Abstract
Background
When compared to the other mismatch repair genes involved in Lynch syndrome, the identification of mutations within PMS2 has been limited (<2% of all identified mutations), yet the immunohistochemical analysis of tumour samples indicates that approximately 5% of Lynch syndrome cases are caused by PMS2. This disparity is primarily due to complications in the study of this gene caused by interference from pseudogene sequences.
Methods
Using a recently developed method for detecting PMS2 specific mutations, we have screened 99 patients who are likely candidates for PMS2 mutations based on immunohistochemical analysis.
Results
We have identified a frequently occurring frame-shift mutation (c.736_741del6ins11) in 12 ostensibly unrelated Lynch syndrome patients (20% of patients we have identified with a deleterious mutation in PMS2, n = 61). These individuals all display the rare allele (population frequency <0.05) at a single nucleotide polymorphism (SNP) in exon 11, and have been shown to possess a short common haplotype, allowing us to calculate that the mutation arose around 1625 years ago (65 generations; 95% confidence interval 22 to 120).
Conclusion
Ancestral analysis indicates that this mutation is enriched in individuals with British and Swedish ancestry. We estimate that there are >10 000 carriers of this mutation in the USA alone. The identification of both the mutation and the common haplotype in one Swedish control sample (n = 225), along with evidence that Lynch syndrome associated cancers are rarer than expected in the probands’ families, would suggest that this is a prevalent mutation with reduced penetrance.
An important factor in keeping the integrity of the genome is an ability to detect and repair single nucleotide changes which occur as a result of replication errors. In eukaryotes this process is regulated by a set of evolutionarily conserved genes which belong to the mismatch repair family. There are four key genes involved in this process whose products function as two heterodimers (MLH1 with PMS2 and MSH2 with MSH6).1 A predisposition to tumorigenesis, known as Lynch syndrome,2 is associated with mutations in the aforementioned genes.3 This predisposition is dominantly inherited with a heterogeneous spectrum of tissues being affected (primarily colorectal and endometrial). The development of a cancer, however, is recessive in that, analogous to the mechanism in many tumour suppressor genes, a second hit on the wild type allele is required. The mechanism of the second hit is often unknown, although deletions, chromosomal loss and promoter methylation are often cited.4–6
Extensive studies over the last 14 years have identified numerous functional mutations within the mismatch repair genes (http://www.med.mun.ca/mmrvariants). Until recently, the vast majority of Lynch syndrome causing mutations had been identified within either MLH1 or MSH2,7 which suggested a less significant role for MSH6 and PMS2 in the mismatch repair mechanism. It is in our opinion that these results are somewhat misleading in that the majority of studies have avoided looking for PMS2 mutations due to the presence of extensive, highly homologous, pseudogenes,8–10 which have made mutation detection by routine methods difficult and error prone. This theory is supported by the fact that tumour studies show that ~5% of Lynch syndrome samples express MLH1, MSH2 and MSH6, but not PMS2 at the protein level,11 yet the mutation detection rate is currently less than 2% (http://www.med.mun.ca/mmrvariants).
We have recently developed a simple yet effective method of avoiding many of the pseudogene associated problems of PMS2 screening through the use of long range polymerase chain reaction (PCR).12 Via this method we have identified a deleterious mutation in ~62% of patients tested (61 out of 99), with 34 different mutations (11 of which occur in more than one family) being identified (authors’ unpublished data). Of particular interest from this recent study has been the identification of a frequently occurring insertion/ deletion mutation (c.736_741delCCCCCTinsTGT-GTGTGAAG; p.P246CfsX3, referred to herein as indel). Since we first reported this mutation in three Lynch syndrome patients,12 we have subsequently identified the same mutation in a further nine families.
The following work uses these families to identify a common haplotype, which suggests that the indel is a founder mutation that arose some 1625 years ago.
PATIENTS AND METHODS
Patients
For the present study only those cases were included (n = 99) in which the tumour did not stain for PMS2 by immunohistochemistry, while it did stain for the MLH1, MSH2 and MSH6 proteins. The National Cancer Institute (NCI) funded Colon Cancer Family Registries provided 35 anonymous samples from four sites: Australasia, Seattle, Mayo Clinic and Ontario. These samples have been accrued to the registry, either through high risk clinic ascertainment or through population based ascertainment. The remaining cases were either from high risk clinics in which patients with an early onset of cancer and/or a family history of cancer predominate (n = 51), or from series in which unselected patients with colorectal cancer (CRC) were screened for mutations by microsatellite instability analysis (n = 13). All patients provided written consent for genetic testing.
Mutation detection
Variants within the PMS2 locus were detected as described previously,12 with the following modifications. Exons 6, 7, 8, and 10 were individually amplified directly from genomic DNA, and PCR2 was reduced to a more readily amplifiable product (1618 bp), from which exon 9 can be directly sequenced. Primers differing from the original protocol are displayed in table 1.
Table 1.
Oligonucleotides used for sequencing, allele specific polymerase chain reaction (PCR) and genotyping
| Primer name | Sequence | Temp °C | Primer name | Sequence | Temp °C |
|---|---|---|---|---|---|
| Exon 7 seq | GCTCTCAGGATAAAATGTTC | 55 | Clen37 For | GCATTTGGAGCAAGATTTCCCTACT | 60 |
| Exon 6 seq | CCCGCTATAATCACTAGAGC | 55 | Clen37 Rev-tailed | † GCCAACACCTCTAATTAGCTCTGAA | 60 |
| ex9 LR For | TTGCTTGTAATCTGCCAGATGTGGT | 65 | D7S2201For | GGTTCACACCTGAAATCCCAACACT | 60 |
| ex9 LR Rev | ATCTACTTTCTCCCTTGGTTGACAT | 65 | D7S2201Rev-FAM | TCTGTACCCAATGTAGAGCAGGACA | 60 |
| indel wt for | ATTCCTTTTGTTCAGCTGCCCCCTA | 65 | D7S2478 For-tailed | † CAGATTCCATATGCAATCCCCATCA | 60 |
| indel mut for | TTTTGTTCAGCTGTGTGTGTGAAGA | 65 | D7S2478 Rev | CGTGCTCCGCCATTTCTGTATACTT | 60 |
| indel wt rev | TCACACACGGAGTCACTAGGGGGCA | 65 | rs12702460seq | AGAAGTTCAACCACATCTGGCAGAT | 60 |
| indel mut rev | CACGGAGTCACTCTTCACACACACA | 65 | rs12702463seq | GCGTTGAAGCAATTCTCCTACCTTA | 60 |
| exon 5 AS For | CGAAGGTTGGAACTCGACTGATGTT | 65 | rs6949598seq | AGATTCAAGCAATTCTCCTGCCTCA | 60 |
| exon 9 AS Rev | TGGTGTCGATTATACATGTGGTAGA | 65 | rs7793254seq | TTCTTGAGACGGAGTCTTGCTTTGT | 60 |
| Clen36 For | AAGGCTGGGTCCAGTAGTGGGA | 60 | rs7788441seq | TTTCCAGAAGAACACCACCTTCACA | 60 |
| Clen36 Rev-tailed | * CATCTTGCTTCCTTAAGGTCTGTCA | 60 | rs7805798seq | TTTCCAGAAGAACACCACCTTCACA | 60 |
| D7S481 For | TAGCGTCTAGTCAGCTACCGTATTA | 60 | SNP-MC1† For/Seq | GTTTATGGCTACACTCCTGTCTAGT | 60 |
| D7S481 Rev-tailed | * CAAAATAGCTAGACACCACCCCACT | 60 | SNP-MC1† Rev | CCCTTTGTACTCTCCCTCACTGAGA | 60 |
| Clen35 For | AATTAGCTGGGCGTGGTACCAGGCA | 60 | rs1468996 For/Seq | AGAAATACAGTTCTTAGTTGGTGGA | 60 |
| Clen35 Rev-tailed | * CCTGTGGGAAGAACGAAGTGTTTCT | 60 | rs1468996 Rev | TTCTCTGTAGCTGCGTAGCTTGTGT | 60 |
| FAM labelled M13 | TGTAAAACGACGGCCAGT | 60 |
Primers have a common tail sequence 5′-tgtaaaacgacggccagt-3′.
52 bp upstream of rs1974766.
Diploid-to-haploid conversion
Haploid converted clones from patient 1 and the sister of patient 3 were created commercially (Mayo Clinic, Rochester, Minnesota, USA; www.mayoclinic.org) using the conversion technology of Yan et al.13
Allele specific amplifications
Single nucleotide polymorphisms (SNPs) in close proximity to the indel mutation were typed on the mutant allele via long range PCR, using standard protocols and allele specific primers, followed by direct sequencing. Allele discriminating primers were positioned at the site of the indel mutation with the second primers being positioned within exons 5 and 9 (see table 1 for primer sequences).
Haplotype analysis
A combination of three novel (Clen35 (BV725467), Clen36 (BV725468), Clen37 (BV725469)) and three DeCode (D7S481 (Z16478), D7S2201 (G08627), D7S2478 (Z53195)) microsatellite markers (see table 1 for primer sequences) were used to generate genotypes spanning ~1 Mb across the PMS2 locus. Markers were typed in diploid DNA and haploid clones generated from two mutation carriers. Markers were typed either by direct labelling of a PCR primer or by utilising a labelled M13 primer in conjunction with an M13-tailed, amplicon specific, primer in a three primer PCR. Each 25 μl PCR reaction contained 12.5 μl of HotStarTaq PCR mix (Qiagen), 25 ng of genomic DNA, 10 pmoles of each primer (for the three primer PCR, 2 pmoles of tailed primer and 10 pmoles of FAM labelled M13 primer were used). Reactions were multiplexed when possible and cycled using the following profile: 96°C for 15 min, 30 cycles (50 cycles were used for the three primer PCR) of 96°C for 30 s, 60°C for 30 s and 72°C for 30 s, and a final extension at 72°C for 10 min. Products were sized using an ABI7000.
Estimating the age of the indel mutation
The DMLE+2.2 software developed by Reeve and Rannala14 was utilised to estimate the age of the indel mutation. The program, which is freely available from www.dmle.org, uses a Bayesian approach to compare differences in linkage disequilibrium, between the mutation and flanking markers, among DNA samples from mutation carriers and unrelated normal controls. In addition to the genotype data, marker locations, population growth rates, and an estimate for the proportion of disease bearing chromosomes being analysed are used by the software.
RESULTS
The notion that PMS2 mutations are rarely involved in the onset of Lynch syndrome has been advocated for the last 10 years, and this has been supported in some part by the sparsity of PMS2 specific mutations recorded in the literature.15 To address this problem, we developed a relatively simple assay which enabled us to identify PMS2 specific mutations in a way that minimises the risk of making false diagnoses as a result of interference from pseudogenes.
Among the deleterious mutations we identified during this study was a frequently occurring insertion/deletion mutation (c.736_741del6ins11). This mutation causes an alteration of the PMS2 protein sequence from residue 246 with a termination signal being created at residue 249, some 614 amino acids before the wild type stop codon. We initially identified this mutation in two Swedish patients and an American patient who had a grandparent of Swedish origin. This geographical connection along with the identification of a rare polymorphism (rs2228007), within exon 11, in all three individuals led us to hypothesise that this mutation may have prevailed as a result of a founding event among the population of Sweden. In our subsequent studies we have identified this same mutation in an additional nine probands from various geographical locations throughout North America and Australasia.
Clinical overview of indel families
The mean age for developing a Lynch syndrome associated cancer among the probands was 52. Their families did not have significant incidences of Lynch syndrome associated cancers, with only one of the families meeting the Amsterdam criteria and six of them meeting the revised Bethesda criteria. When possible, family members, regardless of their cancer status, were screened for the mutation. Data obtained from these family studies (table 2) suggest that this mutation has reduced penetrance. We were able to obtain patient defined ancestral origins for nine of the 12 probands, which indicates a high prevalence of English as well as Swedish ancestry. Two of the three patients without detailed ancestry were from Australia and the third was an African American. Although we cannot be certain, historical records of these populations, especially in the case of Australians, indicate that their ancestries are significantly more likely to be English rather than Swedish (http://www.censusdata.abs.gov.au).
Table 2.
Clinical attributes of the 12 patients who tested positive for the indel
| Patient | Proband location | Sex/age at diagnosis | Site of cancer | Criteria met‡ | No. of affected* parents | No. of affected* siblings | Microsatellite status unstable/No. typed | de novo status | Ancestral information† |
|---|---|---|---|---|---|---|---|---|---|
| 1 | USA | ♂ 49 years | Rectum | Bethesda | 0 | 0(1) | 6/9 | Unknown | pat—Ger; mat—Swe/Nor |
| 2 | Canada | ♀ 44 years | Ascending | Bethesda | 0 | 1(14) | 2/2 | Unknown | pat—Eng; mat—Eng/USA |
| 3 | USA | ♂ 46 years | Splenic flexure | Bethesda | 1 | 0(9) | 5/5 | Mother was an obligate carrier | African American |
| 4 | Sweden | ♂ 54 years | Ascending | None | 1 | 1(1) | 6/6 | Father carried the mutation | Swedish |
| 5 | Sweden | ♀ 51 years | Descending | None | 0 | 0(2) | 2/5 | Unknown | Swedish |
| 6 | Canada | ♀ 74 years | Ascending | Bethesda | 1 | 1(2) | 5/5 | Sister carries the mutation | English |
| 7 | USA | ♂ 57 years | Caecum | None | 0 | 0(3) | 8/10 | Unknown | pat—Irish/English; mat— n/a |
| 8 | USA | ♂ 58 years | Caecum | None | 0 | 0(2) | 9/10 | Unknown | Swedish |
| 9 | USA | ♀ 51 years | Caecum | None | 0 | 0(5) | 8/10 | Unknown | pat—Swe/Ger; mat—Eng |
| 10 | Canada | ♂ 29 years | Rectum | Bethesda | 0 | 0(1) | 5/6 | Unknown | English/Canadian |
| 11 | Australia | ♀ 67 years | Stomach | Amsterdam II | 1 | 2(8) | 3/4 | Brother carries the mutation | Australian |
| 12 | Australia | ♀ 48 years | Caecum | Bethesda | 0 | 0(2) | 8/9 | Sister carries the mutation | Australian |
Affected with any of the following types of cancer: colorectal, endometrial, stomach, ovarian, pancreas, ureter and renal pelvis, biliary tract, brain, small bowel, sebaceous adenoma and keratoacanthoma.
pat, paternal side; mat, maternal side; Ger, German; Swe, Swedish; Nor, Norwegian; Eng, English; Can, Canadian; n/a, data not available.
Mutation analysis and control screening
Complete sequencing of the PMS2 gene in all probands identified no additional deleterious variants, indicating that the indel mutation is the causative factor for the observed phenotype. Analysis of haploid clones demonstrated that a mutant transcript is generated but no full length protein could be detected (data not shown). We were also unable to detect any truncated protein through the use of an N-terminal antibody which rules out a dominant negative mechanism being responsible for the loss of function of PMS2 (data not shown). In addition to the indel mutation, all 12 probands were shown to have the minor allele variant at codon 1531 (rs2228007; Coriell - A:0.95/G:0.05, n = 96) and this was shown to reside on the same allele as the indel mutation in both haploid clones.
A panel of 399 control DNAs (225 Swedish blood donors, 92 Coriell controls, and 82 African American controls—the latter a gift from Dr Rick Kittles (The University of Chicago)) were screened for the presence of the indel mutation. This control screen showed that a single, anonymous sample from the Swedish blood donor set carried the mutation; it was also shown that the same sample carried the minor allele at rs2228007 (allele frequency was 0.027 in the Swedish control samples).
Haplotype analysis
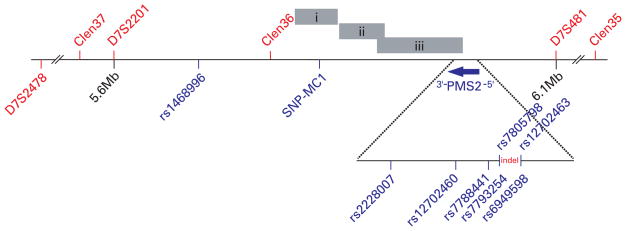
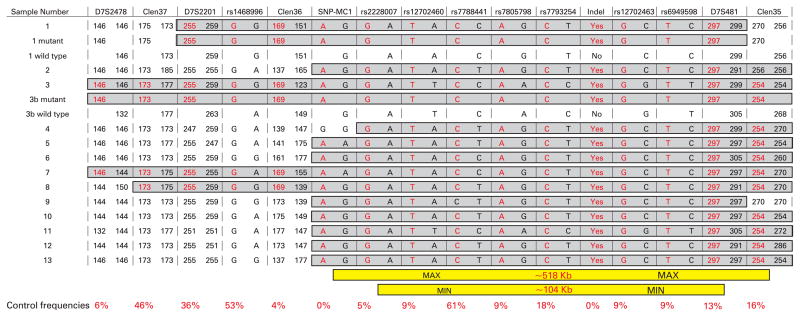
We sought to identify the extent of any putatively conserved haplotype through the typing of additional SNPs and micro-satellite repeat elements. It should be noted that, as with mutation detection, the identification of unique microsatellite sequences is hampered by extensive homologous sequences. The extent of these sequences is so great that no unique sequence, harbouring a microsatellite repeat, could be identified within 200 kb of the 3′ end of PMS2 (fig 1). For the haplotype analysis we utilised three database and three novel microsatellite markers which span ~1 Mb across the PMS2 locus. We also assessed a set of seven SNPs, five of which could be typed using allele specific PCR and as such enabled us to decipher a true mutant haplotype, in all samples, for a region spanning from exon 5 to exon 9 of PMS2. To get a full length haplotype we typed all markers in haploid cell lines obtained from two individuals (patient 1 and the sister of patient 3, who also carried the mutation). Typing of all markers in the 13 individuals (12 patients and one control sample) with the indel mutation identified a common haplotype of between 104 kb (rs2228007 – D7S481) and 518 kb (SNP-MC1 – Clen35) (fig 2). Although the 3′ end of the haplotype is broken by the absence of an SNP in a single sample (patient 4), which could be put down to mutation, the haplotype is lost by eight more of the 13 individuals a further 60 kb downstream as adjudged by the closest 3′ flanking microsatellite (Clen36).
Figure 1.
Genomic region flanking PMS2. Microsatellite and single nucleotide polymorphism (SNP) markers used for the haplotype analysis are shown in red and blue fonts, respectively. Chromosomal fragments which show high levels of homology to the 3′ end of PMS2 are represented by grey bars; (i) chr 7: 5,126,099…5,174,845 bp; (ii) chr 7: 97,435,392…97,486,400 bp; (iii) chr 7: 6,741,350…6,839,016 bp. Physical locations are based on NCBI’s reference build (ver36.2).
Figure 2.
Genotype data spanning the PMS2 locus of 17 DNAs. The 17 DNAs comprise 12 patients, one blood donor control (13), and four haploid clones. Wild type and mutation bearing haploid clones were typed for both patient 1 and the sister of patient 3 (3b). Disease associated genotypes are highlighted in grey, with the disease allele depicted in red. The size of the minimum and maximum conserved haplotypes along with the frequency of the disease associated allele among a panel of control DNAs are shown at the bottom.
Estimating the age of the mutation
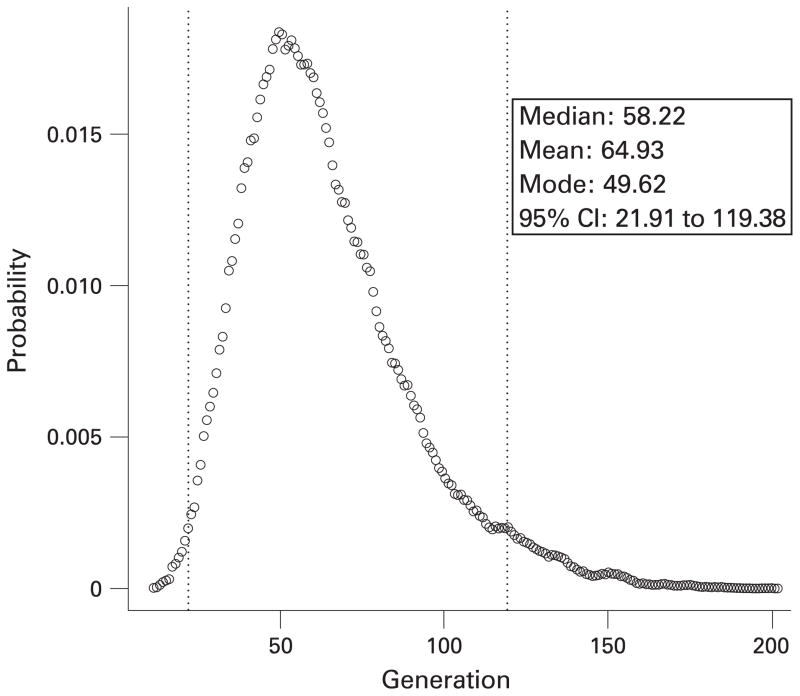
It has been shown that the rate of recombination and population frequency of flanking marker alleles can be used to estimate the age of a conserved haplotype. In this study we utilised a well established method that considers the linkage disequilibrium across all markers in a single analysis, and is implemented in the DMLE+2.2 software. Marker locations were obtained from the human genome reference sequence, and converted to map distances using a conversion factor of 1.67 cM/Mb which was obtained from a comparison of Centimorgan and megabase values for the 7p22 region (based on deCODE mapping data18). We used a population growth rate of 1.05 fold per generation based on European estimates.19 To calculate the proportion of disease bearing chromosomes being studied we extrapolated PMS2 specific data for both the USA and Sweden, from the literature and our own unpublished data (USA: 6% lifetime risk of CRC (American Cancer Society; http://www.cancer.org/downloads/STT/CAFF2006PWSecured.pdf), 2.8% of CRC are Lynch syndrome (authors’ unpublished data)20; Sweden: 5% lifetime risk of CRC, 1.2% of CRC are Lynch syndrome21), to calculate the number of Lynch syndrome cases. Using these figures and our own data for the incidence of the indel we estimated that there would be ~10 874 cases of this mutation in the USA and ~181 cases in Sweden, based on the population sizes of the 2000 censuses. The DMLE+2.2 software predicted an age of ~1625 years (65 generations; 95% confidence interval 22 to 120) (fig 3), which would suggest that this indel mutation arose sometime during the first millennium.
Figure 3.
Age estimates for the haplotype associated with the indel mutation. The posterior probability density of the age (in generations), as estimated by the software DMLE+2.2, is shown when a population growth rate of 1.05-fold per generation (25 years) is assumed. The dotted lines show the 95% confidence intervals for the calculation.
DISCUSSION
We show here that many PMS2 mutations occur, but the determination of the true proportion of all cases of Lynch syndrome that are due to PMS2 mutations must await further, larger, population based studies; nevertheless, our estimate for the present number of carriers of the indel mutation (>10 000 in the USA alone) does implicate PMS2 as a potentially important gene to consider in clinical practice. The clinical and family history data that we were able to assemble and critically evaluate (table 2) do not allow us to assess precisely the penetrance of this mutation. An apparent lack of disease segregation among the affected families (there was no Lynch syndrome associated cancer in either parent for eight of the 12 probands) is a strong indication that this mutation, along with many other truncating mutations within PMS2 (authors’ unpublished data), has reduced penetrance in comparison to the other mismatch repair genes. Lynch syndrome has been defined as a heritable, high penetrance, predisposition to cancer. If some mutations, such as the one described here, carry a lower penetrance, they may be seen not to fall under this definition of Lynch syndrome. Once the penetrance of PMS2 mutations can be more precisely defined, their role in Lynch syndrome will become better established and modifications to the counselling and clinical management of carriers of these mutations may be called for.
A mutation that is seen in many ostensibly unrelated individuals can be either recurrent—that is, it arises repeatedly de novo—or it represents an ancestral event that occurs in the population. In this case the highly conserved haplotype that occurs in all patients virtually excludes a recurrent event. The alternative would be that the associated haplotype somehow predisposes to this particular mutation, a highly unlikely assumption. It follows that even though we were not able to investigate DNA from most parents of the probands, it is likely that in each case one of them carried the mutation.
Our calculations, based on a conserved haplotype of ~520 kb (~0.85 cM), estimated that this mutation originated around 1625 years ago. Although these kinds of estimates tend to have broad margins of error, it is consistent with the ages predicted for similarly sized haplotypes.22,23 Many of the earlier studies along these lines have studied disease in distinct populations such as the Ashkenazi Jews, Icelanders and Finns,23,24 and so they have been able to support their age estimates with historical records of population movements. With our data we are unable to make such reassuring comparisons due to the patient cohort being heterogeneous, with several having limited or no information regarding their ancestral background. Without further data it would be unwise to make firm conclusions as to the geographical origins of this indel mutation; however, our current data would indicate that this mutation originated at a time when there was significant interaction between people of Scandinavian ancestry and those of British ancestry, which could explain our findings that the mutation occurs primarily in people of English and Swedish descent.
The identification of the indel in a control sample (Swedish blood donor) highlights further that this mutation is of significant prevalence; although our clinical findings would indicate that this mutation, like several other reported mutations within PMS2,25,26 has a reduced penetrance, it is in our opinion that this indel could be responsible for a significant proportion of cases of Lynch syndrome that are associated with an aberration in PMS2. It is also worth considering the long term impact of this kind of PMS2 mutation on the affected populations. Over time, if the number of heterozygous carriers increases in the population, we would expect to see a rise in the number of homozygous mutation carriers for PMS2 which presents as a severe childhood cancer syndrome typified by Turcot’s syndrome.27–29
Unlike many other mutations in PMS2, this alteration is located within a region not complicated by pseudogenes; it can therefore be easily screened for by several methods including, but not limited to, direct sequencing, multiplex ligation dependent probe amplification (MRC Holland), and allele size screening, either on an ABI7000 utilising a fluorescently labelled primer or by high resolution agarose electrophoresis.
Acknowledgments
We thank Wendy Frankel MD, Ilene Comeras RN, Shili Lin PhD, and Terry Speed PhD for their contributions to this study. This work was supported by grants CA67941, CA16058, and CA96011 from the National Institutes of Health, grant 05-0264 from the Swedish Cancer Society, and through cooperative agreements with the Ontario Familial Colorectal Cancer Registry, Australasian Colorectal Cancer Registry, Mayo Colorectal Cancer Registry, and the Seattle Familial Colorectal Cancer Family Registry.
Footnotes
Competing interests: None declared.
Patient consent: Informed consent was obtained from the patients for publication of this report.
References
- 1.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci U S A. 1995;92:1950–4. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch HT, Schuelke GS, Kimberling WJ, Albano WA, Lynch JF, Biscone KA, Lipkin ML, Deschner EE, Mikol YB, Sandberg AA, Elston RC, Bailey-Wilson JE, Danes BS. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II). II. Biomarker studies. Cancer. 1985;56:939–51. doi: 10.1002/1097-0142(19850815)56:4<939::aid-cncr2820560440>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki A, Peltomaki P, Mecklin JP, Jarvinen H, Salovaara R, Nystrom-Lahti M, de la Chapelle A, Aaltonen LA. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat Genet. 1994;8:405–10. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, Roylance R, Houlston RS. Two hits revisited again. J Med Genet. 2001;38:81–5. doi: 10.1136/jmg.38.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, Benitez J, Caldes T, Akiyama Y, Yuasa Y, Launonen V, Canal MJ, Rodriguez R, Capella G, Peinado MA, Borg A, Aaltonen LA, Ponder BA, Baylin SB, Herman JG. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–7. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 7.Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–40. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 8.Nicolaides NC, Carter KC, Shell BK, Papadopoulos N, Vogelstein B, Kinzler KW. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 9.De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–64. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa H, Lockman JC, Frankel WL, Hampel H, Steenblock K, Burgart LJ, Thibodeau SN, de la Chapelle A. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–7. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 11.Gill S, Lindor NM, Burgart LJ, Smalley R, Leontovich O, French AJ, Goldberg RM, Sargent DJ, Jass JR, Hopper JL, Jenkins MA, Young J, Barker MA, Walsh MD, Ruszkiewicz AR, Thibodeau SN. Isolated loss of PMS2 expression in colorectal cancers: frequency, patient age, and familial aggregation. Clin Cancer Res. 2005;11:6466–71. doi: 10.1158/1078-0432.CCR-05-0661. [DOI] [PubMed] [Google Scholar]
- 12.Clendenning M, Hampel H, LaJeunesse J, Lindblom A, Lockman J, Nilbert M, Senter L, Sotamaa K, de la Chapelle A. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–5. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, Papadopoulos N, Marra G, Perrera C, Jiricny J, Boland CR, Lynch HT, Chadwick RB, de la Chapelle A, Berg K, Eshleman JR, Yuan W, Markowitz S, Laken SJ, Lengauer C, Kinzler KW, Vogelstein B. Conversion of diploidy to haploidy. Nature. 2000;403:723–4. doi: 10.1038/35001659. [DOI] [PubMed] [Google Scholar]
- 14.Reeve JP, Rannala B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics. 2002;18:894–5. doi: 10.1093/bioinformatics/18.6.894. [DOI] [PubMed] [Google Scholar]
- 15.Liu T, Yan H, Kuismanen S, Percesepe A, Bisgaard ML, Pedroni M, Benatti P, Kinzler KW, Vogelstein B, Ponz de Leon M, Peltomaki P, Lindblom A. The role of hPMS1 and hPMS2 in predisposing to colorectal cancer. Cancer Res. 2001;61:7798–802. [PubMed] [Google Scholar]
- 16.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–6. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 18.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 19.Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol. 1999;16:1791–8. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- 20.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 21.Olsson L, Lindblom A. Family history of colorectal cancer in a Sweden county. Fam Cancer. 2003;2:87–93. doi: 10.1023/a:1025734200635. [DOI] [PubMed] [Google Scholar]
- 22.Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–9. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zabetian CP, Hutter CM, Yearout D, Lopez AN, Factor SA, Griffith A, Leis BC, Bird TD, Nutt JG, Higgins DS, Roberts JW, Kay DM, Edwards KL, Samii A, Payami H. LRRK2 G2019S in families with Parkinson disease who originated from Europe and the Middle East: evidence of two distinct founding events beginning two millennia ago. Am J Hum Genet. 2006;79:752–8. doi: 10.1086/508025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkardottir RB, Sarantaus L, Arason A, Vehmanen P, Bendahl PO, Kainu T, Syrjakoski K, Krahe R, Huusko P, Pyrhonen S, Holli K, Kallioniemi OP, Egilsson V, Kere J, Nevanlinna H. Haplotype analysis in Icelandic and Finnish BRCA2 999del5 breast cancer families. Eur J Hum Genet. 2001;9:773–9. doi: 10.1038/sj.ejhg.5200717. [DOI] [PubMed] [Google Scholar]
- 25.Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, Yurtsever H, Neuweiler J, Riehle HM, Cattaruzza MS, Heinimann K, Schar P, Jiricny J, Marra G. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–71. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 26.Hendriks YM, Jagmohan-Changur S, van der Klift HM, Morreau H, van Puijenbroek M, Tops C, van Os T, Wagner A, Ausems MG, Gomez E, Breuning MH, Brocker-Vriends AH, Vasen HF, Wijnen JT. Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome) Gastroenterology. 2006;130:312–22. doi: 10.1053/j.gastro.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 27.Agostini M, Tibiletti MG, Lucci-Cordisco E, Chiaravalli A, Morreau H, Furlan D, Boccuto L, Pucciarelli S, Capella C, Boiocchi M, Viel A. Two PMS2 mutations in a Turcot syndrome family with small bowel cancers. Am J Gastroenterol. 2005;100:1886–91. doi: 10.1111/j.1572-0241.2005.50441.x. [DOI] [PubMed] [Google Scholar]
- 28.Worthley DL, Walsh MD, Barker M, Ruszkiewicz A, Bennett G, Phillips K, Suthers G. Familial mutations in PMS2 can cause autosomal dominant hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;128:1431–6. doi: 10.1053/j.gastro.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.De Vos M, Hayward BE, Charlton R, Taylor GR, Glaser AW, Picton S, Cole TR, Maher ER, McKeown CM, Mann JR, Yates JR, Baralle D, Rankin J, Bonthron DT, Sheridan E. PMS2 mutations in childhood cancer. J Natl Cancer Inst. 2006;98:358–61. doi: 10.1093/jnci/djj073. [DOI] [PubMed] [Google Scholar]