INTRODUCTION
Alcohol dependence is a complex trait that underlies a range of physiological and behavioral symptoms manifested as tolerance, loss of control, withdrawal, and desire or inability to cut down. Considerable evidence from twin genetic studies1-11 indicates that the trait is heritable with estimates ranging from 40-60% of the risk of alcohol dependence due to genetic factors. A measure of alcohol dependence is alcohol consumption.12-18 Alcohol drinking, as alcohol dependence, has a complex, non-Mendelian pattern of inheritance, indicating involvement of multiple genetic variants.19
Because alcohol use can influence numerous health outcomes, population studies with broad aims typically collect alcohol consumption data, and this data has the potential be investigated in genetic association studies. Grant et al. (2009)20 and Kendler et al. (2010)21 found that it is feasible to closely index the genetic risk for alcohol dependence by collecting relatively simple quantitative data on the alcohol consumption. Fitting a model that included 5 measures of alcohol consumption (lifetime events of heaviest alcohol use including maximum drinks consumed in 24 hours, lifetime maximal tolerance, typical number of drinks per occasion (lifetime), frequency consumed alcohol (heaviest period) and frequency of drinking to intoxication (heaviest period)), Grant and colleagues found a high genetic correlation with Alcohol Dependence symptom scores (>+0.97).20 Evaluating the extent to which each individual measure reflected the genetic risk factor by gender, Kendler et al (2010)21 found that in men maximum drinks consumed in a 24-hour period had the highest loading, followed closely by frequency of drinking to intoxication, while in women frequency of drinking to intoxication loaded most strongly.
These studies show that in theory a lifetime event of heavy alcohol use indexes the genetic risk for alcohol dependence, however no study thus far reflects this concept in practice. Therefore, we used an alcohol consumption phenotype of lifetime history of intake of 5 or more drinks per day almost every day of the week that was collected in cardiovascular cohort studies from the Candidate gene Association Resource (CARe) project. We conducted a genetic association analysis with variants on a genotyping platform that densely covers ~2100 genes. This approach has the advantage of utilizing dense genotyping coverage of a large number of genes without pre-specifying biological hypotheses about the effect of individual genes and genetic variants.
METHODS
We analyzed alcohol consumption from the National Heart, Lung, and Blood Institute (NHLBI)-sponsored CARe project.22 The CARe Project was launched in 2007 to create a resource for association studies of various phenotypes. The CARe project consists of 9 NHLBI cohorts. It is approved by the ethics committees of the participating studies and of the Massachusetts Institute of Technology.
Subjects
Our phenotype of interest was available in three Caucasian cohorts from the CARe project: Atherosclerosis Risk in Communities (ARIC, 1989), Framingham Heart Study (FHS)23-25 and Cardiovascular Health Study (CHS).26 Our sample of subjects in ARIC included 2,138 (632 cases and 1,506 controls) unrelated individuals with a mean 59.8 (SD 5.6) years of age of which 40% were female. The CHS cohort also included unrelated individuals (N=859; 358 cases and 501 controls) with a mean age of 72.2 (SD=+/− 5.2) of which 36.3% were female. Subjects in FHS (Offspring cohort) included 772 related individuals (265 cases and 507 controls) of which 49% were female with the mean 65.1 (+/− 8.9 SD) years of age for the total sample of analyzed individuals.
Phenotype
This analysis was a case-control comparison between light and heavy drinkers. We defined cases as individuals with a lifetime history of drinking five or more drinks per day almost every day of the week. We defined controls as current light drinkers of 1-5 drinks per week to ensure comparison with other drinkers. Among light drinkers, we excluded individuals who may be binge drinkers (four or more drinks per occasion for women and five or more drinks per occasion for men).27
Genotyping Assay
The content of the genotyping array, ITMAT-Broad-CARe or “IBC chip”, is informed by GWAS, expression quantitative trait loci, pathway-based approaches and comprehensive literature searching. It includes loci relevant to alcoholism, such as GABA and alcohol metabolism genes. As an example, it contains densely spaced SNPs from 84 of the 130 genes from the “addiction array”28 and additional genes that are not on the addiction array, but were found to be associated with alcoholism in later genetic association studies.
The loci on the IBC chip are divided into three groups: Group 1: (n = 435 loci) - genes and regions with a high likelihood of functional significance (Tag SNPs selected to capture known variation with minor allele frequency (MAF) > 0.02 and an r2 of at least 0.8 in HapMap populations); Group 2: (n=1,349 loci) - candidate loci that are potentially involved in phenotypes of interest or established loci that required very large numbers of tagging SNPs (Tag SNPs selected to capture known variation with MAF > 0.05 with an r2 of at least 0.5 in HapMap populations); Group 3: (n=232 loci) - composed mainly of the larger genes (100 kb) which were of lower interest a priori to the investigators (includes only non-synonomous SNPs and known functional variants). The average number of SNPs across the Group 1 and Group 2 loci of IBC was compared with GWAS products. The average coverage for Group 1 loci is ~36.5 SNPs per locus on the IBC chip. The Illumina Human1M and Affymetrix 6.0 platform, for comparison, have an average of ~28.0 and ~17.4 SNPs respectively across the equivalent IBC loci. The average number of SNPs observed for the Group 2 loci is ~16.3 SNPs, which is comparable with the current GWAS products.
Additional details regarding the design of the IBC chip have been described in Keating et al (2008).29 In toto, 49,320 SNPs were chosen to map ~2,100 candidate gene loci. For detailed genotyping and QC information, see Musunuru et al (2010).22
Statistical Analysis
For cohorts including unrelated individuals (ARIC, CHS), we used logistic regression to test SNP-phenotype associations. Association analysis was performed in PLINK30 under an additive genetic model. Association results were combined across the two cohorts using an inverse variance meta-analysis approach as implemented in METAL.31 For the FHS cohort for which there were significant numbers of related individuals, we used GWAF (Genome-Wide Association analyses with Family),32 which implements generalized estimating equations in the gee package (http://cran.r-project.org/web/packages/gee/) to test association between the light vs heavy phenotype and each SNP under the additive genetic model. To address population stratification, we conducted principal component analysis as implemented in EIGENSTRAT.33 The first ten principal components were included as covariates in the genetic association analysis. In addition, age and gender were included as covariates in the association analysis. All results were adjusted for residual inflation using the genomic control method. Bonferroni adjustment for multiple comparisons was set at an alpha level of 2.3×10-06. Replication p-significance value was set at p< 0.05.
Imputation of ungenotyped variants was done using a combined CEU+YRI reference panel including SNPs segregating in both CEU and YRI, as well as SNPs segregating in one panel and monomorphic and nonmissing in the other, resulting in ~270,000 total SNPs. The use of the CEU+YRI panel resulted in an allelic concordance rate of ~95.6%, calculated as 1 – 1/2* |imputed_dosage – chip_dosage|. This rate is comparable to rates calculated for individuals of African descent imputed with the HapMap 2 YRI individuals.34 In the first step of imputation, individuals with pedigree relatedness or cryptic relatedness (pi_hat > 0.05) were filtered out. Recombination and error rate estimates for the entire sample were calculated based on a subset of random individuals. Next, these rates were used to impute all sample individuals across the entire reference panel. SNPs with low imputation scores (r2<0.3) and minor allele frequency of <0.01 were filtered out.
RESULTS
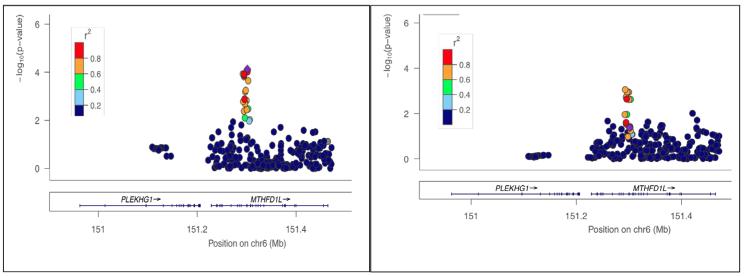
The locus that was the most strongly associated with the lifetime heavy drinking phenotype in ARIC and CHS is presented in Figure 1. Each additional copy of the major rs6933598*C allele (Frequency: HapMap CEU = 0.715) was associated with a decrease in risk of heavy drinking (Results of Combined Meta-analysis: OR=0.75 (CI=0.66-0.86) p=7.46×10−05. Results for Individual Studies: ARIC OR=0.76 (CI=0.64-0.91) p=0.00155. CHS OR=0.75 (CI=0.59-0.95) p=0.017). This locus replicated in FHS OR=0.75 (CI=0.57-0.98) p=0.042.
Figure 1.
Association between the MTHFD1L locus and lifetime incidence of heavy alcohol drinking in the CARe project cohorts. Left panel: Our discovery sample included unrelated individuals from ARIC and CHS. Right panel: Our top SNP rs6933598 (purple diamond) from meta-analysis of ARIC and CHS was also associated with the phenotpe in the Framingham Heart Study (FHS; P = 0.042).
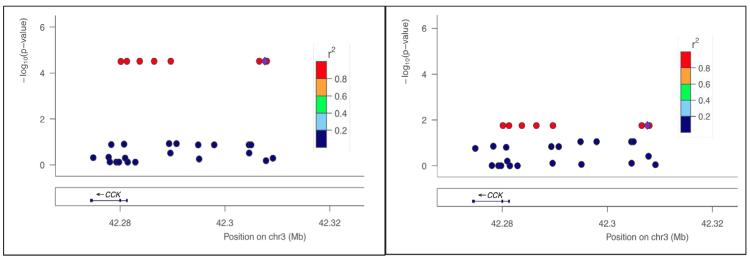
In FHS, the strongest associations of similar significance were located on separate chromosomes. The first SNP, rs12249562, is located on Chromosome 10 in CUBN, where each additional copy of minor rs12249562*A allele (HapMap CEU = 0.195) resulted in a decrease of risk for lifetime heavy drinking OR=0.52 (CI=0.38-0.71) p=3.03×10−05. This association was closely followed by an association in on chromosome 3 near CCK where each additional copy of the minor rs9839267*G allele (HapMap CEU = 0.085) resulted in an increase of risk OR=2.39 (CI=1.58-3.60) p=3.05×10−05. This SNP replicated in CHS at a p value <0.05 OR=1.57 (CI=1.08-2.27) p=0.019 (See Figure 2 for a graphic representation of this locus on our discovery cohort (FHS) and the replication cohort (CHS)). The p value for rs9839267 in ARIC was not <0.05.
Figure 2.
Association between the CCK locus and lifetime incidence of heavy alcohol consumption in the CARe project cohorts. Left panel: Our discovery sample included related individuals from FHS. Right panel: Our top SNP rs9839267 (purple diamond) in FHS was also associated with the phenotype in CHS (P = 0.019).
DISCUSSION
We examined a lifetime heavy alcohol drinking incidence phenotype for association with genetic variants from a large number of candidate genes in three cohorts from the CARe project. We were able to identify variants in CCK and MTHFD1L that modulate risk for heavy alcohol drinking. These results demonstrate the feasibility in evaluating lifetime incidence of heavy alcohol drinking from population-based studies for the purpose of conducting genetic association analyses.
Cholecystokinin was originally found in the gut where it is involved in the secretion of pancreatic enzymes, gall bladder and gut motility. However, it is distributed even more widely in the brain where it is one of the most abundant neuropeptides.35,36 Co-localization of CCK on cell bodies and terminals of classical neurotrasmitters implicated in alcohol abuse potential including gammaaminobutyric acid,37,38 serotinin39 and opiates40 makes the neuropeptide a biologically plausible candidate. However, the role of CCK in regulation of dopamine turnover in the mesoaccumbal projection – a region highly implicated in the primary effects of drugs of abuse and in the process of sensitization - has received the most attention in the relationship of CCK and addiction (see Rotzinger et al., 200341). Indeed, dopaminergic transmission was thought to be implicated in the behavioral finding that antagonism of CCK significantly reduced the intake of ethanol in naïve adult male Wistar rats.42
Very early candidate gene association studies provided initial results regarding the role of CCK in alcoholism. Two previous studies showed that the promoter SNP rs1799923 modulates clinically-diagnosed alcohol dependence in the Japanese population,43,44 but an attempt to show the same in the Caucasian population was not successful.45 This particular SNP was genotyped in CARe and was in fact associated with our phenotype of interest (P= 3.08×10-5 (FHS); p=0.0196 (CHS)) (See Figure 1 – showing 8 SNPs (in red) in high LD, 7 of which were imputed based on the genotyped SNP rs1799923). More recent association studies, evaluating extensive regions of the genome, did not identify gene variant(s) to be implicated in risk for alcoholism.
Our result in the CCK locus allows the following conclusions to be made. First, it demonstrates the significance of CCK beyond the previously-examined single locus analysis when the genome is evaluated more extensively. In addition, it opposes the earlier negative finding in Caucasians and shows the importance of the locus in a population ethnically different from the previously-identified Japanese group.
Folate metabolism is complex and the mechanism(s) by which alcohol inhibits it have not been definitively established. Homocysteine is converted to methionine via tetrahydrofolate (THF). MTHFD1L is involved in tetrahydrofolate (THF) synthesis by catalyzing the reversible synthesis of 10-formyl-THF to formate and THF. Elevated homocysteine levels were first reported by Hultberg et al (1993)46 in patients hospitalized for detoxification after severe alcohol abuse. This finding has since been independently replicated.47,48 Plasma homocysteine levels predict alcohol withdrawal.47 The association between alcohol intake and raised plasma homocysteine levels, in fact, has also been observed in moderate alcohol consumers, with plasma homocysteine levels increasing over a 6-week drinking period.49
The MTHFDL1 gene resides on 6q25.1 and spans ~235 kBs. As shown in Figure 1, the associated locus is in very low LD with the remaining variants in the region, permitting localization of the region of functional significance. There are no published reports of rs6933598 in genetic association analyses. However, a proxy of rs6933598, rs6922269 (r2=0.924 between the two variants in CEU) was associated with coronary heart disease in The Wellcome Trust Case Control Consortium50 and German MI Family Study.51 The same narrow region of a cluster of SNPs in high LD was also recently associated with late-onset Alzheimer’s disease.52
Because alcohol consumption is implicated in a variety of chronic conditions, population studies most commonly collect data to derive the number of grams of alcohol a person consumes per week (or per day). However, this phenotype, usually collected as part of a dietary survey, is a cross-sectional snapshot of alcohol consumption and the data tend to have an extreme violation of normality in distribution. The same problems are encountered with the “Max Drinks” phenotype sometimes collected in population studies (the maximum number of drinks in a 24- hour period in the last 30 days). Our results show that data that summarize lifetime heaviest use of alcohol in cases may be used successfully in future population studies attempting to identify variants implicated in alcoholism risk.
This is a retrospective analysis based on self report which we could not objectively verify. As comorbidities are not generally not included in published genetic association studies of alcoholism, they were not incorporated here either. We are guided to believe that our approach is valid because we have shown that the same locus in CCK, previously associated with clinically-diagnosed alcohol dependence in a single locus analysis of the Japanese population, also mediates heavy alcohol consumption in the ethnically- different Caucasian population. Here, though, we have demonstrated the importance of this gene when the genome is evaluated more extensively, thereby aligning results of previous pharmacological studies that stressed the importance of CCK in alcohol consumption.
Our primary goal in this analysis was to show that in genetic association studies simple measurements of lifetime heaviest use of alcohol may serve in place of more laborious assessment of alcohol dependence, especially in ongoing cohort studies in which diagnostic batteries are, at best, impractical. Provided that cases and controls are defined carefully, this approach is convenient because it allows for collection of sample sizes that may be difficult to obtain with clinical assessment of alcohol dependence. Future studies should evaluate the how the variants discussed here modify expression/structure in order to provide information regarding their influence on the final phenotype.
Acknowledgments
All sources of support: MD Scientist Fellowship in Genetic Medicine (Northerstern Memorial Foundation; PI: A. Hamidovic), National Research Service Award F32DA024920 (NIH/NIDA; PI: A. Hamidovic), Dr. Bonnie Spring’s Professional Account at Northwestern Feinberg School of Medicine, KL2 RR024130-02 (support for E. Jorgenson). The Candidate gene Association Resource (CARe) wishes to acknowledge the support of the National Heart, Lung and Blood Institute and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research (NHLBI contract number HHSN268200960009C). The following eight parent studies have contributed parent study data, ancillary study data, and DNA samples through the Broad Institute (N01-HC-65226) to create this genotype/phenotype database for wide dissemination to the biomedical research community: the Atherosclerosis Risk in Communities (ARIC) study, the Cardiovascular Health Study (CHS), the Cleveland Family Study (CFS), the Coronary Artery Risk Development in Young Adults (CARDIA) study, the Framingham Heart Study (FHS), the Jackson Heart Study (JHS), the Multi-Ethnic Study of Atherosclerosis (MESA), and the Sleep Heart Health Study (SHHS). Individual study funding attributions can be obtained at http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx
References
- 1.Heath AC, Bucholz KK, Madden PA, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27(6):1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 2.Cadoret RJ, O’Gorman TW, Troughton E, et al. Alcoholism and antisocial personality. Interrelationships, genetic and environmental factors. Arch Gen Psychiatry. 1985;42(2):161–167. doi: 10.1001/archpsyc.1985.01790250055007. [DOI] [PubMed] [Google Scholar]
- 3.Cadoret RJ, Troughton E, O’Gorman TW. Genetic and environmental factors in alcohol abuse and antisocial personality. J Stud Alcohol. 1987;48(1):1–8. doi: 10.15288/jsa.1987.48.1. [DOI] [PubMed] [Google Scholar]
- 4.Cloninger CR, Bohman M, Sigvardsson S. Inheritance of alcohol abuse. Cross-fostering analysis of adopted men. Arch Gen Psychiatry. 1981;38(8):861–868. doi: 10.1001/archpsyc.1981.01780330019001. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin DW, Schulsinger F, Hermansen L, et al. Alcohol problems in adoptees raised apart from alcoholic biological parents. Arch Gen Psychiatry. 1973;28(2):238–243. doi: 10.1001/archpsyc.1973.01750320068011. [DOI] [PubMed] [Google Scholar]
- 6.Hrubec Z, Omenn GS. Evidence of genetic predisposition to alcoholic cirrhosis and psychosis: twin concordances for alcoholism and its biological end points by zygosity among male veterans. Alcoholism: Clinical & Experimental Research. 1981;5(2):207–215. doi: 10.1111/j.1530-0277.1981.tb04890.x. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Heath AC, Neale MC, et al. A population-based twin study of alcoholism in women. JAMA. 1992;268(14):1877–1882. [PubMed] [Google Scholar]
- 8.Kendler KS, Prescott CA, Neale MC, et al. Temperance board registration for alcohol abuse in a national sample of Swedish male twins, born 1902 to 1949. Arch Gen Psychiatry. 1997;54(2):178–184. doi: 10.1001/archpsyc.1997.01830140090015. [DOI] [PubMed] [Google Scholar]
- 9.McGue M, Pickens RW, Svikis DS. Sex and age effects on the inheritance of alcohol problems: a twin study. J Abnorm Psychol. 1992;101(1):3–17. doi: 10.1037//0021-843x.101.1.3. [DOI] [PubMed] [Google Scholar]
- 10.Pickens RW, Svikis DS, McGue M, et al. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Arch Gen Psychiatry. 1991;48(1):19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- 11.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156(1):34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 12.Agrawal A, Grant JD, Littlefield A, et al. Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs. 2009;70(2):157–168. doi: 10.15288/jsad.2009.70.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansell NK, Agrawal A, Whitfield JB, et al. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Research & Human Genetics: the Official Journal of the International Society for Twin Studies. 2008;11(3):287–305. doi: 10.1375/twin.11.3.287. [DOI] [PubMed] [Google Scholar]
- 14.Heath AC, Meyer J, Jardine R, et al. The inheritance of alcohol consumption patterns in a general population twin sample: II. Determinants of consumption frequency and quantity consumed. J Stud Alcohol. 1991;52(5):425–433. doi: 10.15288/jsa.1991.52.425. [DOI] [PubMed] [Google Scholar]
- 15.Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57(1):69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 16.Kaprio J, Rose RJ, Romanov K, et al. Genetic and environmental determinants of use and abuse of alcohol: the Finnish Twin Cohort studies. Alcohol Alcohol Suppl. 1991;1:131–136. [PubMed] [Google Scholar]
- 17.Prescott CA, Hewitt JK, Truett KR, et al. Genetic and environmental influences on lifetime alcohol-related problems in a volunteer sample of older twins. J Stud Alcohol. 1994;55(2):184–202. doi: 10.15288/jsa.1994.55.184. [DOI] [PubMed] [Google Scholar]
- 18.Whitfield JB, Zhu G, Madden PA, et al. The genetics of alcohol intake and of alcohol dependence. Alcoholism: Clinical & Experimental Research. 2004;28(8):1153–1160. doi: 10.1097/01.alc.0000134221.32773.69. [DOI] [PubMed] [Google Scholar]
- 19.Goldman D, Oroszi G, O’Malley S, et al. COMBINE genetics study: the pharmacogenetics of alcoholism treatment response: genes and mechanisms. Journal of Studies on Alcohol - Supplement. 2005;15:56–64. doi: 10.15288/jsas.2005.s15.56. discussion 33. [DOI] [PubMed] [Google Scholar]
- 20.Grant JD, Agrawal A, Bucholz KK, et al. Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry. 2009;66(8):795–800. doi: 10.1016/j.biopsych.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendler KS, Myers J, Dick D, et al. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcoholism: Clinical & Experimental Research. 2010;34(6):1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musunuru K, Lettre G, Young T, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3(3):267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawber TR, Meadors GF, Moore FE., Jr. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41(3):279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinleib M, Kannel WB, Garrison RJ, et al. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4(4):518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 25.Splansky GL, Corey D, Yang Q, et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165(11):1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 26.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 27.Centers for Disease Control and Prevention [Accessed April 26, 2011];Alcohol and Public Health Fact Sheets: Binge Drinking. 2010 http://www.cdc.gov/alcohol/fact-sheets/binge-drinking.htm.
- 28.Hodgkinson CA, Yuan Q, Xu K, et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43(5):505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3(10):e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen M-H, Larson MG, Hsu Y-H, et al. A three-stage approach for genome-wide association studies with family data for quantitative traits. BMC Genetics. 2010;11:40. doi: 10.1186/1471-2156-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Huang L, Li Y, Singleton AB, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84(2):235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawley JN. Comparative distribution of cholecystokinin and other neuropeptides. Why is this peptide different from all other peptides? Ann N Y Acad Sci. 1985;448:1–8. doi: 10.1111/j.1749-6632.1985.tb29900.x. [DOI] [PubMed] [Google Scholar]
- 36.Moran TH, Schwartz GJ. Neurobiology of cholecystokinin. Crit Rev Neurobiol. 1994;9(1):1–28. [PubMed] [Google Scholar]
- 37.Alho H, Ferrarese C, Vicini S, et al. Subsets of GABA-ergic neurons in dissociated cell cultures of neonatal rat cerebral cortex show co-localization with specific modulator peptides. Brain Res. 1988;467:193–204. doi: 10.1016/0165-3806(88)90023-5. [DOI] [PubMed] [Google Scholar]
- 38.Hendry SH, Jones EG, DeFelipe J, et al. Neuropeptide-containing neurons of the cerebral cortex are also GABAergic. Proc Natl Acad Sci U S A. 1984;81(20):6526–6530. doi: 10.1073/pnas.81.20.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Kooy D, Hunt SP, Steinbusch HW, et al. Separate populations of cholecystokinin and 5-hydroxytryptamine-containing neuronal cells in the rat dorsal raphe, and their contribution to the ascending raphe projections. Neurosci Lett. 1981;26(1):25–30. doi: 10.1016/0304-3940(81)90420-1. [DOI] [PubMed] [Google Scholar]
- 40.Gall C, Lauterborn J, Burks D, et al. Co-localization of enkephalin and cholecystokinin in discrete areas of rat brain. Brain Res. 1987;403(2):403–408. doi: 10.1016/0006-8993(87)90085-0. [DOI] [PubMed] [Google Scholar]
- 41.Rotzinger S, Vaccarino FJ. Cholecystokinin receptor subtypes: role in the modulation of anxiety-related and reward-related behaviours in animal models. J Psychiatry Neurosci. 2003;28(3):171–181. [PMC free article] [PubMed] [Google Scholar]
- 42.Crespi F. The role of cholecystokinin (CCK), CCK-A or CCK-B receptor antagonists in the spontaneous preference for drugs of abuse (alcohol or cocaine) in naive rats. Methods Find Exp Clin Pharmacol. 1998;20(8):679–697. doi: 10.1358/mf.1998.20.8.487502. [DOI] [PubMed] [Google Scholar]
- 43.Harada S, Okubo T, Tsutsumi M, et al. A new genetic variant in the Sp1 binding cis-element of cholecystokinin gene promoter region and relationship to alcoholism. Alcoholism: Clinical & Experimental Research. 1998;22(3 Suppl):93S–96S. doi: 10.1111/acer.1998.22.s3_part1.93s. [DOI] [PubMed] [Google Scholar]
- 44.Ishiguro H, Saito T, Akazawa S, et al. Association between drinking-related antisocial behavior and a polymorphism in the serotonin transporter gene in a Japanese population. Alcoholism: Clinical & Experimental Research. 1999;23(7):1281–1284. [PubMed] [Google Scholar]
- 45.Ishiguro H, Saito T, Shibuya H, et al. No association between C-45T polymorphism in the Sp1 binding site of the promoter region of the cholecystokinin gene and alcoholism. Psychiatry Res. 1999;85(2):209–213. doi: 10.1016/s0165-1781(98)00127-9. [DOI] [PubMed] [Google Scholar]
- 46.Hultberg B, Berglund M, Andersson A, et al. Elevated plasma homocysteine in alcoholics. Alcoholism: Clinical & Experimental Research. 1993;17(3):687–689. doi: 10.1111/j.1530-0277.1993.tb00820.x. [DOI] [PubMed] [Google Scholar]
- 47.Bleich S, Degner D, Bandelow B, et al. Plasma homocysteine is a predictor of alcohol withdrawal seizures. Neuroreport. 2000;11(12):2749–2752. doi: 10.1097/00001756-200008210-00028. [DOI] [PubMed] [Google Scholar]
- 48.Cravo ML, Gloria LM, Selhub J, et al. Hyperhomocysteinemia in chronic alcoholism: correlation with folate, vitamin B-12, and vitamin B-6 status. Am J Clin Nutr. 1996;63(2):220–224. doi: 10.1093/ajcn/63.2.220. [DOI] [PubMed] [Google Scholar]
- 49.Bleich S, Bleich K, Kropp S, et al. Moderate alcohol consumption in social drinkers raises plasma homocysteine levels: a contradiction to the ‘French Paradox’? Alcohol Alcohol. 2001;36(3):189–192. doi: 10.1093/alcalc/36.3.189. [DOI] [PubMed] [Google Scholar]
- 50.Wellcome Trust Case Control C Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naj AC, Beecham GW, Martin ER, et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6(9) doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]