Abstract
Oligonucleotide-based receptors or aptamers can interact with small molecules, but the ability to achieve high-affinity and selectivity of these interactions depends strongly on functional groups or epitopes displayed by the binding targets. Some classes of targets are particularly challenging: for example, monosaccharides have scarce functionalities and no aptamers have been reported to recognize, let alone distinguish from each other, glucose and other hexoses. Here we report aptamers that differentiate low-epitope targets such as glucose, fructose, or galactose by forming ternary complexes with high-epitope organic receptors for monosaccharides. In a follow-up example, we expand this method to isolate high-affinity oligonucleotides against aromatic amino acids complexed in situ with a non-specific organometallic receptor. The method is general and enables broad clinical use of aptamers for detection of small molecules in mix-and-measure assays, as demonstrated by monitoring postprandial waves of phenylalanine in human subjects.
Aptamers are often pursued as recognition elements for biosensors1,2. While the development of new protocols3,4 has enabled isolation of high-affinity protein-binding aptamers in general, similar advances in recognition of small molecules have not been forthcoming. This is because many small molecules of analytical interest lack epitopes that would elicit strong interactions with nucleic acids5 or other biomolecular receptors, such as antibodies. The introduction of modified oligonucleotides6 and organic receptors as cofactors7 have been suggested as helpful in isolating aptamers for challenging small molecules, but with no practical impact as of yet. While the approaches using modified oligonucleotides might indeed improve affinity for many targets, these may still fail if targets inherently lack epitopes. Furthermore, such modifications increase the production costs of sensors. The problem with previous attempts7 to incorporate organic receptors for small molecules within aptamers was that the binding of receptors to oligonucleotides competed with binding to targets, leading to drastic drops in affinity. We now propose to avoid all these issues with low-epitope displaying targets by pursuing unmodified oligonucleotides that will be selected to interact with the in-situ formed complex that low-epitope targets form with organic or organometallic receptors. This would lead to formation of ternary complexes of oligonucleotides, organic receptors, and targets, with oligonucleotides avoiding productive interactions with organic receptors on their own. Therefore, if successfully isolated, sensors formed from these oligonucleotides will respond with high sensitivity and selectivity to the presence of small molecules in the presence of an excess of organic/organometallic receptors, acting as in situ derivatization agents. A successful approach like this could change clinical chemistry, enabling rapid and sensitive mix-and-measure assays for almost any class of small molecule guest that can interact with organic/organometallic hosts.
Results
Differentiation of monosaccharaides
Glucose might be the most prominent example of a small molecule that has up to now resisted attempts to isolate aptameric reagents that bind to it, despite a universally recognized need for improved sensors in the context of the real-time glucose monitoring in patients with diabetes9. Our approach to sensing glucose or other monosaccharides with aptamers is based on the idea that we can improve the epitope presentation in glucose during selection of oligonucleotides by targeting the in situ formed complexes with an organic receptor, as an example of which we chose Shinkai’s receptor 8,10 (1, Fig. 1a). Shinkai’s receptor is an example of a successful bis-boronic-acid-based fluorescent glucose sensor designed to overcome the natural preference of mono-boronic acids for fructose over glucose. Based on its mechanism of action, we expected Shinkai’s receptor to drastically change its conformation depending on the monosaccharide target, presenting spatially a different arrangement of epitopes to aptamers, thus enabling their selectivity (Supplementary Fig. 1).
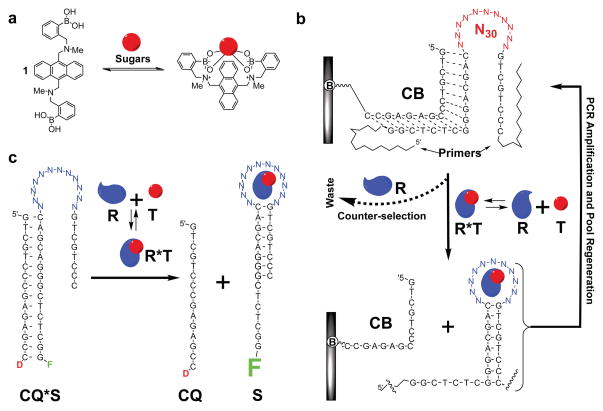
Figure 1. Basic principles of glucose recognition with boronic acid receptors and of isolation of aptamers against complexes with receptors.
a, A schematic representation of complexation of Shinkai’s receptor 1 with glucose (presented as a sphere). b, A schematic representation of selections used in this work: A library (e.g., a randomized region N30 flanked with primers) is attached to an agarose-streptavidin column via a biotinylated complementary oligonucleotide (CB). Upon binding to a complex between a receptor (R, e.g., Shinkai’s sensor) and a target molecule (T, e.g., glucose or other monosaccharides), aptameric structures are preferentially released from the column due to stabilization of the stem formation, PCR amplified, and, therefore, evolutionary favored to survive from multiple selection cycles; counter-selection against receptor itself is introduced to minimize competition between aptamer and target for the receptor. c, An example of a structure-switching aptameric sensor that comes from the selection procedure: The aptameric structure that has been selected in C is labeled with fluorescein (F), while the capture strand is labeled with a quencher (D in CQ stands for quencher dabcyl, or 4-([4-(dimethylamino)phenyl]-azo)-benzoic acid) to yield a sensor; upon binding to a complex, CQ is displaced, leading to an increase in fluorescence signal.
Our selection of oligonucleotides binding to monosaccharide-receptor complexes started with a recently reported11 variant of early solution-phase selection and amplification protocols (Fig. 1b)12,13. This protocol was chosen for two reasons: First, Shinkai’s receptor, or any other organic host molecules, can be used without modifying it in order to attach it to a solid state matrix, as it was done previously in reports using organic receptors as cofactors for aptamers7. And, second, at the end of the procedure, isolated oligonucleotides which bind complexes (aptamers) are already in their sensor forms (Fig. 1c), thereby simplifying the post-selection analysis. We added 1 to the elution buffer in the presence of an excess of a targeted monosaccharide (e.g., glucose 2, fructose 3, or galactose 4 in Fig. 2; excess sugar makes the receptor fully soluble), ensuring that the target for the aptamers is a complex of monosaccharide with 1. We also introduced counter-selections against: (a) Shinkai’s receptor itself, to the extent it is soluble on its own (Supplementary Fig. 5) in the presence of oligonucleotides, thereby focusing selection on a receptor serving as an in situ derivatization agent; (b) mixture of steroids in order to eliminate hydrophobic three-way junctions that would otherwise overwhelm the selection while providing no useful selectivity for a complex; and, (c) in the case of galactose and fructose, two non-targeted sugars added in an excess to the Shinkai’s receptor (for glucose, the third type of counter-selection was not performed); this counter-selection reduces the affinity of the organic receptor for non-targets in ternary complexes or, in other words, eliminates the cross-reactivity of 1.
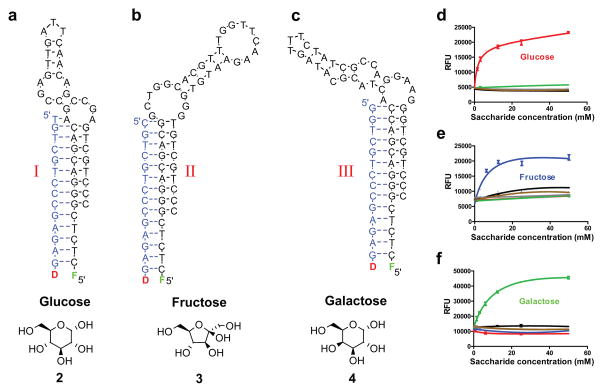
Figure 2. Results of three selection procedures against monosaccharides.
a–c, I, II, and III are structure-switching aptameric sensors responding to glucose (2), fructose (3), and galactose (4) (all monosaccharides are shown as a single, dominant, form, which does not necessarily represent the form that is bound by Shinkai’s receptor when recognized by aptamers). d–f, The response of sensors I–III (relative fluorescence units or RFUs – as read out from the plate reader – vs. concentration of monosaccharides in mM; initial concentration of Shinkai’s receptor, 50 μM with 2% methanol used). The tested monosaccharides are glucose (
 ), fructose (
), fructose (
 ), galactose (
), galactose (
 ), mannose (●), allose (
), mannose (●), allose (
 ), and altrose (
), and altrose (
 ). Each aptameric sensor is primarily responsive to a single monosaccharide, the one used in selection. All measurements were performed in triplicates with standard deviations (SDs) shown.
). Each aptameric sensor is primarily responsive to a single monosaccharide, the one used in selection. All measurements were performed in triplicates with standard deviations (SDs) shown.
As a result of these three selections, we isolated oligonucleotides in the form of aptameric structure-switching11,12 sensors that respond with high specificity to each of the three targeted monosaccharides (e.g., I–III). The evolutionary pressure in these selections is based on the selective ability of receptor*target complexes to enhance closures of the stems displacing complementary oligonucleotide (C) from aptamers; thus, the mechanism of reporting glucose concentrations as shown in Fig. 1c/2a directly results from the selection11. The combination of positive and multiple negative (counter-) selections was able to even completely overcome the initial preferences of 1 (for glucose) or mono-boronic acids (for fructose) (Fig. 2). For example, while the original Shinkai’s sensor is more responsive to both glucose and fructose over galactose (Supplementary Fig. 3), the aptameric sensor III against galactose complex specifically responds to the addition of galactose. Mid-points of sensitivity of sensors I–III to monosaccharides were similar to dissociation constants of Shinkai’s receptor, corrected for the competition with complementary oligonucleotide C, indicating that the aptamer and target did not compete for the receptor. Additionally, sensor I was in this example adjusted, by choosing the length of C, to respond to a concentration below 3 mM glucose, which is important for hypoglycemia and a region in which traditional glucometers do not perform well9. Of note, the aptamer on which sensor I is based will actually interact with a glucose complex in the range of 1–10 μM (this is a competitive assay; solubility issues and multiple binding sites of the receptor to oligonucleotides interfere with more precise characterization, cf., Supplementary Fig. 3 and Fig. 4). One more benefit of using aptameric sensors is that they shift fluorescence response of Shinkai’s receptor to whatever fluorophore we choose for them (i.e., fluorescein can be substituted with other fluorophores), thereby allowing multicolor detection.
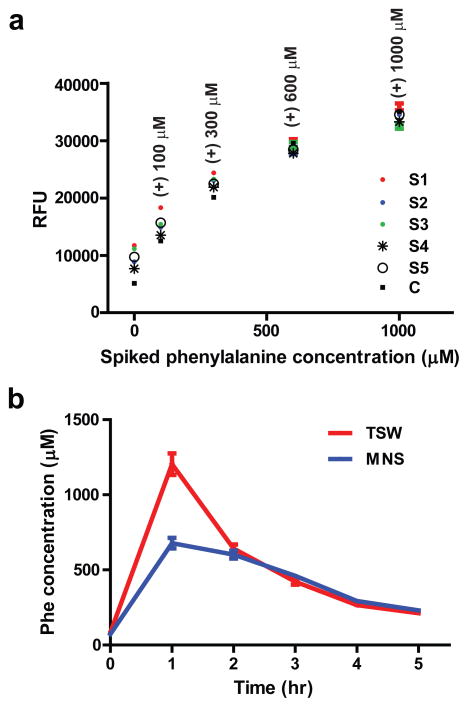
Figure 4. Measurement in human serum and real time monitoring of postprandial waves.
a. Five samples of healthy human sera (e.g., range 30–100 μM) were diluted 1:200 in buffer containing 100 μM ‘Cp*Rh(III)’ and fluorescence intensity measured (in relative fluorescence units or RFUs – as read out directly from the plate reader). Samples were also spiked with clinically relevant concentrations of Phe, thus, demonstrating the clear stratification based on concentrations. C (black squares) is a standard curve of the response of the sensor to these concentrations of Phe, but without sera present. b. Observation of a postprandial wave in the serum of the two senior investigators after a Phe oral load (100 mg/kg); concentration was read out at 1:400 dilution of a serum from a calibration curve without serum and with no further corrections; All measurements were performed in triplicates with standard deviations (SDs) shown.
Aptamers for aromatic amino acids
Our work on monosaccharides was designed to demonstrate the ability of aptamers to recognize analytes that have no epitopes suitable for tight interactions with nucleic acids while at the same time imposing selectivity on organic receptors for analytes that are challenging because of their structural similarity. We next decided to address the issue whether this approach can lead to significant improvements to affinities for less challenging targets that have, e.g., aromatic epitopes, while at the same time addressing an unmet need, e.g., the rapid measurement of amino acids in body fluids. Elevated blood amino acids in newborns are indicative for inborn errors of amino acid metabolism that require immediate and often life-long treatment with a diet restricted in the specific amino acid to prevent disability and serious motor-mental sequelae. Inborn errors of amino acid metabolism are detected in newborn screenings by analysis of whole blood obtained from a heel-prick about 24 h after birth using mass spectrometry. Consecutive follow up and monitoring of blood levels is carried out in specialized laboratories with a usual turnaround time between two and 21 days. There are currently no methods to determine amino acids rapidly and accurately outside of specialized laboratories. A simple test based on a measurement in a highly diluted (to avoid interferences) droplet of fasting blood or serum could be used to detect excessive amino acid and allow adjustment of therapy in real time.
As our primary targets we chose three aromatic amino acids: (a) phenylalanine (Phe, 5), the measurement of which is important in the detection, confirmation, and therapy monitoring for the most common inborn metabolic disease, phenylketonuria (PKU)14; (b) tyrosine (Tyr, 6), the measurement of which is indicative of various types of tyrosinemia15; and (c) tryptophan (Trp, 7), a physiological precursor of serotonin) that is increased in familial tryptophanemia and tryptophanuria16. Numerous RNA aptamers interacting with amino acids have been reported in the context of studies of the origins of life and translational machinery17, but these would be challenging to adapt in fluorescent assays for clinical applications, due to their affinity being mismatched with actual needs (assuming that the intrinsic instability of RNA can be addressed). Specifically, RNA aptamers with mid-micromolar affinities for phenylalanine18 and tyrosine19, and a DNA aptamer with low-micromolar affinity for tryptophan20, were reported before, and these could serve as benchmarks for our method as well.
For an in situ derivatization of amino acids we chose a pentamethylcyclopentadienyl-rhodium(III) (‘Cp*Rh(III)’) complex (8)21. The complex interacts preferentially with bidentate and tridentate ligands, usually with micromolar affinities, but with no useful selectivity; it was introduced by Severin and colleagues22 as the amino-acid complexation reagent in cross-reactive arrays of indicator-displacement sensors for identification of amino acids. Our goal was to isolate aptamers tightly binding to complexes of individual amino acids in order to be able to dilute body fluid in an excess of ‘Cp*Rh(III)’ (globally derivatizing all ligands) and directly measure the concentration of an amino acid with an aptamer. Our idea was that this complex would be suitable because it could effectively crosslink nucleic acids with amino acids, by providing additional electrophilic and aromatic epitopes for binding to nucleic acids, while at the same time complexing amino acids.
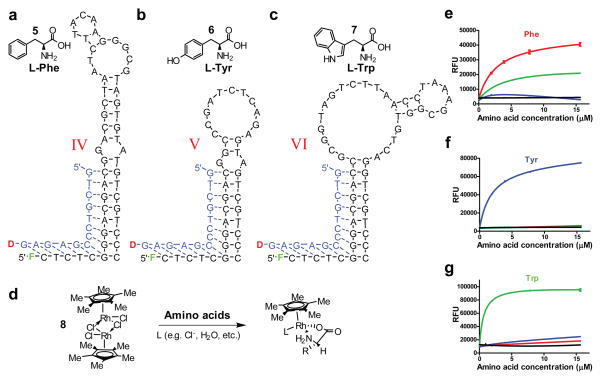
We followed an aptamer selection procedure similar to the one used for receptor 1 and isolated a number of sensors for targeted amino acids. In the case of phenylalanine counter-selections individually with both ‘Cp*Rh(III)’ and ‘Cp*Rh(III)-Tyr’ were performed, in the case of Tyr only ‘Cp*Rh(III)’ was used in counter-selection, while in the case of Trp both ‘Cp*Rh(III)-Tyr’ and ‘Cp*Rh(III)-Phe’ were used. The aptameric sensors for Tyr (V) showed selectivity over complexes with other amino acids at the low concentrations, with no observed response to ‘Cp*Rh(III)’ in the absence of other tested potential ligands, at higher concentrations it showed some response to Phe/Trp (Supplementary Fig. 7). Targeted counter-selection was successful in eliminating the cross-reactivity of Phe sensor IV with Tyr, but this sensor showed response at low concentrations with Trp, against which no counter-selection was performed (Supplementary Fig. 7; subsequent counter-selection against Trp eliminated response to this amino acid at low concentrations, cf. Supplementary Fig. 13). Aptamer VI against Trp complex had exquisite selectivity against other tested amino acids. The dissociation constants of adducts of aptamers with Phe, Tyr, and Trp complexes were 180, 60, and 120 nM, respectively (Supplementary Fig. 8), in each case between one and two orders of magnitude higher affinity than what was reported with best aptamers without addition of the ‘Cp*Rh(III)’ complex.
Next, sensors for Phe and Tyr (IV–V) were tested for their ability (Supplementary Fig. 9) to differentiate healthy sera (fasting) diluted up to 1:512 in a solution containing 100 μM ‘Cp*Rh(III)’ from the same sera spiked (prior to dilution) with 0.25, 0.5 and 1 mM amino acids, indicating that these sensors can be used in actual clinical assays. We performed additional confirmatory assays with the Phe sensor; clinically, in monitoring dietary intervention, for a Phe sensor it would be important to distinguish at least three critical cut-off concentrations: (i) in the adult PKU patient population, the goal is to keep the Phe concentration in blood below 600 μM14; (ii) in children with PKU, the goal is to keep the Phe concentration below 360 μM14; while (iii) the targeted concentration in patients adhering to diet should be also kept above 120 μM, to ensure the sufficient biosynthesis of neurotransmitters14. When we took five sera from healthy humans, spiking them with 100, 300, 600 μM and 1 mM of Phe (Fig. 4a), and diluting them 1:200 in a buffer with reagents, the sensor IV was able to clearly stratify samples spiked at all these concentrations and the unspiked samples, indicating potential for clinical and at home use. Second, in an IRB-approved self-experiment, two senior investigators (TSW and MNS) ingested 100 mg/kg of phenylalanine and observed transient postprandial waves of phenylalanine in serum obtained from capillary blood (Fig. 4b). While full clinical validation with a large number of samples, comparison to standard clinical methods, studies of interferences, matrix effects, and longitudinal monitoring of actual patient population will be necessary to establish firmly the suitability of these sensors to address patients’ needs, the postprandial waves we observed in two human subjects corresponds to what is expected a real life situation22, except that in PKU patients these would persist longer due to the defective phenylalanine catabolism that defines disease.
Discussion
It is instructive to compare the high affinities observed after our selection to early work of Anslyn and Ellington in which they used a bisorganoboronic receptor as a cofactor and isolated aptamers selective for tartarate over citrate7. Their characterized aptamer responded to tartarate, but not citrate, in the presence of the bisorganoboronic receptor; however, the affinity of tartarate for the receptor within the aptamer dropped significantly in comparison to the free receptor (the authors estimate >30-fold, Kd changed from ~7 μM to >200 μM), that is, the aptamer binding to the receptor seems to have interfered with the receptor binding to targets. This observation is in sharp contrast to our results, in which oligonucleotides with closed stems are stabilized in comparison to its complex with competitor oligonucleotide by binding to receptor*target complexes at low concentrations (Supplementary Fig. 4), but not by the receptors themselves. This leads to a system that is more, not less, sensitive to an analyte under conditions in which there is an excess of derivatization agent, it enables quantification of targeted analytes beyond what is possible with a receptor or just an aptamer against a target alone, and enhances potential for clinical applications. We attribute our success in generating useful analytical reagents to primarily two factors that were different from previous attempts to isolate similar aptamers: (1) Our selection was performed in solution phase, with a receptor as an in situ derivatization agent, coupled with a successful counter-selection against the receptor itself, instead of against a dissimilar analog; these select against incorporation of a receptor as a cofactor; and (2) Our elution step was not based on displacement of aptamer candidates from their complexes with targets by a competitor binding to a free receptor; such a procedure disfavors low koff values, thereby further reducing the impact of affinity as the criteria in selection pressure. Another perspective to analyse this difference is to look at the order of events in the formation of ternary complex: in our case, small molecule receptors have to bind their ligands (sugars or amino acids), and only thereafter the complex will favor closing of the stem of the aptamer in the ternary complex.
The importance of our results goes beyond a purely commercial potential in monitoring sugar and amino acid concentrations using newly generated sensors and their nonfluorescent analogs: First, we offer a general solution to a long standing problem that prevented aptameric sensors for small molecules being generally applicable, by demonstrating that, with the judicious selection of in situ derivatization agents and tailored counter-selection conditions, even traditionally difficult small molecule targets are not off-limits for sensors based on oligonucleotides. Second, our work supports earlier proposals7 that oligonucleotide-based receptors can be used to tailor specificity of synthetic receptors for small molecules in a way that is not fully accessible through traditional synthetic organic (or organometallic) chemistry, and vice versa, that an organic receptor endows oligonucleotides with binding and sensing properties that are not readily accessible otherwise. In contrast to this pioneering work7, we show that under the right conditions, we do not have to lose affinity of the organic receptor in the tradeoff for enabling oligonucleotide binding (of note, we also lose affinity of receptors for non-targets through counterselection). Third, for those small molecules for which aptamers have already been isolated, we demonstrate that the affinity of oligonucleotide-based sensors to small molecules can be increased by approximately two orders of magnitude, thus adjusted to concentrations that are closer to being useful for addressing actual patient needs. Finally, the isolation of nucleic acid based sensors for glucose opens possibilities to integrate nucleic acid based devices into molecular-level closed loop systems for the delivery of insulin23 when this advance is combined with the progress in DNA nanotechnology24.
Methods
All technical details, procedures, and sequences are provided in the Supporting Information.
Supplementary Material
Figure 3. Results of three selection procedures against amino acids.
a–c, IV, V, and VI are structure-switching aptameric sensors against L-Phe (5), L-Tyr (6), L-Trp (7), respectively; d, Structure of [(Cp*RhCl2)2] complex 8 that in solution provides the ‘Cp*Rh(III)’ moiety that complexes amino acids (shown as a generic amino acid) and other nucleophilic ligands present in solution (L). e–f, the response of sensors IV–VI at low concentrations of amino acids, representative for measurements in diluted blood or serum (relative fluorescence units or RFUs – as read out from the plate reader – vs. concentration of amino acids in μM, with complex added at 100 μM). The tested amino acids are: phenylalanine (
 ), tyrosine (
), tyrosine (
 ), tryptophan (
), tryptophan (
 ) and a mixture of eight amino acids (●). All measurements were performed in triplicates with standard deviations (SDs) shown.
) and a mixture of eight amino acids (●). All measurements were performed in triplicates with standard deviations (SDs) shown.
Article summary.
Recognition, differentiation, and sensing with artificial receptors of small molecules displaying only sparse functionalities are some of the important traditional challenges of supramolecular chemistry. The authors address this problem by forming ternary complexes between small molecule targets, their non-specific organic (or organometallic) receptors, and oligonucleotides selected in an evolutionary process.
Acknowledgments
We are grateful to the NSF (CBET-1033288 and CBET-1026592), NIH (RGM104960), and JDRF (Innovative program) for funding this research.
Footnotes
Competing financial interests
The authors declare competing financial interests in the form of patent application(s).
Author contributions
M. N. S. proposed and led the project.. K.-A. Y. isolated aptamers listed in this report. S. P. and S. T. performed synthetic and characterization work on organic receptors. T. S. W. provided clinical context and organized self-experiments, with B. K. providing technical support in these. M. B., M. H., P. P., D. K performed earlier experiments on glucose aptamers that eventually led to the development of the method described here. All co-authors participated in the design of their own experiments and analysis of data. M. N. S., K.-A. Y., and S. T. wrote the manuscript, with the exception of synthetic procedures written by S. P.
References
- 1.Keefe AD, Pai S, Ellington AD. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 3.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimoto M, Yamashige R, Matsunaga KI, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 5.McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012:Article id 748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imaizumi Y, et al. Efficacy of base-modification on target binding of small molecule DNA aptamers. J Am Chem Soc. 2013;135:9412–9419. doi: 10.1021/ja4012222. [DOI] [PubMed] [Google Scholar]
- 7.Manimala JC, Wiskur SL, Ellington AD, Anslyn EV. Tuning the specificity of a synthetic receptor using a selected nucleic acid receptor. J Am Chem Soc. 2004;126:16515–16519. doi: 10.1021/ja0478476. [DOI] [PubMed] [Google Scholar]
- 8.James TD, Samankumara Sandanayake KRA, Shinkai S. A glucose-selective molecular fluorescence sensor. Angew Chem Int Edit. 1994;33:2207–2209. [Google Scholar]
- 9.Klonoff DC. The need for separate performance goals for glucose sensors in the hypoglycemic, normoglycemic, and hyperglycemic ranges. Diabetes Care. 2004;27:834–836. doi: 10.2337/diacare.27.3.834. [DOI] [PubMed] [Google Scholar]
- 10.Larkin JD, Frimat KA, Fyles TM, Flower SE, James TD. Boronic acid based photoinduced electron transfer (PET) fluorescence sensors for saccharides. New J Chem. 2010;34:2922–2931. [Google Scholar]
- 11.Yang KA, Pei R, Stefanovic D, Stojanovic MN. Optimizing cross-reactivity with evolutionary search for sensors. J Am Chem Soc. 2012;134:1642–1647. doi: 10.1021/ja2084256. [DOI] [PubMed] [Google Scholar]
- 12.Nutiu R, Li Y. In vitro selection of structure-switching signaling aptamers. Angew Chem Int Edit. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 13.Rajendran M, Ellington AD. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal Chem. 2008;390:1067–1075. doi: 10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]
- 14.Blau N, van Spronsen FJ, Levy HL. Phenylketonuria. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- 15.Hanley WB, et al. “Hypotyrosinemia” in phenylketonuria. Mol Genet Metab. 2000;69:286–294. doi: 10.1006/mgme.2000.2985. [DOI] [PubMed] [Google Scholar]
- 16.Snedden W, Mellor CS, Martin JR. Familial hypertryptophanemia, tryptophanuria and indoleketonuria. Clin Chim Acta. 1983;131:247–256. doi: 10.1016/0009-8981(83)90094-3. [DOI] [PubMed] [Google Scholar]
- 17.Yarus M, Widmann JJ, Knight R. RNA-amino acid binding: A stereochemical era for the genetic code. J Mol Evol. 2009;69:406–429. doi: 10.1007/s00239-009-9270-1. [DOI] [PubMed] [Google Scholar]
- 18.Illangasekare M, Yarus M. Phenylalanine-binding RNAs and genetic code evolution. J Mol Evol. 2002;54:298–311. doi: 10.1007/s00239-001-0045-6. [DOI] [PubMed] [Google Scholar]
- 19.Manironi C, Scerch C, Fruscoloni P, Tocchini-Valentini GP. Molecular recognition of amino acids by RNA aptamers: The evolution into an L-tyrosine binder of a dopamine-binding RNA motif. RNA. 2000;6:520–527. doi: 10.1017/s1355838200991763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, et al. Characterization and application of a DNA aptamer binding to L-tryptophan. Analyst. 2011;136:577–585. doi: 10.1039/c0an00550a. [DOI] [PubMed] [Google Scholar]
- 21.Buryak A, Severin K. A chemosensor array for the colorimetric identification of 20 natural amino acids. J Am Chem Soc. 2005;127:3700–3701. doi: 10.1021/ja042363v. [DOI] [PubMed] [Google Scholar]
- 22.Rampini S, Anders PW, Curtius HC, Marthaler T. Detection of heterozygotes for phenylketonuria by column chromatography and discriminatory analysis. Pediat Res. 1969;3:287–297. doi: 10.1203/00006450-196907000-00004. [DOI] [PubMed] [Google Scholar]
- 23.Taylor S, Stojanovic MN. Is there a future for DNA-based molecular devices in diabetes management? J Diabetes Sci Technol. 2007;1:440–444. doi: 10.1177/193229680700100319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.