Abstract
Collectin liver 1 (CL-L1, alias CL-10) and collectin kidney 1 (CL-K1, alias CL-11), encoded by the COLEC10 and COLEC11 genes, respectively, are highly homologous soluble pattern recognition molecules in the lectin pathway of complement. These proteins may be involved in anti-microbial activity and in tissue development as mutations in COLEC11 are one of the causes of the developmental defect syndrome 3MC. We studied variations in COLEC10 and COLEC11, the impact on serum concentration and to what extent CL-L1 and CL-K1 serum concentrations are correlated. We sequenced the promoter regions, exons and exon-intron boundaries of COLEC10 and COLEC11 in samples from Danish Caucasians and measured the corresponding serum levels of CL-L1 and CL-K1. The median concentration of CL-L1 and CL-K1 was 1.87 μg/ml (1.00–4.14 μg/ml) and 0.32 μg/ml (0.11–0.69 μg/ml), respectively. The level of CL-L1 strongly correlated with CL-K1 (ρ = 0.7405, P <0.0001). Both genes were highly conserved with the majority of variations in the non-coding regions. Three non-synonymous variations were tested: COLEC10 Glu78Asp (rs150828850, minor allele frequency (MAF): 0.003), COLEC10 Arg125Trp (rs149331285, MAF: 0.007) and COLEC11 His219Arg (rs7567833, MAF: 0.033). Carriers of COLEC10 Arg125Trp had increased CL-L1 serum levels (P = 0.0478), whereas promoter polymorphism COLEC11-9570C>T (rs3820897) was associated with decreased levels of CL-K1 (P = 0.044). In conclusion, COLEC10 and COLEC11 are highly conserved, which may reflect biological importance of CL-L1 and CL-K1. Moreover, the strong inter individual correlation between the two proteins suggests that a major proportion are found as heterooligomers or subjected to the same regulatory mechanisms.
Introduction
Collectins are C-type lectins, a family of pattern recognition molecules involved in innate immunity. Collectins share a common multimeric structure where a basic subunit composed of three polypeptide chains undergoes varying degrees of oligomerization [1]. The polypeptide chains are composed of a collagen-like region, an alpha helical neck domain and a carbohydrate recognition domain (CRD). Through the clustering of the CRDs the collectins are capable of recognizing and binding to glycoconjugates on the surface of pathogens or to altered host cells, thereby facilitating their clearance [2]. Well-characterized collectins comprise mannose-binding lectin (MBL), surfactant protein A (SP-A) and D (SP-D), whose functions have been extensively studied [2]. MBL is a recognition molecule in the lectin pathway of complement along with Ficolin-1, Ficolin-2 and Ficolin-3. More recently described collectins comprises CL-L1 (alias collectin-10, collectin liver 1 or CL-10) [3], CL-K1 (alias collectin-11, collectin kidney 1 or CL-11) [4], and CL-P1 (alias collectin-12, collectin placenta 1, CL-12 or SRCL). Both CL-L1 and CL-K1 are soluble molecules that have been found circulating in blood in complex with lectin complement pathway associated serine proteases (MASPs) [5–8]. Two genes, MASP1 and MASP2, encode MASP-1/-3 (alias MASP1 isoform 1 and 2) and MASP-2 (alias MASP2 isoform 1), respectively, and encode also the alternatively spliced non-catalytic products, MAP-1 (alias MAp44 or MASP1 isoform 3) and sMAP (alias MAp19, MAP-2 or MASP2 isoform 2), respectively [9].
The COLEC10 gene encoding CL-L1 comprises six exons, is located on chromosome 8q23-q24.1 and is primarily expressed in the liver, placenta, and adrenal glands [3]. The protein-coding transcript gives rise to a 277 amino acid long protein with four defined regions: N-terminal segment (19 aa), collagen-like region (72 aa), alpha-helical coiled-coil neck region (34 aa), and the CRD (125 aa). The N-terminal segment and the first Gly-Xaa-Yaa repeat of the collagen-like region are encoded by exon 1 [10]; the rest of the collagen-like region by exons 2–4 (see also Fig. 1). Exon 5 encodes the neck region and exon 6 the CRD. Trimer assembly is stabilized by non-covalent interactions in the collagen-like and neck region, and inter-chain disulphide bonds between cysteine residues in the N-terminal segment (Cys12) and neck region (Cys119, Cys121) [7].
Fig 1. Distribution of polymorphisms and pair wise LD analysis (r2) observed in COLEC10.
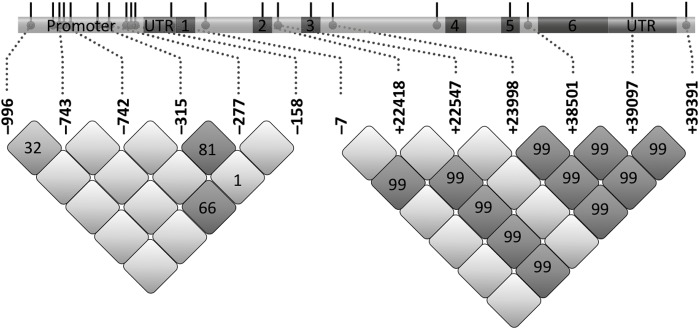
A schematic drawing of the gene COLEC10 is given at the top. Exons are numbered and promoter and non-translated regions are indicated. The numbers in the grid refer to the r2 values of a given pair of SNPs. Darker colours of the boxes means higher r2 values.
The COLEC11 gene encoding CL-K1 comprises seven exons and is located on chromosome 2p25.3. The protein has a domain organization identical with CL-L1. CL-K1 has been found forming oligomers ranging from monomers to hexamers of the trimeric subunit, stabilized by inter chain disulfide bonds [5;8]. COLEC11 is predominantly expressed in the liver, fetal liver, adrenals gland, small intestine, thymus, spinal cord, placenta, pancreas and kidney but low expression levels are found in many other tissues [4].
The median circulating levels of CL-L1 and CL-K1 have been estimated to be 3 and 0.3 μg/ml respectively [5;11;7]. Recently, it was shown that CL-L1 and CL-K1 form heterooligomers, and that the heterooligomers comprise a major proportion of all circulating CL-L1 and CL-K1 [8]. This feature bears resemblance with C1q, the recognition molecule of the classical pathway of complement and with other C1q-related proteins [12]. The heteromeric trimers of C1q have a composition similar to CL-L1, CL-K1 heterooligomers, with heteromeric trimers consisting of polypeptide chains originating from different genes. Formation of the collectin heterooligomers leads, in comparison with CL-L1 and CL-K1 homooligomers alone, to a substantial higher degree of oligomerization and complement activation via interaction with MASP-2 [7;8].
Carbohydrate inhibition studies have shown that CL-K1 in addition to binding to mannose-like carbohydrates (e.g. L-fucose, alpha-D-methyl-mannose and D-mannose) also bind to bacterial and yeast extracts, as well as to apoptotic cells and intact microorganisms [4;5]. Moreover, murine CL-K1 was observed to reduce influenza virus A infectivity and to bind to DNA and heparin [5;8]. Native CL-L1 present in serum showed binding to L-fucose, D-mannose but these observations may be influenced by heteromeric complex formation with CL-K1 [7].
Recently it was shown that mutations in COLEC11 or in MASP1 are the common underlying causes of the Carnevale, Mingarelli, Malpuech and Michels syndromes, united in the syndrome now termed 3MC [13–17]. The clinical effect of variations in the serum concentration of CL-L1 and CL-K1 is more or less unknown. However, elevated CL-K1 plasma levels have been associated with the presence of disseminated intravascular coagulation (DIC), suggesting a possible pathophysiological role for CL-K1 in uncontrolled clotting and bleeding [18].
In the present report we have studied to what extent variations in the COLEC10 and COLEC11 may exist and how these variations affect the circulating levels of CL-L1 and CL-K1 in healthy individuals. To provide new insight into the presence of heterooligomeres in circulation we also aimed to evaluate the degree of correlation between serum levels of CL-L1 and CL-K1.
Materials and Methods
Donor samples
DNA was isolated from 296 unrelated Danish Caucasian blood donors using King Fisher (Thermo Scientific). Serum from 96 of the 296 blood donors was obtained for quantification of levels of CL-L1 and CL-K1. Samples were obtained with written informed consent, and the study was approved by the Regional Ethical Committee of the Capital Region of Denmark (H2–2011–133).
Sequencing of COLEC10 and COLEC11
COLEC10 and COLEC11 promoters and exonic regions were amplified using PCR (for sequences of the primers see Table 1). Promoters were sequenced as three partially overlapping fragments spanning 1000 bp upstream the translation start site ATG. All forward primers included a 5′-T7 sequence (5′-TAATACGACTCACTATAGGG-3′). PCR amplifications were carried out in 12 μl volumes containing 50 ng of genomic DNA, 0.25 μM of each primer, 2.5 mM MgCl2, 0.2 mM dNTP, 50 mM KCl, 10 mM Tris-HCl, pH 8.4, and 0.4 units of Platinum Taq DNA Polymerase (Invitrogen). The PCR reactions were performed according to the following setup: 120s94°C, 35x(10s94°C; 30s60°C; 45s72°C), 120s72°C. The PCR products were sequenced using a T7 biotinylated primer with the ABI BigDye cycle sequencing terminator kit (Applied Biosystems). Sequencing products were purified on the PyroMark Vacuum Prep Workstation (Biotage) using streptavidin beads (Genovision), and sequence analysis was performed on an ABI Prism 3100 Genetic Analyser (Applied Biosystems). The base-called sequences were aligned using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and Chromas Lite (http://technelysium.com.au/) software. All chromatograms were confirmed visually.
Table 1. COLEC10 and COLEC11 primer sets.
| Forward primer | Reverse primer | Internal primer | |
|---|---|---|---|
| COLEC10 | |||
| Promoter region | |||
| Fragment 1 | 5′-cctttcacctaactcttagttga-3′ | 5′-gcctttctggtacactatctc-3′ | |
| Fragment 2 | 5′-ttcgcctttctgccacatcc-3′ | 5′-gaactccagtgtgcatgtcag-3′ | |
| Fragment 3 | 5′-tcaagtactcagcagaaggca-3′ | 5′-caaagccattcattgctgtgg-3′ | |
| Exonic regions | |||
| Exon 1 | 5′-ggtccatttggagcactgag-3′ | 5′-ataagtaacagaccagatctgg-3′ | |
| Exon 2 | 5′-aagtgcatgtgaacatacatcc-3′ | 5′-atgcaacttgagaaacatatgag-3′ | |
| Exon 3 | 5′-tagtatatattcatactcactcac-3′ | 5′-ccaccatgcccagctaattc-3′ | |
| Exon 4 | 5′-ttatgactcatttaattagttgcac-3′ | 5′-actgccagagttttctcttgg-3′ | |
| Exon 5 | 5′-tttagaattgtcttatagttcattc-3′ | 5′-cagacttgaggtgtcctagg-3′ | |
| Exon 6 | 5′-tctgtgactacctcataatagg-3′ | 5′-tgtaacctggcgagttattgg-3′ | |
| 5′-gagtgccatcttaccatgtac-3′ | 5′-aaggagaacttccaagtatagc-3′ | ||
| Pyrosequencing | |||
| –7C>T | 5′-tgtgtgttccaaatacttccc-3′ | 5′-[B]gactctgaatttgcaaaagaaatag-3′ | 5′-tgggagacccttttctgagg-3′ |
| Glu78Asp | 5′-acattagccacatttcaaggga-3′ | 5′-[B]acctcggtctctgagtgttg-3′ | 5′-gtttgcttttcaggaattaaagg-3′ |
| Arg125Trp | 5′-[B]cgtgtcttcatcggcatca-3′ | 5′-ctccttgtcaaactcacacatga-3′ | 5′-ccacttgttgaaggtccgca-3′ |
| COLEC11 | |||
| Promoter region | |||
| Fragment 1 | 5′-tgtctggttttggtatcagggt-3′ | 5′-gcttctatgcagcccgaatc-3′ | |
| Fragment 2 | 5′-tcacctttgaacctagcttctta-3′ | 5′-cctggggagtgtgggaac-3′ | |
| Fragment 3 | 5′-ttgggatgttcgggttggag-3′ | 5′-atcaaactttggaactctggtg-3′ | |
| Exonic regions | |||
| Exon 1 | 5′-ttgggatgttcgggttggag-3′ | 5′-atcaaactttggaactctggtg-3′ | |
| Exon 2 | 5′-ttggtgcctggtgctgacc-3′ | 5′-ggaattccatggccgaagac-3′ | |
| Exon 3 | 5′-aaacacaggccctttctaagag-3′ | 5′-ccacaggtcagcacaaggag-3′ | |
| Exon 4 | 5′-ctccattctatagtgtgtatgac-3′ | 5′-agctgcagcccctccatcc-3′ | |
| Exon 5 | 5′-aaagaatgatcactcgataatcc-3′ | 5′-gtgtccaccgtgggatgtg-3′ | |
| Exon 6 | 5′-ttgaaatacatgtgtctgccct-3′ | 5′-gcacagccacaggcagcag-3′ | |
| Exon 7 | 5′-acctcccagccctgtcctg-3′ | 5′-cattttctcagtttagacaacagc-3′ | |
| 5′-cccaacaatgcctacgacg-3′ | 5′-catggataatagtgtgaaggaacc-3′ | ||
| Pyrosequencing | |||
| His219Arg | 5′-[B]tggaaagagagcaaatgttcct-3′ | 5′-actggaattgtagtgttgatgga-3′ | 5′-actaatatccagttgtccaacaaa-3′ |
Forward and reverse primers were used for PCR amplification, internal primer was used for pyrosequencing. All forward primers for the promoter and exonic regions include a 5′-T7 sequence (5′-TAATACGACTCACTATAGGG-3′). Biotinylated oligos are indicated as [B].
Pyrosequencing-based genotyping assay
Pyrosequencing was carried out as described by Munthe-Fog et al. [19]. In brief, specific primer sets were designed to amplify the three non synonymous SNPs found in COLEC10 and COLEC11, (rs150828850, rs149331285 and rs7567833) and the SNP located in the 5′UTR of COLEC10 (ss749616235) (for primer list see Table 1). DNA was PCR amplified and purified as described above. Purified PCR products were incubated 5 min at 85°C with 10 μM of internal primer and annealing buffer (magnesium acetate 2 mM, Tris 20 mM, pH 7.6). The pyrosequencing reaction and sequence analysis were carried out in the PSQ 96MA (Biotage).
SNP identification
The observed polymorphisms (Tables 2 and 3) can be found in NCBI’s SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) through accession numbers (rs) or submission numbers (ss).
Table 2. Genetic variation observed in the COLEC10 gene.
| Variation | dbSNP | Region | Amino acid change | AA | Aa | aa | MAF |
|---|---|---|---|---|---|---|---|
| −996C>T | rs117863403 | Promoter | - | 93 (96.9%) | 3 (3.1%) | 0 | 0.016 |
| −796delT | ss991382312 | Promoter | - | 95 (99%) | 1 (1%) | 0 | 0.005 |
| −743C>T | ss991382313 | Promoter | - | 95 (99%) | 1 (1%) | 0 | 0.005 |
| −742G>A | rs3812490 | Promoter | - | 94 (98%) | 2 (2%) | 0 | 0.010 |
| −663C>G | rs1485297 | Promoter | - | 81(84.4%) | 14 (14.6%) | 1 (1%) | 0.083 |
| −315G>C | rs1485298 | Promoter | - | 62 (64.6%) | 28 (29.2%) | 6 (6.3%) | 0.208 |
| −277T>C | rs2450048 | Promoter | - | 56 (58.3%) | 33 (34.4%) | 7 (7.3%) | 0.245 |
| −161_−157 delAAAAT | rs148350292 | Promoter | - | 95 (99%) | 1 (1%) | 0 | 0.005 |
| −150C>T | rs3829048 | Promoter | - | 93 (96.9%) | 3 (3.1%) | 0 | 0.016 |
| −145G>A | ss991382317 | Promoter | - | 95 (99%) | 1 (1%) | 0 | 0.005 |
| −7C>T | ss749616235 | 5′ UTR | - | 299 (99.7%) | 1 (0.3%) | 0 | 0.005 |
| +228A>G | rs2465383 | Intron 1 | - | 26 (26%) | 55 (55%) | 19 (19%) | 0.465 |
| +22418A>G | rs16891987 | Exon 2 | - | 97 (97%) | 3 (3%) | 0 | 0.015 |
| +22547T>C | rs149290883 | Intron 2 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +23881A>C | rs150828850 | Exon 3 | Glu78Asp | 298 (99.3%) | 2 (0.7%) | 0 | 0.005 |
| +23998A>T | rs4512407 | Intron 3 | - | 97 (97%) | 3 (3%) | 0 | 0.015 |
| +36545C>T | rs149331285 | Exon 5 | Arg125Trp | 296 (98.7%) | 4 (1.3%) | 0 | 0.005 |
| +38501C>A | rs11987106 | Intron 5 | - | 97 (97%) | 3 (3%) | 0 | 0.015 |
| +39097A>G | rs1064556 | 3′ UTR | - | 97 (97%) | 3 (3%) | 0 | 0.015 |
| +39391C>G | rs1064557 | 3′ downstream | - | 97 (97%) | 3 (3%) | 0 | 0.015 |
The base position refers to the location of the polymorphism considering the A from the translation start site ATG as +1, and the adjacent 5′ base as -1. All variations adhered to the Hardy-Weinberg equilibrium. ‘AA’, individuals homozygote for the major allele; ‘Aa’, heterozygous individuals; ‘aa’, individuals homozygous for the minor allele; MAF, minor allele frequency.
Table 3. Genetic variation observed in the COLEC11 gene.
| Base position | dbSNP | Region | Amino acid change | AA | Aa | aa | MAF |
|---|---|---|---|---|---|---|---|
| −10007_− 10004delCATT | ss991382311 | Promoter | - | 95 (99%) | 1 (1%) | 0 | 0.005 |
| −9970T>C | rs1864480 | Promoter | - | 53 (55.2%) | 40 (41.7%) | 3 (3%) | 0.240 |
| −9766T>C | rs4849953 | Promoter | - | 44 (45.8%) | 44 (45.8%) | 8 (8.4%) | 0.313 |
| −9591G>T | rs34596301 | Promoter | - | 86 (89.6%) | 10 (10.4%) | 0 | 0.052 |
| −9570C>T | rs3820897 | Promoter | - | 52 (54.2%) | 41 (42.7%) | 3 (3%) | 0.245 |
| −9175T>C | rs77246730 | 5′ UTR | - | 193 (98.5%) | 3 (1.5%) | 0 | 0.010 |
| +8822C>T | rs3811531 | Intron 2 | - | 36 (36%) | 46 (46%) | 18 (18%) | 0.410 |
| +8926G>A | rs7567724 | Intron 2 | - | 77 (77%) | 20 (20%) | 3 (3%) | 0.130 |
| +8939G>A | rs17017752 | Intron 2 | - | 95 (95%) | 4 (4%) | 1 (1%) | 0.030 |
| +9072G>A | rs67826307 | Intron 3 | - | 95 (95%) | 4 (4%) | 1 (1%) | 0.030 |
| +9091C>T | rs72769325 | Intron 3 | - | 97 (97%) | 3 (3%) | 0 | 0.015 |
| +33064T>G | ss749616236 | Intron 3 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +33116C>T | rs3811528 | Intron 3 | - | 49 (49%) | 40 (40%) | 11 (11%) | 0.310 |
| +33119G>T | rs36024497 | Intron 3 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +33125G>A | ss749616237 | Intron 3 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +33144A>G | rs3811527 | Intron 3 | - | 49 (49%) | 40 (40%) | 11 (11%) | 0.310 |
| +33145C>T | rs148763047 | Intron 3 | - | 98 (98%) | 2 (2%) | 0 | 0.010 |
| +33233T>C | rs34347318 | Exon 4 | - | 98 (98%) | 2 (2%) | 0 | 0.010 |
| +33245T>C | rs17017791 | Exon 4 | - | 99 (99%) | 1 (1%) | 0 | 0005 |
| +35844C>T | rs17017804 | Intron 4 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +35846C>T | rs144784212 | Intron 4 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +39141C>T | rs34436491 | Exon 6 | - | 99 (99%) | 1 (1%) | 0 | 0.005 |
| +39618C>G | rs7567833 | Exon 7 | His219Arg | 280 (93.3%) | 20 (6.7%) | 0 | 0.025 |
| +39739C>T | rs114716171 | Exon 7 | - | 96 (96%) | 4 (4%) | 0 | 0.020 |
The base position refers to the location of the polymorphism considering the A from the translation start site ATG as +1, and the adjacent 5′ base as -1. All variations adhered to the Hardy-Weinberg equilibrium. ‘AA’; individuals homozygote for the major allele, ‘Aa’; heterozygous individuals, ‘aa’; individuals homozygous for the minor allele; MAF, minor allele frequency.
In silico prediction of the biological consequences of variations
In silico analysis of functional effects was performed on the observed nsSNPs. PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) evaluates physical properties, proximity to functional structures, and the evolutive conservation to predict the functional significance of a mutation using a trained Naïve Bayes classifier [20]. BLOSUM62 (http://www.ncbi.nlm.nih.gov) is an amino acid substitution matrix that assigns a score to each aligned pair of residues based on the odds of finding both amino acids in an alignment on purpose rather than by chance. SIFT (http://sift.bii.a-star.edu.sg/) is a multi-step sequence alignment comparison algorithm that estimates whether an amino acid substitution may have an effect on protein function, based upon the premise that highly conserved residues are more intolerant to substitution than those less conserved [21;22]. Align-GVGD (http://agvgd.iarc.fr/) classifies a substitution from most likely to least likely to interfere with protein function based on multiple sequence alignments (MSA) and the combined Grantham Variation (GV) and Grantham Deviation (GV) scores, which measure the biochemical distance between aminoacids [23]. Protein MSA was constructed using ClustalW included in the program Bioedit (www.mbio.ncsu.edu/BioEdit/bioedit.html). CL-L1 and CL-K1 protein sequences were aligned with their orthologues from 17 different species in order to build a dataset with a sufficient size and alignment depth to compensate for the appearance of constrained positions due to chance [24;25]. PhD-SNP is a trained support vector machine-based predictor that classifies mutations as neutral polymorphisms or disease related based on sequence and profile information [26]. Furthermore, an evolutionary conservation profile was generated by Consurf [27]. This web-based program calculates the degree of conservation (from most variable to most conserved) for every position of the protein using MSA and an empirical Bayesian algorithm.
CL-L1 and CL-K1 levels in serum
The serum levels of CL-L1 and CL-K1 was quantified using validated double sandwich immuno assays as previously described in detail [7] and [28].
Statistical analysis
Hardy-Weinberg was calculated applying the χ2 test to the simple gene counting results, implementing Yates’ correction when considered appropriate. Linkage disequilibrium (LD), expressed as r2, and observed haplotypes were assessed by SHEsis software [29;30]. The statistical significance of the genetic impact on serum levels was tested with Mann-Whitney U test, Kruskal-Wallis test and Spearman Rank correlation. Whiskers and outliers were calculated according to Tukey’s method.
Results
Genetic variation in COLEC10 and COLEC11
We sequenced the promoter region spanning 1000 bp upstream the translation start site, the exon and intron/exon boundaries in COLEC10 and COLEC11.
COLEC10. A total of 10 known and novel variations were observed in the promoter region (Table 2) including a novel variation located in the 5’UTR. Another 10 variations were observed in the exons and the flanking regions. Two low frequent variations (MAF < 0.01) resulted in an amino acid change: Glu78Asp (rs150828850) and Arg125Trp (rs149331285). The Glu78Asp variation in exon 3 occurs in a variable residue and was predicted to be benign, in contrast to Arg125Trp in exon 6, which is located in an evolutionary constrained position in the neck domain and was by in silico analysis predicted to be potentially critical for the structure (Table 4). A complete list of observed variations and linkages are given in Table 2 and Fig. 1. To test whether any of the identified variations were located within a regulatory domain, all promoter variations were analyzed in silico (SwissRegulon Database). COLEC10 161_-157AAAATdel overlaps with a SRY and several Forkhead box (FOX) binding sites. SRY and the FOX family of transcription factors are known regulators of multiple cellular and developmental processes such as liver differentiation [31;32]. Of interest, additional perfect LD was observed between seven loci; COLEC10–7 (ss749616235), +22418 (rs16891987), +22547 (rs149290883), +23998 (rs4512407), +38501 (rs11987106), +39097 (rs1064556), and +39391 (rs1064557), however, these minor alleles were clustered in 4 individuals only.
Table 4. Non-synonymous variations in COLEC10 and COLEC11.
| Variation | Region | PolyPhen-2 | Blosum62 | SIFT | Align-GVGD | PhD-SNP | Consurf |
|---|---|---|---|---|---|---|---|
| COLEC10 Glu78Asp (rs150828850) | Exon 3 | Benign | 2 | 1 | C0 | Neutral (7) | Variable(3) |
| COLEC10 Arg125Trp (rs149331285) | Exon 5 | Probably damaging | -3 | 0 | C65 | Disease (3) | Conserved (9) |
| COLEC11 His219Arg (rs7567833) | Exon 7 | Benign | 0 | 0.462 | 29 | Neutral (3) | Variable(3) |
PolyPhen-2 classifies qualitatively a mutation as benign, possibly damaging, or probably damaging. BLOSUM62 matrix values range from 4 (more likely) to -4 (less likely substitutions). SIFT values >0.05 indicate substitutions with little effect on protein function, while values <0.05 point out possible deleterious substitutions. Align-GVGD categorizes a mutation in seven graded classifiers from less likely (class C0) to most likely to interfere with protein function (class C65). PhD-SNP produces a binary prediction (neutral vs disease-related polymorphism), together with the reliability score of the prediction (0–9). Consurf position-specific conservation scores are divided in 9 grades, from the most variable (grade 1) to the most conserved positions (grade 9).
COLEC11. Four known variations and a novel 4-bp deletion were observed in the promoter region (Fig. 2, Table 3). In the 5’UTR and the intron/exon boundaries 12 known and two novel variations were identified. Further five variations were found within the exons, one resulting in an amino acid change: His219Arg (rs7567833) located in exon 7, which encodes the carbohydrate recognition domain of CL-K1. Only individuals heterozygous for His219Arg were identified (N = 5, MAF 0.025). His219Arg showed a moderate linkage with +33245 in exon 4 (rs17017791), +35844 in intron 4 (rs17017804), +39141 in exon 4 (rs34436491); and more robust linkage with +9091 in intron 3 (rs72769325). In addition, two SNPs in intron 3, rs3811528 and rs3811527 (+8939 and +9072), with MAFs of 0.31 had a perfect LD.
Fig 2. Distribution of polymorphisms and pair wise LD analysis (r2) observed in COLEC11.
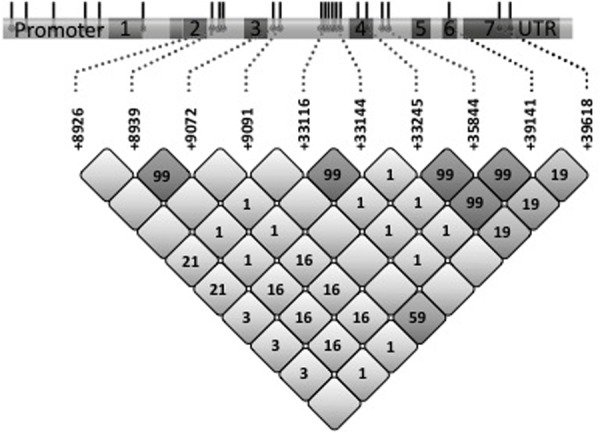
A schematic drawing of the gene COLEC11 is given at the top. Exons are numbered and promoter and non-translated regions are indicated. The numbers in the grid refer to the r2 values of a given pair of SNPs. Darker colours of the boxes means higher r2 values.
Serum levels
CL-L1 mean serum concentration was estimated to be 1.87 μg/ml (range 1.00—l4.14 μg/ml). CL-K1 serum concentration ranged from 0.11 to 0.69 μg/ml, with a mean concentration of 0.32 μg/ml. None of the highly frequent polymorphisms in the COLEC10 promoter region were associated with CL-L1 serum levels. Interestingly, although no individuals homozygous for the minor allele were found, individuals heterozygous for Arg125Trp (rs149331285) (N = 3) had significantly higher levels of CL-L1 in serum (P = 0.0478, Fig. 3A).
Fig 3. Impact of allelic variants on serum levels.
A-C, the serum levels of CL-L1 and CL-K1 and the corresponding genotype. Horizontal bars indicate median values; number in parenthesis, number of individuals and white circles, outliers. Statistical significance was determined by non-parametric Mann-Whitney and Kruskal-Wallis tests. D, the distribution of CL-L1 and CL-K1 levels in the cohort. The strength of the correlation was calculated using Spearman Rank.
In COLEC11 the promoter polymorphism—9570C>T (rs3820897) was significantly associated with higher levels of CL-K1 (P = 0.044, Fig. 3B). The non-synonymous SNP; His219Arg had no significant effect on the level of CL-K1 (Fig. 3C). The allele count for several of the genotyped positions was too low to significantly determine the impact on the level of circulating proteins. A strong correlation, independent of age and gender (not shown) between CL-L1 and CL-K1 (ρ = 0.7405, P <0.0001) was observed (Fig. 3D). However, none of the identified genetic variations in COLEC10 were significantly associated with the circulating levels of CL-K1 and vice versa (data not shown), thus COLEC10 Arg125Trp was not associated with the level of CL-K1 (P = 0.916) and neither was COLEC11–9570C>T with the level of CL-L1 (P = 0.938).
Discussion
To study how the genetic variation affects structure and concentrations in the background population we examined the COLEC10 and COLEC11 in healthy individuals and determined the circulating levels of CL-L1 and CL-K1 in corresponding serum samples.
Screening COLEC10 and COLEC11, we found two variations with a putative impact on either serum level and/or protein structure. COLEC10 Arg125Trp (rs149331285) was predicted by in silico analysis to have a significant effect on protein structure. This nsSNP is located in the neck domain, an alpha helical coiled-coil region involved in the trimerization of the collagen chains. The helical coil region consists of three parallel right-handed alpha helices with hydrophobic residues repeated in a heptad pattern. The association of the alpha helices aligns the collagen chains allowing them to fold in a zipper-like fashion. There is extensive evidence that oligomerization of SP-D monomers is required for high-affinity binding to carbohydrates [33] and for many of its biological functions [34;35]. Thus, COLEC10 Arg125Trp may be of considerable importance if the substitution disrupts the coil structure causing the trimers to fall apart or affect the assembly of heteromeric complexes. Curiously, the level of CL-L1 was significantly higher in the three heterozygous individuals we identified. With a calculated MAF of 0.005, this variation would be interesting to characterize further.
Although only present in three individuals the promoter polymorphism COLEC11–9570C>T (rs3820897) was significantly associated with the level of CL-K1 (P = 0.044). We identified several other variations that potentially could influence the protein levels in serum. However, due to limitations in our cohort size the impact of the genetic variation on the circulating levels require further analyses in larger cohorts. Among the more interesting variations is the COLEC10–161–157AAAATdel expected to interrupt the binding site of several transcription factors that regulate an array of events ranging from liver development (e.g. FOXA2) [31] to immune response modulation (FOXJ1, FOXO3, FOXQ1) [36]. Additional studies into COLEC10 and COLEC11 regulatory cassettes could resolve these questions.
No effect on serum levels was observed for the polymorphism COLEC11 His219Arg (rs7567833, MAP 0.033), located in the carbohydrate recognition domain. Phylogenetic analysis revealed that the G allele (the minor allele) is indeed the ancestral allele. Although located adjacent to the ligand binding site in the CRD, this substitution could affect the binding affinity of the variant molecule towards its ligand as seen for Ficolin-2 [37]. Amino acid substitutions in the pathogen recognition domain affecting the ligand binding have previously been reported for Ficolin-2. Two non-synonymous polymorphisms in FCN2 positioned near the binding site markedly alter the binding capacity for GlcNAc and thus the complement activation potential [37]. Several Ficolin-2 clinical associations have been reported and genotypes conferring low lectin activity have been associated with increased risk of infections [38]. Thus, COLEC11 His219Arg could be of clinical relevance.
Recently, heteromeric complexes between CL-L1 and CL-K1, stabilized by disulfide bonds, were observed in circulation by Henriksen et al [8]. The observation was further supported by co-transfections of the two proteins in CHO cells in which the ratio of CL-L1 and CL-K1 in the formed heteromeric complexes was estimated to 1:2 in favor of CL-K1. Both recombinant CL-K1 and CL-L1 and the heterocomplexes from serum are able to form complexes with the MASPs leading to activation of the complement cascade. In addition, the complement activation potential of the heterocomplexes was reported to be more potent than the potential observed for CL-K1 homocomplexes [8].
In the present study we observed a strong correlation between CL-L1 and CL-K1 serum levels (ρ = 0.7405, Fig. 3C), independent of age or gender lending support to the above observation that a major proportion of CL-L1 and CL-K1 exist as heterooligomeric complexes in the circulation whereas only a minor part of CL-L1 and CL-K1 circulates as homocomplexes. A more definite prediction of the composition of the heterocomplexes i.e. the CL-L1:CL-K1 ratio would be highly speculative as the relatively large difference in levels between CL-L1 and CL-K1 could be due to differences in the techniques for estimating the protein concentration.
In conclusion, we report the finding of three gene variations in COLEC10 and COLEC11 with putative effect on the circulating levels and function of CL-L1 and CL-K1. COLEC10 Arg125Trp (rs149331285) was predicted to have a significant effect on the protein structure however carriers of the variant had significant higher levels of circulating CL-L1. In COLEC11 the promoter polymorphism COLEC11–9570C>T (rs3820897) was significantly associated with the level of CL-K1. Furthermore, located in the CRD of CL-K1 the polymorphism COLEC11 His219Arg (rs7567833) could potentially affect the binding capacity of CL-K1 towards its ligand thereby altering the lectin pathway activation potential of CL-K1. Thus, our study offers an overview of COLEC10 and COLEC11 sequence-variant footprint in the Caucasian population and the observations made here will serve to establish more detailed footprint for genotype-phenotype studies in the Caucasian population as well as in other ethnic groups.
Acknowledgments
The authors wish to thank Vibeke Witved and Linda Schneider for excellent technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
Funding was provided by The Danish Research Council for Independent Research, The Novo Nordisk Research Foundation, The Lundbeck Research Foundation, The Svend Andersen Research Foundation, Rigshospitalet and The University of Copenhagen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hakansson K, Reid KB (2000) Collectin structure: a review. Protein Sci 9:1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van de Wetering JK, van Golde LM, Batenburg JJ (2004) Collectins: players of the innate immune system. Eur J Biochem 271:1229–1249. [DOI] [PubMed] [Google Scholar]
- 3. Ohtani K, Suzuki Y, Eda S, Kawai T, Kase T, Yamazaki H, et al. (1999) Molecular cloning of a novel human collectin from liver (CL-L1). J Biol Chem 274:13681–13689. [DOI] [PubMed] [Google Scholar]
- 4. Keshi H, Sakamoto T, Kawai T, Ohtani K, Katoh T, et al. (2006) Identification and characterization of a novel human collectin CL-K1. Microbiol Immunol 50:1001–1013. [DOI] [PubMed] [Google Scholar]
- 5. Hansen S, Selman L, Palaniyar N, Ziegler K, Brandt J, et al. (2010) Collectin 11 (CL-11, CL-K1) is a MASP-1/3-associated plasma collectin with microbial-binding activity. J Immunol 185:6096–6104. 10.4049/jimmunol.1002185 [DOI] [PubMed] [Google Scholar]
- 6. Ma YJ, Skjoedt MO, Garred P (2012) Collectin-11/MASP Complex Formation Triggers Activation of the Lectin Complement Pathway—The Fifth Lectin Pathway Initiation Complex. J Innate Immun 5:242–250. 10.1159/000345356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Axelgaard E, Jensen L, Dyrlund TF, Nielsen HJ, Enghild JJ, et al. (2013) Investigations on collectin liver 1. J Biol Chem 288:23407–23420. 10.1074/jbc.M113.492603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henriksen ML, Brandt J, Andrieu JP, Nielsen C, Jensen PH, et al. (2013) Heteromeric complexes of native collectin kidney 1 and collectin liver 1 are found in the circulation with MASPs and activate the complement system. J Immunol 191:6117–6127. 10.4049/jimmunol.1302121 [DOI] [PubMed] [Google Scholar]
- 9. Yongqing T, Drentin N, Duncan RC, Wijeyewickrema LC, Pike RN (2012) Mannose-binding lectin serine proteases and associated proteins of the lectin pathway of complement: two genes, five proteins and many functions? Biochim Biophys Acta 1824:253–262. 10.1016/j.bbapap.2011.05.021 [DOI] [PubMed] [Google Scholar]
- 10. Selman L, Hansen S (2012) Structure and function of collectin liver 1 (CL-L1) and collectin 11 (CL-11, CL-K1). Immunobiology 217:851–863. 10.1016/j.imbio.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 11. Yoshizaki T, Ohtani K, Motomura W, Jang SJ, Mori K, et al. (2012) Comparison of human blood concentrations of collectin kidney 1 and mannan-binding lectin. J Biochem 151(1):57–64 10.1093/jb/mvr114 [DOI] [PubMed] [Google Scholar]
- 12. Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, et al. (2008) Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J 416:161–177. 10.1042/BJ20081240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carnevale F, Krajewska G, Fischetto R, Greco MG, Bonvino A, (1989) Ptosis of eyelids, strabismus, diastasis recti, hip defect, cryptorchidism, and developmental delay in two sibs. Am J Med Genet 33:186–189. [DOI] [PubMed] [Google Scholar]
- 14. Rooryck C, Diaz-Font A, Osborn DP, Chabchoub E, Hernandez-Hernandez V, et al. (2011) Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat Genet 43:197–203. 10.1038/ng.757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malpuech G, Demeocq F, Palcoux JB, Vanlieferinghen P (1983) A previously undescribed autosomal recessive multiple congenital anomalies/mental retardation (MCA/MR) syndrome with growth failure, lip/palate cleft(s), and urogenital anomalies. Am J Med Genet 16:475–480. [DOI] [PubMed] [Google Scholar]
- 16. Michels VV, Hittner HM, Beaudet AL (1978) A clefting syndrome with ocular anterior chamber defect and lid anomalies. J Pediatr 93:444–446. [DOI] [PubMed] [Google Scholar]
- 17. Mingarelli R, Castriota SA, Dallapiccola B (1996) Two sisters with a syndrome of ocular, skeletal, and abdominal abnormalities (OSA syndrome). J Med Genet 33:884–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi K, Ohtani K, Larvie M, Moyo P, Chigweshe L, et al. (2014) Elevated plasma CL-K1 level is associated with a risk of developing disseminated intravascular coagulation (DIC). J Thromb Thrombolysis 38:331–338 10.1007/s11239-013-1042-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munthe-Fog L, Hummelshøj T, Hansen BE, Koch C, Madsen HO, et al. (2007) The impact of FCN2 polymorphisms and haplotypes on the Ficolin-2 serum levels. Scand J Immunol 65:383–392. [DOI] [PubMed] [Google Scholar]
- 20. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, et al. (2003) Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci U S A 100:5896–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, et al. (2003) Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci U S A 100:5902–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, et al. (2006) Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cooper GM, Brudno M, Green ED, Batzoglou S, Sidow A, (2003) Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res 13:813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB (2008) In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat 29:1327–1336. 10.1002/humu.20892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Capriotti E, Calabrese R, Casadio R (2006) Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22:2729–2734. [DOI] [PubMed] [Google Scholar]
- 27. Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, et al. (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33:W299–W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selman L, Henriksen ML, Brandt J, Palarasah Y, Waters A, et al. (2012) An enzyme-linked immunosorbent assay (ELISA) for quantification of human collectin 11 (CL-11, CL-K1). J Immunol Methods 375:182–188. 10.1016/j.jim.2011.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Zhang Z, He Z, Tang W, Li T, et al. (2009) A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res 19:519–523. 10.1038/cr.2009.33 [DOI] [PubMed] [Google Scholar]
- 30. Shi YY, He L (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15:97–98. [DOI] [PubMed] [Google Scholar]
- 31. Benayoun BA, Caburet S, Veitia RA (2011) Forkhead transcription factors: key players in health and disease. Trends Genet. 27:224–232. 10.1016/j.tig.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 32. Kakhki SA, Shahhoseini M, Salekdeh GH (2013) Comparative SRY incorporation on the regulatory regions of pluripotency/differentiation genes in human embryonic carcinoma cells after retinoic acid induction. Mol Cell Biochem 376:145–150. 10.1007/s11010-013-1562-5 [DOI] [PubMed] [Google Scholar]
- 33. Hoppe HJ, Barlow PN, Reid KB (1994) A parallel three stranded alpha-helical bundle at the nucleation site of collagen triple-helix formation. FEBS Lett 344:191–195. [DOI] [PubMed] [Google Scholar]
- 34. Brown-Augsburger P, Hartshorn K, Chang D, Rust K, Fliszar C, et al. (1996) Site-directed mutagenesis of Cys-15 and Cys-20 of pulmonary surfactant protein D. Expression of a trimeric protein with altered anti-viral properties. J Biol Chem 271:13724–13730. [DOI] [PubMed] [Google Scholar]
- 35. Hartshorn KL, Reid KB, White MR, Jensenius JC, Morris SM, et al. (1996) Neutrophil deactivation by influenza A viruses: mechanisms of protection after viral opsonization with collectins and hemagglutination-inhibiting antibodies. Blood 87:3450–3461. [PubMed] [Google Scholar]
- 36. Jonsson H, Peng SL (2005) Forkhead transcription factors in immunology. Cell Mol Life Sci 62:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hummelshoj T, Munthe-Fog L, Madsen HO, Fujita T, Matsushita M, et al. (2005) Polymorphisms in the FCN2 gene determine serum variation and function of Ficolin-2. Hum Mol Genet 14:1651–1658. [DOI] [PubMed] [Google Scholar]
- 38. Kilpatrick DC, Chalmers JD (2012) Human L-Ficolin (Ficolin-2) and Its Clinical Significance. J Biomed Biotechnol 2012: 138797 10.1155/2012/138797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.



