manually drilled osteotomy was followed by manual insertion of the implant rather than machine drilling. Therefore, a saline injection for cooling was not needed. All SLA-treated surfaces were submerged under the bone, and tooth roots or nerves were not injured.
C-implants were implanted under general anesthesia with an intramuscular injection of Zoletil 50 (1.5 cc; Virbac, Carros, France) and Rompun (0.7 cc; Bayer Korea, Ltd., Seoul, Korea) 1 and 4 weeks before euthanization. After the surgery, the antibiotic gentamicin (Komipharm International Co., Ltd., Shinheung, Korea) and the anti-inflammatory agent ketoprofen (Uni Biotech Co., Ltd., Chungnam, Korea) were administered by intramuscular injections. As a control, two surgical defects were created with the same manual drill 1 week before euthanization in the mandibular basal bone. There was no contact between the titanium implant and the bone in the control group.
Samples were retrieved after soft tissue was removed with a trephine bur with an inner diameter of 6.0 mm. The study protocol was approved by the Institutional Animal Care and Use Committee of Kyung Hee University (KHMC-IACUC-2012-026).
2.2. RNA Preparation and Quality Check
After the implant and remaining soft tissue were carefully removed, RNA was prepared from the samples with TRIzol reagent (750 μL per sample; Life Technologies, Grand Island, NY, USA) and a RNeasy mini kit (QIAGEN Inc., Valencia, CA, USA). Hard tissue was homogenized with a TissueLyser (QIAGEN Inc.) at 25 Hz for 10 min. The plate was incubated at room temperature and then centrifuged at 12,000 ×g for 1 min. Chloroform (150 μL) was added to the well, and the plate was then vortexed extensively. The plate was incubated at room temperature for 2-3 min and then centrifuged at 12,000 ×g for 1 min. The supernatant (350 μL) was mixed with an equal volume of 70% ethanol in a new well. Then, the RNA was extracted with the RNeasy mini Kit according to the manufacturer's instructions. RNA integrity was measured with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and a sample with an RNA Integrity Number greater than or equal to 8 was considered acceptable.
2.3. Transcriptome Sequencing
Transcriptome Sequencing was initiated by transforming the mRNA in the total RNA samples into a template library, which was followed by cluster generation with the components provided in the TruSeq Sample Preparation RNA Kit (Illumina, Inc., San Diego, CA, USA). First, the poly-A-containing mRNA molecules were purified with magnetic beads that were attached to poly-T oligo indicators. Subsequently, at an increased temperature, the mRNA was fragmented into small parts with divalent cations. The cleaved RNA fragments were transcribed into first-strand cDNA with reverse transcriptase and random primers. Subsequently, the second strand was synthesized with RNase H and DNA polymerase. The cDNA fragments were end-repaired by adding a single A base, to which adapters were ligated. Then, the cDNAs were refined and enriched. On the surface of the flow cell, a distinctive bridged amplification reaction occurred with the Illumina kit. Therefore, a flow cell containing numerous unique clusters was placed into the HiSeq 2000 (Illumina, Inc.), which enabled imaging and extension in automated cycles. Sequences were produced by 100-bp paired-end technology. All four nucleotides needed to be present in each sequencing cycle, and this resulted in higher accuracy than the other methods that use only one nucleotide at a time in the reaction mix. The cycles were repeated one base at a time, and a series of images, each of which represented a single-base extension at a unique cluster, was generated.
2.4. Gene Ontology (GO) Enrichment Analysis
GO term annotations for Canis familiaris were obtained from Ensembl (release 75) at Biomart (http://www.ensembl.org/biomart/martview). In order to determine whether a GO term was enriched in a set of genes (upregulated or downregulated), the number of genes within a set that had a specific GO term was compared to the number in the control gene set. The control gene set was generated by randomly choosing genes from all of the annotated genes (gene length-controlled). Therefore, each test gene had a corresponding control gene. The P value for each enriched GO category in the test gene set was calculated as the fraction of times that the F test was lower than or equal to the F control, where F test and F control represent the fraction of genes in the test set or random control set, respectively, and were linked with the present GO term based on 10,000 randomly chosen control sets. In order to choose significantly enriched GO terms, a P value threshold (1/total number of GO terms considered) was applied.
2.5. Real-Time RT-PCR
The samples used in the RNA-Seq analysis were quantified by real-time RT-PCR. cDNA was synthesized with 100 ng of RNA and the Superscript II RT-PCR System (Life Technologies) at 42°C according to the manufacturer's recommendations for oligo(dT)20-primed cDNA synthesis. Then, the cDNA was diluted 1 : 2 prior to RT-PCR.
For the quantitative TaqMan PCR, the reactions were performed in 384-well microtiter plates in a final volume of 10 μL with a QuantStudio 12 K Flex Real-time PCR System (Life Technologies). Optimum results were obtained with the following reaction conditions: 5 μL of Universal Master Mix (containing dNTPs, MgCl2, reaction buffer, and AmpliTaq Gold; Life Technologies), 90 nM of primers, and 250 nM of fluorescence-labeled TaqMan probe (Life Technologies). Finally, 2 μL of template cDNA was added to the reaction mixture. The primer/TaqMan probe combinations were designed based on each target sequence. The amplification cycling conditions were as follows: a 10-min template denaturation step at 95°C, which was followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. All of the samples were amplified in triplicate, and the data were analyzed with Sequence Detector software (Life Technologies).
The comparative Ct method was used for relative quantification. The target genes were angiopoietin-4 (ANGPT4), platelet-derived growth factor receptor α (PDGFRA), phosphatidylinositol-5-phosphate 4 kinase type-2 α (PIP4K2A), and WNT1-inducible-signaling pathway protein 2 (WISP2).
3. Results
3.1. Sample Analysis for Quality Control
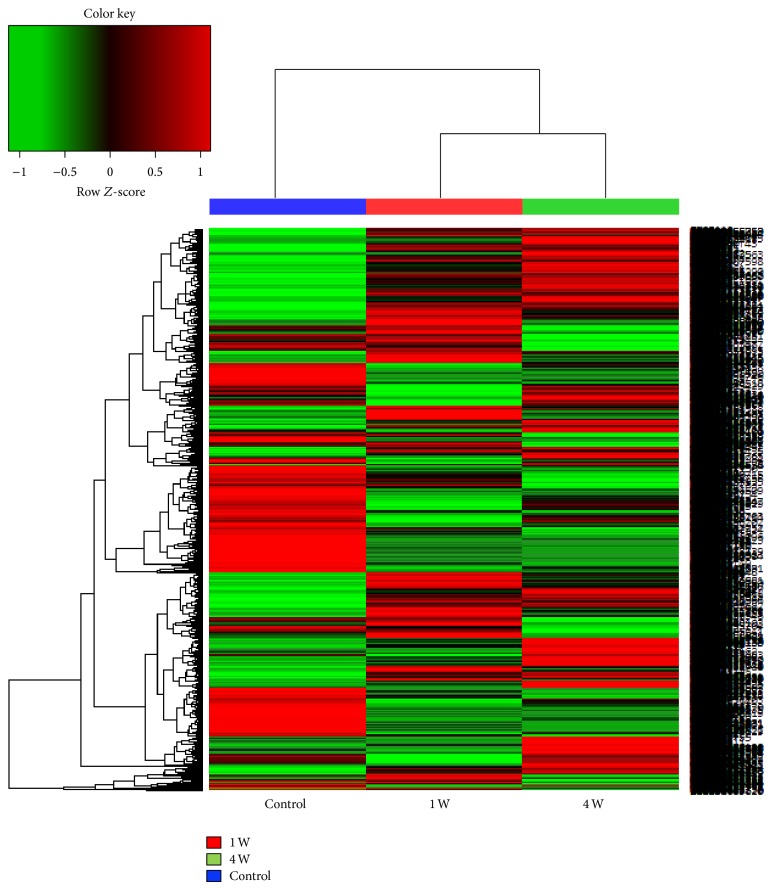
In the first step of the principal gene analysis, 13,690 (of the 21,744 total) transcripts with zero fragments per kilobase of exon per million fragments mapped (FPKMs) were excluded. Of the remaining 8,054 transcripts, 1,643 and 1,839 transcripts had a >2-fold difference in gene expression in the tissues around the implant at 1 week and 4 weeks, respectively, compared with the control surgical defect sites (Figure 1). Among the differentially expressed genes, 773 transcripts were upregulated and 870 were downregulated at 1 week, whereas 937 transcripts were upregulated and 902 were downregulated at 4 weeks.
Figure 1.
Heatmap of the samples. The red lines represent upregulated genes, and the green lines represent downregulated genes at 1 week and 4 weeks after implantation.
3.2. Functional Annotation of the Transcriptome
Based on the GO classifications in the implant group compared with the surgical defect group at the defined time points of 1 week or 4 weeks after implantation, the principally regulated functions were determined, including the GO terms, gene numbers, and the P values. Briefly, in the 1-week implant group, the upregulated GO categories included cell growth (CYR61, IGFBP6, LOC476202, ESM1, IGFBP2, CRIM1, and WISP2), DNA helicase activity (MCM4, MCM2, MCM6, MCM5, RECQL, and MCM3), and calcium ion binding (CCBE1, SMOC1, EHD2, CLEC3B, LOC100856635, and FKBP10). The downregulated GO categories included the oxidation-reduction process (FMO3, AHCYL2, LOC484867, ALDH3A1, GSR, and FTH1), intermediate filaments (KRT6A, KRT86, KRT18, KRT6B, and KRT14), and structural molecule activity (CLDN10, KRT13, DSP, KRT17, VAPA, OCLN, and CLDN1) (Table 1). In the 4-week implant group, the upregulated GO categories included ECM binding (DCN, VTN, SMOC1, BGN, NID1, and FBLN2), stem cell fate specification (SOX17 and SOX18), and intramembranous ossification (MMP2, MN1, and FGF18), and the downregulated categories included keratin filaments (KRT6A, KRT86, KRT6B, and KRT14), oxidoreductase activity (FAM81A, LOC484867, GSR, GPX2, and SOD1), and superoxide metabolic process (PCDP1, CYB5R4, NOXO1, and SOD2) (Table 2). There was some overlap in the differentially expressed genes between the 1-week and 4-week implant groups. There were 299 upregulated genes and 481 downregulated genes at both time points.
Table 1.
Main genes that were up- or downregulated based on the gene ontology (GO) analysis at week 1 compared with the levels of expression in the control.
| Upregulated genes | Downregulated genes | ||||
|---|---|---|---|---|---|
| GO category | Numbers of genes | P value | GO category | Numbers of genes | P value |
| Cellular component | |||||
| Basement membrane | 14 | <0.0001 | Apical plasma membrane | 15 | <0.0001 |
| Extracellular matrix | 38 | <0.0001 | Glutamate-cysteine ligase complex | 2 | <0.0001 |
| Extracellular region | 53 | <0.0001 | Hemoglobin complex | 2 | <0.0001 |
| Extracellular space | 47 | <0.0001 | Intermediate filament | 12 | <0.0001 |
| Proteinaceous extracellular matrix | 17 | 0.0001 | Keratin filament | 8 | <0.0001 |
|
| |||||
| Biological process | |||||
| Cartilage morphogenesis | 3 | <0.0001 | Adhesion to symbiont | 2 | <0.0001 |
| Collagen fibril organization | 9 | <0.0001 | Cellular aldehyde metabolic process | 3 | <0.0001 |
| DNA replication | 13 | <0.0001 | Cilium movement | 4 | 0.0003 |
| DNA replication initiation | 5 | <0.0001 | Desmosome organization | 2 | <0.0001 |
| DNA unwinding involved in DNA replication | 4 | <0.0001 | Fucose metabolic process | 2 | <0.0001 |
| Extracellular fibril organization | 3 | 0.0003 | Glutathione metabolic process | 9 | <0.0001 |
| Extracellular matrix organization | 11 | 0.0003 | Male sex differentiation | 2 | <0.0001 |
| Regulation of cell growth | 7 | <0.0001 | Neutrophil aggregation | 2 | <0.0001 |
| Oxidation-reduction process | 49 | <0.0001 | |||
| Positive regulation of cation channel activity | 2 | <0.0001 | |||
| Protein homotetramerization | 8 | <0.0001 | |||
| Regulation of cell death | 3 | 0.0003 | |||
| Response to metal ion | 2 | <0.0001 | |||
|
| |||||
| Molecular function | |||||
| Calcium ion binding | 42 | 0.0002 | Aldehyde dehydrogenase (NAD) activity | 3 | <0.0001 |
| DNA helicase activity | 6 | <0.0001 | Aldehyde dehydrogenase [NAD(P)+] activity | 2 | <0.0001 |
| Extracellular matrix binding | 7 | <0.0001 | Alpha-L-fucosidase activity | 2 | <0.0001 |
| Extracellular matrix structural constituent | 7 | <0.0001 | Flavin adenine dinucleotide binding | 12 | <0.0001 |
| Heparin binding | 11 | <0.0001 | Glutamate-cysteine ligase activity | 2 | <0.0001 |
| Insulin-like growth factor binding | 7 | <0.0001 | Oxidoreductase activity | 44 | <0.0001 |
| Platelet-derived growth factor binding | 5 | <0.0001 | Oxidoreductase activity, acting on CH-OH group of donors | 5 | <0.0001 |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | 5 | 0.0003 | |||
| Structural molecule activity | 16 | 0.0001 | |||
| Superoxide dismutase activity | 3 | 0.0003 | |||
| UDP-N-acetylmuramate dehydrogenase activity | 4 | <0.0001 | |||
The values were compared according to fold change.
Table 2.
Main genes that were up- or downregulated based on the GO analysis at week 4 compared with the levels of expression in the control.
| Upregulated genes | Downregulated genes | ||||
|---|---|---|---|---|---|
| GO category | Numbers of genes | P value | GO category | Numbers of genes | P value |
| Cellular component | |||||
| Extracellular matrix | 27 | <0.0001 | Basolateral plasma membrane | 12 | 0.0001 |
| Extracellular region | 55 | <0.0001 | Cell surface | 27 | 0.0002 |
| Extracellular space | 50 | <0.0001 | Cell-cell junction | 13 | 0.0001 |
| Ribosome | 38 | <0.0001 | Glutamate-cysteine ligase complex | 2 | <0.0001 |
| Keratin filament | 8 | <0.0001 | |||
|
| |||||
| Biological process | |||||
| Angiogenesis | 16 | 0.0002 | Chaperone mediated protein folding requiring cofactor | 5 | <0.0001 |
| Bone mineralization | 6 | 0.0002 | Desmosome organization | 2 | <0.0001 |
| Cartilage morphogenesis | 3 | <0.0001 | Glutathione metabolic process | 8 | <0.0001 |
| Heterophilic cell-cell adhesion | 5 | 0.0003 | Glycerol-3-phosphate metabolic process | 3 | <0.0001 |
| Intramembranous ossification | 3 | 0.0001 | Male sex differentiation | 2 | <0.0001 |
| Negative regulation of mesodermal cell fate specification | 3 | <0.0001 | Neutrophil aggregation | 2 | <0.0001 |
| Regulation of stem cell division | 2 | <0.0001 | Oxidation-reduction process | 49 | <0.0001 |
| Stem cell fate specification | 2 | <0.0001 | Smooth muscle cell migration | 2 | <0.0001 |
| Translation | 40 | <0.0001 | Superoxide metabolic process | 5 | 0.0001 |
| Termination of signal transduction | 2 | <0.0001 | |||
| Ventricular system development | 4 | 0.0001 | |||
|
| |||||
| Molecular function | |||||
| Ceramide kinase activity | 2 | <0.0001 | Catalytic activity | 52 | <0.0001 |
| Extracellular matrix binding | 6 | <0.0001 | Chemokine binding | 2 | <0.0001 |
| Retinoid binding | 3 | <0.0001 | Glutamate-cysteine ligase activity | 2 | <0.0001 |
| Structural constituent of ribosome | 37 | <0.0001 | Glycerol channel activity | 2 | <0.0001 |
| Interleukin-8 binding | 2 | <0.0001 | |||
| NAD binding | 9 | <0.0001 | |||
| Oxidoreductase activity | 44 | <0.0001 | |||
| Oxidoreductase activity, acting on CH-OH group of donors | 5 | <0.0001 | |||
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 7 | 0.0001 | |||
| Structural molecule activity | 18 | <0.0001 | |||
| UDP-N-acetylmuramate dehydrogenase activity | 4 | 0.0001 | |||
The values were compared according to fold change.
3.3. Differential Expression of Selected Gene Categories
A number of different gene categories were examined in the RNA-Seq analysis, and four major functional groupings of differentially expressed genes were identified across the time course of peri-implant bone healing, including (1) cell proliferation, (2) ECM, (3) growth factors, and (4) osteogenic-related genes.
3.3.1. Highly Expressed Genes Involved in Cell Proliferation
The genes and signaling pathways that were related to cell proliferation are listed in Table 3. A number of genes were differentially regulated at 1 week and 4 weeks. At 1 week, the levels of expression of CDK14, PLCD1, and ZBTB16 were 2-fold higher than those in the control. Similarly, at 4 weeks, the levels of expression of FGF18, HES4, and FYN were 2-fold higher than those in the control.
Table 3.
Differentially expressed genes associated with cell proliferation.
| Gene symbol | Description | 1 W | 4 W |
|---|---|---|---|
| AIF1 | Allograft inflammatory factor 1 isoform 1 | 2.81 | 4.23 |
| ANGPT1 | Angiopoietin-1 precursor | 3.21 | 2.18 |
| APOA1 | Apolipoprotein A-I | 2.12 | 4.42 |
| BLOC1S2 | Biogenesis of lysosome-related organelles complex-1 subunit 2 | 3.35 | 2.98 |
| CDK14 | Cyclin-dependent kinase 14 isoform 3 | 4.91 | 1.45 |
| CLEC11A | C-type lectin domain family 11 member A | 4.33 | 5.44 |
| CXCL12 | Stromal cell-derived factor 1 precursor | 2.52 | 4.36 |
| DLC1 | Rho GTPase-activating protein 7 | 4.75 | 2.67 |
| DPT | Dermatopontin | 3.84 | 2.01 |
| FGF18 | Fibroblast growth factor 18 | 2.37 | 2.81 |
| FYN | Tyrosine-protein kinase Fyn isoform 2 | 2.05 | 2.62 |
| HES4 | Transcription factor HES-4 | 2.16 | 2.18 |
| LECT1 | Leukocyte cell-derived chemotaxin 1 | 3.68 | 9.13 |
| MAP2K1 | Dual specificity mitogen-activated protein kinase 1 | 2.42 | 3.64 |
| MSX1 | Homeobox protein MSX-1 | 3.49 | 2.94 |
| NFIB | Nuclear factor 1 B-type isoform 3 | 2.73 | 2.94 |
| PDGFRA | Platelet-derived growth factor receptor alpha | 3.94 | 2.42 |
| PLCD1 | 1-Phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 isoform 3 | 4.40 | 3.63 |
| RBP4 | Retinol-binding protein 4 | 4.08 | 6.30 |
| SERPINF1 | Pigment epithelium-derived factor | 4.82 | 3.27 |
| SMYD2 | N-lysine methyltransferase SMYD2 | 2.29 | 2.76 |
| TBX2 | T-box transcription factor TBX2 | 2.22 | 5.48 |
| VEGFC | Vascular endothelial growth factor C | 3.00 | 2.82 |
| VSIG4 | V-set and immunoglobulin domain-containing protein 4 | 4.73 | 4.18 |
| ZBTB16 | Zinc finger and BTB domain-containing protein 16 isoform 2 | 3.25 | 5.25 |
The values were compared according to fold change. W: week.
3.3.2. Genes Related to ECM Pathways
Many of the genes that were related to the ECM are shown in Table 4. FN1, COL1A2, and FBN1 were upregulated at 1 week compared to their levels in the control group. In contrast, at 4 weeks, the levels of expression of other genes, such as LAMB2, VTN, and MMP11, were 2-fold higher than those in the control.
Table 4.
Differentially expressed genes associated with the extracellular matrix (ECM) pathway.
| Gene symbol | Description | 1 W | 4 W |
|---|---|---|---|
| BGN | Biglycan precursor | 12.26 | 5.86 |
| COL1A2 | Collagen α-2(I) chain precursor | 9.27 | 1.28 |
| COL5A3 | Collagen α-3(28V)29 chain | 6.95 | 7.00 |
| DCN | Decorin precursor | 6.23 | 4.29 |
| DPT | Dermatopontin | 3.84 | 2.01 |
| EFEMP1 | EGF containing fibulin-like extracellular matrix protein 1 | 3.33 | 2.58 |
| FBLN1 | Fibulin-1 | 3.14 | 1.53 |
| FBLN5 | Fibulin-5 | 5.21 | 4.02 |
| FBN1 | Fibrillin-1 isoform 3 | 6.67 | −2.52 |
| FMOD | Fibromodulin | 3.18 | 3.05 |
| FN1 | Fibronectin | 3.59 | −1.19 |
| LAMB2 | Laminin subunit beta-2 | 1.21 | 2.21 |
| LUM | Lumican | 6.28 | 2.55 |
| MMP11 | Stromelysin-3 precursor | 1.51 | 2.06 |
| NID1 | Nidogen-1 isoform 1 | 4.87 | 3.02 |
| OGN | Mimecan | 12.21 | 3.05 |
| PCOLCE | Procollagen C-endopeptidase enhancer 1 isoform 3 | 3.17 | 2.00 |
| PDGFRA | Platelet-derived growth factor receptor alpha | 3.94 | 2.42 |
| POSTN | Periostin | 5.61 | 2.39 |
| PRELP | Prolargin | 7.41 | 6.22 |
| SERPINF1 | Pigment epithelium-derived factor | 4.82 | 3.27 |
| SMOC2 | SPARC related modular calcium binding 2 precursor | 7.68 | 11.27 |
| TIMP3 | Metalloproteinase inhibitor 3 isoform 1 | 4.19 | 2.08 |
| TNC | Tenascin precursor | 6.19 | 6.87 |
| VTN | Vitronectin isoform 2 | 1.06 | 4.33 |
The values were compared according to fold change. W: week.
3.3.3. Genes Related to Growth Factors
Other gene groups, such as genes that were related to growth factor pathways, including the platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), transforming growth factor β (TGF-β ), vascular endothelial growth factor (VEGF), and insulin-like growth factor (IGF), were analyzed across the two time points with a cut-off of a 2-fold difference (Table 5). The selected genes were upregulated during the early and late stages of the bone healing process.
Table 5.
Differentially expressed genes associated with growth factors.
| Gene symbol | Description | 1 W | 4 W |
|---|---|---|---|
| ANGPT4 | Angiopoietin-4 | 4.65 | 9.81 |
| CLEC11A | C-type lectin domain family 11 member A | 4.33 | 5.44 |
| CLEC3B | Tetranectin | 3.31 | 6.98 |
| CRIM1 | Cysteine-rich motor neuron 1 protein | 4.27 | 3.51 |
| EFEMP1 | EGF containing fibulin-like extracellular matrix protein 1 | 3.33 | 2.58 |
| ESM1 | Endothelial cell-specific molecule 1 isoform 2 | 3.37 | 7.04 |
| FGF13 | Fibroblast growth factor 13 isoform 1 | 1.67 | 2.33 |
| FGF18 | Fibroblast growth factor 18 | 2.37 | 2.81 |
| FGF23 | Fibroblast growth factor 23 | −1.12 | 3.93 |
| FYN | Tyrosine-protein kinase Fyn isoform 2 | 2.05 | 2.62 |
| IGFBP2 | Insulin-like growth factor binding protein 2 | 2.89 | −1.44 |
| IGFBP6 | Insulin-like growth factor binding protein 6 isoform 3 | 3.11 | 2.45 |
| LECT1 | Leukocyte cell-derived chemotaxin 1 | 3.68 | 9.13 |
| LRP1 | Prolow-density lipoprotein receptor-related protein 1 | 2.25 | 3.28 |
| LTBP3 | Latent-transforming growth factor beta-binding protein 3 | 1.76 | 2.79 |
| NRP2 | Neuropilin-2 isoform 4 | 8.79 | 3.50 |
| PDGFRA | Platelet-derived growth factor receptor α | 3.94 | 2.42 |
| PTN | Pleiotrophin | 3.28 | 2.34 |
| SIRT1 | NAD-dependent deacetylase sirtuin-1 isoform 1 | 2.67 | 2.93 |
| SMAD6 | Mothers against decapentaplegic homolog 6 | 2.36 | 6.82 |
| TGFB1 | Transforming growth factor beta-1 precursor | 4.07 | 8.44 |
| TWF2 | Twinfilin-2 | 2.49 | 2.92 |
| VEGFC | Vascular endothelial growth factor C | 3.00 | 2.82 |
| WISP2 | WNT1-inducible-signaling pathway protein 2 | 8.61 | 6.15 |
| YWHAG | 14-3-3 protein γ | 3.26 | 2.45 |
The values were compared according to fold change. W: week.
3.3.4. Identification of the Genes That Were Related to Bone Remodeling Pathways
Finally, the genes that were related to osteogenic-corresponding pathways, including the regulation of osteoblasts or osteoclast proliferation and development, were categorized (Table 6). Groups of genes that were involved in these pathways, including PTH1R, DLX5, and SMOC1, were upregulated at both time points. Moreover, the genes that were differentially expressed at 1 week and 4 weeks were classified according to their osteogenic differentiation pathway, including the TGF-β/BMP and Wnt signaling pathways. At both time points, several genes that encoded proteins that were involved in the TGF-β/BMP signaling pathway, which is a critical regulator of skeletal development, were identified, including TGFB1, BMP5, MSX1, and SMAD6. The Wnt signaling pathways also appeared to play a critical role during the early and late stages of bone healing. Genes belonging to this pathway, such as WISP2, FZD1, FZD2, LRP1, and LRP5, were upregulated at both time points.
Table 6.
Differentially expressed genes associated with osteogenesis.
| Gene symbol | Description | 1 W | 4 W |
|---|---|---|---|
| Osteogenesis | |||
| CYR61 | Protein CYR61 | 3.03 | 1.67 |
| DLX5 | Homeobox protein DLX-5 isoform 2 | 3.03 | 9.05 |
| FGF23 | Fibroblast growth factor 23 | −1.12 | 3.93 |
| ID3 | DNA-binding protein inhibitor ID-3 | 2.55 | 3.22 |
| ILK | Integrin-linked protein kinase isoform 1 | 2.49 | 1.38 |
| OSR2 | Protein odd-skipped-related 2 isoform 1 | 4.71 | 1.74 |
| PTH1R | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor | 1.95 | 20.12 |
| SFRP2 | Secreted frizzled-related protein 2 precursor | 3.34 | 4.35 |
| SMAD3 | Mothers against decapentaplegic homolog 3 | 2.10 | −1.15 |
| SMOC1 | SPARC-related modular calcium-binding protein 1 isoform 1 | 3.83 | 4.36 |
| SNAI1 | Zinc finger protein SNAI1 | 2.26 | 2.66 |
| SNAI2 | Zinc finger protein SNAI2 | 2.81 | 2.61 |
| TYROBP | TYRO protein tyrosine kinase-binding protein precursor | 1.44 | 2.12 |
|
| |||
| TGF-β | |||
| BMP5 | Bone morphogenetic protein 5 isoform 1 | 2.68 | 2.34 |
| BMPER | BMP-binding endothelial regulator protein | 1.77 | 3.14 |
| CYR61 | Protein CYR61 | 3.03 | 1.67 |
| DLX5 | Homeobox protein DLX-5 isoform 2 | 3.03 | 9.05 |
| GATA3 | Trans-acting T-cell-specific transcription factor GATA-3 isoform 1 | 2.19 | 1.01 |
| HES4 | Transcription factor HES-4 | 2.16 | 2.18 |
| MSX1 | Homeobox protein MSX-1 | 3.49 | 2.94 |
| SFRP2 | Secreted frizzled-related protein 2 precursor | 3.34 | 4.35 |
| SMAD6 | Mothers against decapentaplegic homolog 6 | 2.36 | 6.82 |
| TGFB1 | Transforming growth factor beta-1 precursor | 4.07 | 8.44 |
|
| |||
| WNT | |||
| BARX1 | BARX homeobox 1 | −1.03 | 25.85 |
| CDK14 | Cyclin-dependent kinase 14 isoform 3 | 4.91 | 1.45 |
| DKK1 | Dickkopf-related protein 1 | 12.93 | 2.09 |
| DLX5 | Homeobox protein DLX-5 isoform 2 | 3.03 | 9.05 |
| FZD1 | Frizzled-1 | 4.08 | 2.10 |
| FZD2 | Frizzled-2 | 2.05 | 1.30 |
| HIC1 | Hypermethylated in cancer 1 protein isoform 1 | 2.62 | 2.03 |
| LATS2 | Serine/threonine-protein kinase LATS2 | 4.14 | 2.15 |
| LRP1 | Prolow-density lipoprotein receptor-related protein 1 | 2.25 | 3.28 |
| LRP5 | Low-density lipoprotein receptor-related protein 5 | 2.89 | 1.43 |
| NXN | Nucleoredoxin | 3.35 | 2.48 |
| SFRP2 | Secreted frizzled-related protein 2 precursor | 3.34 | 4.35 |
| SFRP4 | Secreted frizzled-related protein 4 isoform 1 | 2.90 | 4.82 |
| SNAI2 | Zinc finger protein SNAI2 | 2.81 | 2.61 |
| WISP2 | WNT1-inducible-signaling pathway protein 2 | 8.61 | 6.15 |
The values were compared according to fold change. W: week.
3.4. Confirmation of the Differential Expression of Selected Genes by Real-Time RT-PCR
The RNA-Seq results were validated by evaluating the levels of expression of selected genes with real-time RT-PCR analyses. These analyses demonstrated that the selected genes were up- or downregulated in a similar way to those in the RNA-Seq analysis, and the fold changes of four genes (ANGPT4, PDGFRA, PIP4K2A, and WISP2) were confirmed with both methods. The expression levels of these genes that were determined by these two methods were generally consistent, indicating that the RNA-Seq data were valid (Table 7).
Table 7.
Real-time polymerase chain reaction (PCR) validation.
| Symbol | Description | Assay ID | Time | RNA-sequencing | Real-time PCR |
|---|---|---|---|---|---|
| ANGPT4 | Angiopoietin-4 | Cf02656885_m1 | 1 w | 2.22 | 1.76 |
| 4 w | 3.29 | 1.52 | |||
| PDGFRA | Platelet-derived growth factor receptor alpha | Cf02687293_m1 | 1 w | 1.98 | 3.39 |
| 4 w | 1.27 | 0.74 | |||
| PIP4K2A | Phosphatidylinositol-5-phosphate 4-kinase type-2 alpha | Cf02703943_mH | 1 w | 1.09 | 1.98 |
| 4 w | 1.57 | 1.20 | |||
| WISP2 | WNT1-inducible-signaling pathway protein 2 | Cf02702369_g1 | 1 w | 3.11 | 1.65 |
| 4 w | 2.62 | 4.06 |
The values were compared according to fold change.
4. Discussion
Initial stability is important for the good prognosis of dental prosthetic implants and TSADs in orthodontics. Implant design, surface-treatment method, bone-to-implant contact area, and bone quality are related to initial stability. Secondary stability is mainly related to the osteogenesis and osseointegration that occurs on the surface of the bone that is in contact with the titanium. Favorable initial stability between bone and the titanium fixture immediately after its insertion does not guarantee continuous stability under load, such as occlusal forces or orthodontic forces. This is because the bone-healing processes surrounding the titanium implant differ in each individual. In order to understand bone healing around titanium implants, characteristic gene expression at specific time points in the healing process needs to be investigated.
Implant bone healing has been studied by microscopic analyses, histomorphometric evaluations, immunological identifications, and quantitative PCR [17, 27, 28]. The molecular mechanisms underlying implant-bone healing have been examined by microarray and RNA-Seq analyses [29, 30]. Ivanovski et al. [19] used microarray in order to perform transcriptional profiling of the peri-implant bone. Their study showed that the expression of genes that were related to cell proliferation and immunoinflammatory processes dominated on day 4 after insertion of an SLA-treated titanium implant in a human model. In contrast, on day 14, osteogenesis-, angiogenesis-, and neurogenesis-related genes were predominantly expressed.
Peri-implant bone healing is different from normal bone healing [31]. Kojima et al. [18] studied the gene expression patterns around titanium implants with a microarray containing 20,000 rat genes. They showed that 86 genes were upregulated in the implant group compared with the control group at one or more time points (1 week, 2 weeks, or 4 weeks) after implantation. ECM-related genes, bone resorption-related genes, and growth factor-regulating genes were differentially expressed compared to the osteotomy-healing group.
The present study showed upregulation or downregulation in the levels of expression of a set of gene transcripts that were associated with the presence of an implant in bone. Since we intended to classify the gene transcripts that were potentially responsible for the osteogenic aspects of osseointegration, this study focused on the expression of genes that were induced by peri-implant bone healing. The success of a dental implant depends on predictable biological responses to xenobiotic materials, and further studies of these complex cellular and molecular mechanisms are needed to improve clinical outcomes.
RNA-Seq technology provides information on the expression of thousands of genes in a single experiment [32]. The present study used RNA-Seq technology to analyze the gene expression profile during in vivo bone healing around implants that were placed in the mandible. This study was designed to study implant-inducible gene expression patterns at two time points: 1 week (early stage) and 4 weeks (late stage) after implantation. When the gene profile of each implant group at each time was compared with that of a surgical defect control group, we found that a number of gene transcripts were highly upregulated only at 1 week, which suggested the importance of these genes at this early stage in the processes of peri-implant bone healing. Genes encoding ECM constituents were upregulated at 1 week. In contrast, genes associated with bone mineralization, ossification, and regulation of stem cell fate were upregulated at 4 weeks. Therefore, these results suggested that the molecular processes of peri-implant bone healing can be characterized by determining the sets of upregulated genes with transcriptional analyses.
Interestingly, among the genes that were downregulated at both time points were the genes that were related to the oxidation-reduction reaction (redox reaction). Redox reactions are known to play an important role in cell proliferation and differentiation, which are the key processes in the construction of new tissues [33, 34]. Consistent with the present results, a previous study showed that manipulation of the cellular redox state, which results in reduced reactive oxygen species levels, enhances tissue production and increases bone mineralization during osteogenesis [35]. Despite the differences in redox potentials under various biological conditions, the present findings suggested that the modulation of redox reactions increased bone mineralization during implant bone healing.
In the present study, the implant-bone relationships of genes that were related to four categories were assessed, including cell proliferation, ECM, growth factors, and osteogenesis, and these processes have been widely identified as biological mechanisms that are related to bone formation. Cell proliferation-related genes were identified at both time points in peri-implant bone healing. As previously mentioned, cell proliferation is a key biological process in tissue development and regeneration [36, 37]. The present study identified several genes, such as CDK14, PLCD1, and ZBTB16, which were upregulated during peri-implant bone healing. It is well known that cyclin-dependent kinase 14, which is encoded by CDK14, is a cell cycle regulator, and its upregulation indicates an increased cell proliferation that increased during peri-implant bone healing [38]. PLCD1 (phospholipase C-delta1), which is a key enzyme in phosphoinositide turnover, is involved in a variety of physiological functions [39]. A previous study showed that PLCD1 could be compartmentalized as a function of cell cycle progression [40]. Moreover, zinc finger and BTB domain-containing protein 16 (ZBTB16) is located in the nucleus and is linked to cell cycle progression [41]. Therefore, the upregulation of these potential implant-inducible genes suggested that cell cycle progression was a critical mechanism during peri-implant bone formation.
Many ECM-related genes were upregulated in the implant group up to 4 weeks after implant (late stage), including FN1 (fibronectin), COL1A2 (type I collagen), FBN1 (fibrillin1), LAMB2 (laminin subunit β2), and VTN (vitronectin). These genes are known to support mineral deposition and bone formation [42–45]. Among the upregulated groups of genes were bone resorption-related genes, such as matrix metalloproteinases. The balance between the deposition and resorption of bone is crucial for the formation and maintenance of the peri-implant bone mass [46]. Thus, the present study clearly showed that the regulation of ECM genes was involved in the process of implant bone healing.
The present transcriptome analysis also identified various growth factors that were responsive to the titanium implants. Bone tissue repair is known to involve complex biological events that are regulated by a number of cytokines and growth factors, such as PDGF, FGF, IGF, EGF, and VEGF, which induce the migration of osteoprogenitor cells to damaged sites, their subsequent differentiation towards specific cell lineages, their cell proliferation and revascularization, and the production of ECM [47]. In the present study, these growth factor-related genes were upregulated during peri-implant bone healing. Thus, they could be powerful therapeutic agents in bone formation and regeneration.
Finally, the signaling pathways that were associated with osteogenic development were profiled. The various osteogenic genes that were upregulated during peri-implant bone healing represented the biological and biomechanical establishment of osseointegration. The expression of one of these upregulated genes, PTH1R (parathyroid hormone 1 receptor), was 20-fold higher at the late stage of implant healing than that in the control. Despite the debate surrounding the effect of PTH/PTH1R on osteogenesis, a previous study showed that a PTH1R signaling agonist promotes osteoblastic bone formation without stimulating bone resorption [48]. In the present study, other genes, including SMOC1 (SPARC-related modular calcium-binding protein 1), were upregulated at both time points. The protein encoded by this gene was reported to substantially increase the expression of osteoblast differentiation-related genes in bone marrow-derived mesenchymal stem cells [49]. DLX5 (homeobox protein DLX-5) is another transcription factor that is important for osteoblast differentiation and fracture healing [50]. It has been reported that DLX5 increases the expression of the osteogenic transcription factor Runx2 after BMP2 stimulation, indicating that DLX5 is a BMP-responsive transcriptional activator [51]. The TGF-β/BMP and Wnt pathways are the major pathways in osteoblast differentiation and bone formation. Previous studies have shown that TGF-β/BMP is the most potent regulator of osteogenesis in various cell types and experimental conditions [52–55]. The present study confirmed the overexpression of TGF-β/BMP-related gene transcripts at both the early and late stages of peri-implant bone healing. These results suggested that TGF-β/BMP signaling contributed to the repair of impaired skeletogenesis during development. Wnt signaling is another potential target mechanism in the regulation of osteogenesis. Studies have shown that Wnt pathway activation increases osteoblast/osteocyte survival and bone regeneration [56, 57]. In the present study, the RNA-Seq analysis identified differentially expressed genes that were involved in Wnt signaling (WISP2, FZD1, FZD2, LRP1, and LRP5). During implant-responsive osteogenesis, the Wnt1 gene was upregulated concomitant with the overexpression of Frizzled and LRP, which act as specific receptors for Wnt, suggesting the important role of Wnt machinery in peri-implant bone formation [58, 59].
In conclusion, these findings clearly demonstrated changes in the gene transcripts that were related to redox reactions, cell proliferation, ECM regulation, growth factors, and osteogenic-related TGF-β/BMP and Wnt signaling during implant-bone healing in a transcriptome analysis. This study only compared the levels of gene transcript expression between surgical defect and peri-implant healing processes, and a comparison of gene modulation that occurs in response to implants with different properties is planned in future studies. The RNA-Seq analysis with functional gene classification provided an understanding of the biological processes and signaling pathways that were involved in peri-implant bone formation. In addition, the biological data obtained here may help predict the outcomes of clinical strategies that are aimed at promoting osseointegration and bone regeneration.
One limitation of this study was that more bone than just the actual interfacial contact area between the titanium implant and the bone was included in order to meet the minimum bone volume required for the RNA-Seq analysis. Furthermore, if additional studies are done on SLA-treated implants compared with smooth-surface implant controls, significant information will be obtained on implant bone healing according to surface treatment for orthodontic procedures that use TSADs.
5. Conclusions
Bone healing at 1 and 4 weeks after the placement of orthodontic mini-implants showed characteristic gene expression in RNA-Seq analyses compared with the healing of surgical defects without mini-implants.
Gene profiling analyses showed that gene transcripts that were related to redox reactions, cell proliferation, ECM regulation, growth factors, and the osteogenic-related TGF-β/BMP and Wnt signaling pathways were significantly changed.
Genes encoding ECM constituents were upregulated at the early stage of healing and genes that were associated with bone mineralization, ossification, and the regulation of stem cell fate were upregulated at the late stage of healing.
Acknowledgments
This paper is partially from the thesis of K.-Y. Nahm of Kyung Hee University. The authors would like to thank K. Y. Seo for editing the paper. This work was supported by the National Research Foundation of Korea (NRF), which is funded by the Korean government (MEST) (no. 2012R1A5A2051388).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Hyo-Won Ahn and Seong-Hun Kim contributed equally to this work.
References
- 1.Chung K.-R., Kim S.-H., Kook Y.-A. The C-orthodontic micro-implant. Journal of Clinical Orthodontics. 2004;38(9):478–488. [PubMed] [Google Scholar]
- 2.Lee S.-J., Ahn S.-J., Lee J. W., Kim S.-H., Kim T.-W. Survival analysis of orthodontic mini-implants. American Journal of Orthodontics and Dentofacial Orthopedics. 2010;137(2):194–199. doi: 10.1016/j.ajodo.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Jee J.-H., Ahn H.-W., Seo K.-W., et al. En-masse retraction with a preformed nickel-titanium and stainless steel archwire assembly and temporary skeletal anchorage devices without posterior bonding. The Korean Journal of Orthodontics. 2014;44(5):236–245. doi: 10.4041/kjod.2014.44.5.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharif M. O., Waring D. T. Contemporary orthodontics: the micro-screw. British Dental Journal. 2013;214(8):403–408. doi: 10.1038/sj.bdj.2013.376. [DOI] [PubMed] [Google Scholar]
- 5.Kim J.-H., Park Y.-C. Evaluation of mandibular cortical bone thickness for placement of temporary anchorage devices (TADs) Korean Journal of Orthodontics. 2012;42(3):110–117. doi: 10.4041/kjod.2012.42.3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papageorgiou S. N., Zogakis I. P., Papadopoulos M. A. Failure rates and associated risk factors of orthodontic miniscrew implants: a meta-analysis. American Journal of Orthodontics & Dentofacial Orthopedics. 2012;142(5):577.e7–595.e7. doi: 10.1016/j.ajodo.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Cheng S.-J., Tseng I.-Y., Lee J.-J., Kok S.-H. A prospective study of the risk factors associated with failure of mini-implants used for orthodontic anchorage. The International Journal of Oral and Maxillofacial Implants. 2004;19(1):100–106. [PubMed] [Google Scholar]
- 8.Viwattanatipa N., Thanakitcharu S., Uttraravichien A., Pitiphat W. Survival analyses of surgical miniscrews as orthodontic anchorage. The American Journal of Orthodontics and Dentofacial Orthopedics. 2009;136(1):29–36. doi: 10.1016/j.ajodo.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Kim J. S., Choi S. H., Cha S. K., et al. Comparison of success rates of orthodontic mini-screws by the insertion method. The Korean Journal of Orthodontics. 2012;42(5):242–248. doi: 10.4041/kjod.2012.42.5.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deguchi T., Takano-Yamamoto T., Kanomi R., Hartsfield J. K., Jr., Roberts W. E., Garetto L. P. The use of small titanium screws for orthodontic anchorage. Journal of Dental Research. 2003;82(5):377–381. doi: 10.1177/154405910308200510. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Kim J. Y., Choi Y. J., Kim K.-H., Chung C. J. Effects of placement angle and direction of orthopedic force application on the stability of orthodontic miniscrews. The Angle Orthodontist. 2013;83(4):667–673. doi: 10.2319/090112-703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.-H., Kook Y.-A., Lee W., Kim I., Chung K.-R. Two-component mini-implant as an efficient tool for orthognathic patients. American Journal of Orthodontics and Dentofacial Orthopedics. 2009;135(1):110–117. doi: 10.1016/j.ajodo.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 13.O'Sullivan D., Sennerby L., Meredith N. Measurements comparing the initial stability of five designs of dental implants: a human cadaver study. Clinical Implant Dentistry and Related Research. 2000;2(2):85–92. doi: 10.1111/j.1708-8208.2000.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura H., Shim J., Butz F., Aita H., Gupta V., Ogawa T. Glycosaminoglycan degradation reduces mineralized tissue-titanium interfacial strength. Journal of Biomedical Materials Research—Part A. 2006;77(3):478–486. doi: 10.1002/jbm.a.30624. [DOI] [PubMed] [Google Scholar]
- 15.Heimann R. B., Wirth R. Formation and transformation of amorphous calcium phosphates on titanium alloy surfaces during atmospheric plasma spraying and their subsequent in vitro performance. Biomaterials. 2006;27(6):823–831. doi: 10.1016/j.biomaterials.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 16.Chou B.-Y., Chang E. Microstructural characterization of plasma-sprayed hydroxyapatite-10wt% ZrO2 composite coating on titanium. Biomaterials. 1999;20(19):1823–1832. doi: 10.1016/s0142-9612(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 17.Omar O., Svensson S., Zoric N., et al. In vivo gene expression in response to anodically oxidized versus machined titanium implants. Journal of Biomedical Materials Research A. 2010;92(4):1552–1566. doi: 10.1002/jbm.a.32475. [DOI] [PubMed] [Google Scholar]
- 18.Kojima N., Ozawa S., Miyata Y., Hasegawa H., Tanaka Y., Ogawa T. High-throughput gene expression analysis in bone healing around titanium implants by DNA microarray. Clinical Oral Implants Research. 2008;19(2):173–181. doi: 10.1111/j.1600-0501.2007.01432.x. [DOI] [PubMed] [Google Scholar]
- 19.Ivanovski S., Hamlet S., Salvi G. E., et al. Transcriptional profiling of osseointegration in humans. Clinical Oral Implants Research. 2011;22(4):373–381. doi: 10.1111/j.1600-0501.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa T., Nishimura I. Different bone integration profiles of turned and acid-etched implants associated with modulated expression of extracellular matrix genes. The International Journal of Oral & Maxillofacial Implants. 2003;18(2):200–210. [PubMed] [Google Scholar]
- 21.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mooney M., Bond J., Monks N., et al. Comparative RNA-Seq and microarray analysis of gene expression changes in B-cell lymphomas of Canis familiaris. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0061088.e61088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagalakshmi U., Wang Z., Waern K., et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Zhu K., Liu F., et al. Identification of somatic mutations in human prostate cancer by RNA-Seq. Gene. 2013;519(2):343–347. doi: 10.1016/j.gene.2013.01.046. [DOI] [PubMed] [Google Scholar]
- 25.Wakasa Y., Oono Y., Yazawa T., et al. RNA sequencing-mediated transcriptome analysis of rice plants in endoplasmic reticulum stress conditions. BMC Plant Biology. 2014;14, article 101 doi: 10.1186/1471-2229-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X., Zhang Y., Williams J., et al. Parallel comparison of Illumina RNA-Seq and Affymetrix microarray platforms on transcriptomic profiles generated from 5-aza-deoxy-cytidine treated HT-29 colon cancer cells and simulated datasets. BMC Bioinformatics. 2013;14(supplement 9):p. S1. doi: 10.1186/1471-2105-14-s9-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Santis E., Lang N. P., Cesaretti G., Mainetti T., Beolchini M., Botticelli D. Healing outcomes at implants installed in sites augmented with particulate autologous bone and xenografts. An experimental study in dogs. Clinical Oral Implants Research. 2013;24(1):77–86. doi: 10.1111/j.1600-0501.2012.02456.x. [DOI] [PubMed] [Google Scholar]
- 28.Mengatto C. M., Mussano F., Honda Y., Colwell C. S., Nishimura I. Circadian rhythm and cartilage extracellular matrix genes in osseointegration: a genome-wide screening of implant failure by vitamin D deficiency. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0015848.e15848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopwood B., Tsykin A., Findlay D. M., Fazzalari N. L. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-β/bone morphogenic protein signalling. Arthritis Research & Therapy. 2007;9(5, article R100) doi: 10.1186/ar2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton-Wurster N., Mateescu R. G., Todhunter R. J., et al. Genes in canine articular cartilage that respond to mechanical injury: gene expression studies with Affymetrix canine GeneChip. Journal of Heredity. 2005;96(7):821–828. doi: 10.1093/jhered/esi105. [DOI] [PubMed] [Google Scholar]
- 31.Grizon F., Aguado E., Huré G., Baslé M. F., Chappard D. Enhanced bone integration of implants with increased surface roughness: a long term study in the sheep. Journal of Dentistry. 2002;30(5-6):195–203. doi: 10.1016/s0300-5712(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 32.Jääger K., Islam S., Zajac P., Linnarsson S., Neuman T. RNA-seq analysis reveals different dynamics of differentiation of human dermis- and adipose-derived stromal stem cells. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0038833.e38833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M. H., Kang J. H., Lee S. W. The effect of surface microgrooves and anodic oxidation on the surface characteristics of titanium and the osteogenic activity of human periodontal ligament cells. Archives of Oral Biology. 2013;58(1):59–66. doi: 10.1016/j.archoralbio.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Imhoff B. R., Hansen J. M. Differential redox potential profiles during adipogenesis and osteogenesis. Cellular and Molecular Biology Letters. 2011;16(1):149–161. doi: 10.2478/s11658-010-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid B., Afzal J. M., Mccartney A. M., Abraham M. R., O'Rourke B., Elisseeff J. H. Enhanced tissue production through redox control in stem cell-laden hydrogels. Tissue Engineering Part A. 2013;19(17-18):2014–2023. doi: 10.1089/ten.tea.2012.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moisenovich M. M., Arkhipova A. Y., Orlova A. A., et al. Composite scaffolds containing silk fibroin, gelatin, and hydroxyapatite for bone tissue regeneration and 3D cell culturing. Acta Naturae. 2014;6(20):96–101. [PMC free article] [PubMed] [Google Scholar]
- 37.Penha E. M., Meira C. S., Guimarães E. T., et al. Use of autologous mesenchymal stem cells derived from bone marrow for the treatment of naturally injured spinal cord in dogs. Stem Cells International. 2014;2014:8. doi: 10.1155/2014/437521.437521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidson G., Niehrs C. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends in Cell Biology. 2010;20(8):453–460. doi: 10.1016/j.tcb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura Y., Hamada Y., Fujiwara T., et al. Phospholipase C-delta1 and -delta3 are essential in the trophoblast for placental development. Molecular and Cellular Biology. 2005;25(24):10979–10988. doi: 10.1128/mcb.25.24.10979-10988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stallings J. D., Tall E. G., Pentyala S., Rebecchi M. J. Nuclear translocation of phospholipase C-δ1 is linked to the cell cycle and nuclear phosphatidylinositol 4,5-bisphosphate. The Journal of Biological Chemistry. 2005;280(23):22060–22069. doi: 10.1074/jbc.m413813200. [DOI] [PubMed] [Google Scholar]
- 41.Kikugawa T., Kinugasa Y., Shiraishi K., et al. PLZF regulates Pbx1 transcription and Pbx1—HoxC8 complex leads to androgen-independent prostate cancer proliferation. The Prostate. 2006;66(10):1092–1099. doi: 10.1002/pros.20443. [DOI] [PubMed] [Google Scholar]
- 42.Tang C.-H., Yang R.-S., Huang T.-H., Liu S.-H., Fu W.-M. Enhancement of fibronectin fibrillogenesis and bone formation by basic fibroblast growth factor via protein kinase C-dependent pathway in rat osteoblasts. Molecular Pharmacology. 2004;66(3):440–449. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 43.Nistala H., Lee-Arteaga S., Smaldone S., et al. Fibrillin-1 and -2 differentially modulate endogenous TGF-beta and BMP bioavailability during bone formation. The Journal of Cell Biology. 2010;190(6):1107–1121. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min S.-K., Kang H. K., Jang D. H., et al. Titanium surface coating with a Laminin-derived functional peptide promotes bone cell adhesion. BioMed Research International. 2013;2013:8. doi: 10.1155/2013/638348.638348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seiffert D. Detection of vitronectin in mineralized bone matrix. The Journal of Histochemistry and Cytochemistry. 1996;44(3):275–280. doi: 10.1177/44.3.8648088. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki K., Aoki K., Ohya K. Effects of surface roughness of titanium implants on bone remodeling activity of femur in rabbits. Bone. 1997;21(6):507–514. doi: 10.1016/S8756-3282(97)00204-4. [DOI] [PubMed] [Google Scholar]
- 47.Caplan A. I., Correa D. PDGF in bone formation and regeneration: new insights into a novel mechanism involving MSCs. Journal of Orthopaedic Research. 2011;29(12):1795–1803. doi: 10.1002/jor.21462. [DOI] [PubMed] [Google Scholar]
- 48.Gesty-Palmer D., Flannery P., Yuan L., et al. A beta-arrestin-biased agonist of the parathyroid hormone receptor (PTH1R) promotes bone formation independent of G protein activation. Science Translational Medicine. 2009;1(1, article ra1) doi: 10.1126/scitranslmed.3000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi Y.-A., Lim J., Kim K. M., et al. Secretome analysis of human BMSCs and identification of SMOC1 as an important ECM protein in osteoblast differentiation. Journal of Proteome Research. 2010;9(6):2946–2956. doi: 10.1021/pr901110q. [DOI] [PubMed] [Google Scholar]
- 50.Acampora D., Merlo G. R., Paleari L., et al. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126(17):3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 51.Lee M. H., Kim Y. J., Kim H. J., et al. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. The Journal of Biological Chemistry. 2003;278(36):34387–34394. doi: 10.1074/jbc.m211386200. [DOI] [PubMed] [Google Scholar]
- 52.Luo G., Hofmann C., Bronckers A. L. J. J., Sohocki M., Bradley A., Karsenty G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes and Development. 1995;9(22):2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 53.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 54.Feng X.-Y., Zhao Y.-M., Wang W.-J., Ge L.-H. Msx1 regulates proliferation and differentiation of mouse dental mesenchymal cells in culture. European Journal of Oral Sciences. 2013;121(5):412–420. doi: 10.1111/eos.12078. [DOI] [PubMed] [Google Scholar]
- 55.Oka K., Oka S., Sasaki T., et al. The role of TGF-beta signaling in regulating chondrogenesis and osteogenesis during mandibular development. Developmental Biology. 2007;303(1):391–404. doi: 10.1016/j.ydbio.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hens J. R., Wilson K. M., Dann P., Chen X., Horowitz M. C., Wysolmerski J. J. TOPGAL mice show that the canonical Wnt signaling pathway is active during bone development and growth and is activated by mechanical loading in vitro. Journal of Bone and Mineral Research. 2005;20(7):1103–1113. doi: 10.1359/JBMR.050210. [DOI] [PubMed] [Google Scholar]
- 57.Silkstone D., Hong H., Alman B. A. β-catenin in the race to fracture repair: in it to Wnt. Nature Clinical Practice Rheumatology. 2008;4(8):413–419. doi: 10.1038/ncprheum0838. [DOI] [PubMed] [Google Scholar]
- 58.MacDonald B. T., He X. Frizzled and LRp5/6 receptors for wnt/β-catenin signaling. Cold Spring Harbor Perspectives in Biology. 2012;4(12) doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niehrs C. The complex world of WNT receptor signalling. Nature Reviews Molecular Cell Biology. 2012;13(12):767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]