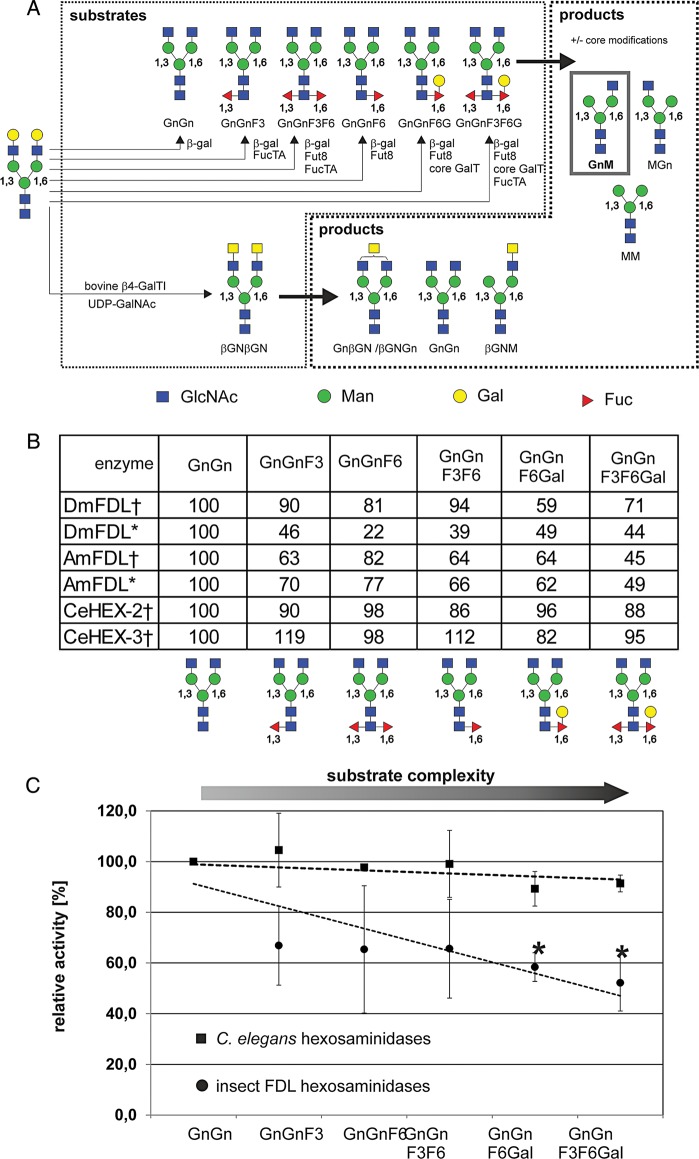
Fig. 5.
Production of substrates used in this study and the sensitivity of insect FDL enzymes to N-glycan core modifications. (A) Substrates with and without complex core modifications. Dabsylated-GalGal glycopeptide was incubated with the Aspergillus nidulans galactosidase rlacA (Dragosits et al. 2014) in order to remove the galactose residues, followed by treatment with the A. thaliana FucTA (Wilson et al. 2001), C. elegans FUT-8 (Paschinger et al. 2005) and the recombinant N-glycan core β1,4-galactosyltransferase (Titz et al. 2009) as described in the Materials and methods section. An aliquot of the dabsylated-GnGn glycopeptide was treated with bovine β1,4-galactosyltransferase I in the presence of UDP-GalNAc to create the dabsylated-βGNβGN glycopeptide. Enzymes were used sequentially as indicated in order to obtain several complex core modifications. (B) Efficiency of Caenorhabditis and insect FDL(-like) enzymes for dabsylated-GnGn glycopeptide substrates with core modifications was tested. Values show relative efficiency with dabsylated-GnGn glycopeptide set to 100% efficiency in product formation (dabsylated-GnM) as estimated by the area of the relevant peaks in MALDI-TOF/TOF MS spectra. Average values of duplicate measurements are shown. (C) Overall impact of core modifications (substrate complexity) on the activity of C. elegans hexosaminidases (HEX-2/3, squares) when compared with insect FDL enzymes (DmFDL and AmFDL, circles). Values represent averages±standard deviation. Linear regressions (dashed lines) were used to depict the trend of substrate conversion. Activities on substrates with a significant decrease (p < 0.005) as compared with GnGn are marked with an asterisk. Dagger indicates the enzyme produced in P. pastoris and asterisk indicates the enzyme produced in High Five insect cells.