Abstract
A fundamental goal in understanding the mechanisms of autoimmune disease is the characterization of autoantigens that are targeted by autoreactive antibodies and T cells. Unfortunately, the identification of autoantigens is a difficult problem. We have begun to explore a novel route to the discovery of autoantibody/autoantigen pairs that involves comparative screening of combinatorial libraries of unnatural, synthetic molecules for compounds that bind antibodies present at much higher levels in the serum of individuals with a given autoimmune disease than in the serum of control individuals. We have shown that this approach can yield “antigen surrogates” capable of capturing disease-specific autoantibodies from serum. In this report, we demonstrate that the synthetic antigen surrogates can be used to affinity purify the autoantibodies from serum and that these antibodies can then be used to identify their cognate autoantigen in an appropriate tissue lysate. Specifically, we report the discovery of a peptoid able to bind autoantibodies present in about one-third of nonobese diabetic (NOD) mice. The peptoid-binding autoantibodies were highly enriched through peptoid affinity chromatography and employed to probe mouse pancreatic and brain lysates. This resulted in identification of murine GAD65 as the native autoantigen. GAD65 is a known humoral autoantigen in human type 1 diabetes mellitus (T1DM), but its existence in mice had been controversial. This study demonstrates the potential of this chemical approach for the unbiased identification of autoantigen/autoantibody complexes.
Introduction
A central issue in the study of autoimmune disease is the identification of autoantigens recognized by the humoral or cellular adaptive immune responses. This is often a difficult problem. Many efforts directed toward the discovery of autoantibody–autoantigen complexes focus on mixing serum samples from case or control individuals with some panel of autoantigen candidates, then identifying which of these candidates retain far more antibody from the case samples than from the controls. These panels can be proteome arrays,1 peptide arrays, lipid arrays,2 phage-displayed cDNA libraries,3 or other collections of biomolecules formatted in a variety of ways. Obviously, such experiments will work only if the autoantigen is among the candidates included in the panel, and this will not always be the case.
We have begun to explore an alternative strategy that substitutes large numbers of synthetic, unnatural molecules for the autoantigen candidate panel.4,5 It has long been known that antibodies can bind selectively to ligands that are structurally distinct from their native antigen partners, for example peptide “mimotopes” of carbohydrate antigens.6 Our efforts are an extension of this concept to far more chemically diverse combinatorial libraries containing many different motifs not found in nature. The hope is that differential screening of case and control serum samples against such a library would result in the identification of synthetic “antigen surrogates” that bind disease-linked antibodies well enough to pull them out of the serum, even though the compound could not possibly act as a structural mimic of the bona fide autoantigen. The antigen surrogate, or more likely an optimized derivative, could be employed as a “capture agent” in ELISA-like assays of potential diagnostic utility. Moreover, it might be possible to employ the synthetic compound to affinity purify the antibodies it recognizes which could, in turn, be mixed with an appropriate tissue lysate to pull out the native autoantigen, providing a “back door” route to the discovery of disease-specific autoantigens.
We have demonstrated the feasibility of the differential screening step in a study using serum samples obtained from patients with neuromyelitis optica (NMO), an autoimmune disease in which autoantibodies against aquaporin 4 (AQP4) drive demyelination of the optic nerve. From a library of 100 000 hexameric peptoids, a compound was isolated that bound antibodies present at much higher levels in the sera of most NMO patients than in serum obtained from control individuals. It was then shown that the peptoid-binding antibodies were indeed anti-AQP4 IgGs.4
In this study, we apply this technology to type 1 diabetes mellitus (T1DM). T1DM is a chronic autoimmune disease characterized by a T cell mediated immune response to pancreatic β-cells.7,8 There is also a humoral response. Over the past four decades, intense research efforts have uncovered a few major islet cell antigens (ICAs) such as the 65 kDa isoform of glutamic acid decarboxylase (GAD65);9 protein tyrosine phosphatase, receptor type, N (PTPRN, also known as insulinoma antigen-2 (IA-2));10−14 and zinc transporter 8 (ZnT8).15 The nonobese diabetes (NOD) mouse has been adopted as a popular model of spontaneous diabetes.16 NOD mice are an inbred Swiss strain that harbor mutations within an ortholog to the human T1DM-susceptibility locus and therefore share key pathological hallmarks with human T1DM.
We report here the isolation of a peptoid17 from a comparative screen that binds antibodies present at much higher levels in the serum of some NOD mice than most control mice. Most importantly, we demonstrate that this peptoid can be employed as an affinity reagent to enrich its antibody binding partner from serum. When this enriched antibody population was incubated with murine pancreatic and brain extracts, we found that it bound to the 65 kDa isoform of glutamic acid decarboxylase (GAD65). Interestingly, GAD65 is a known humoral autoantigen in human T1DM patients but was not thought to be so in NOD mice,18−22 a difference that has led to suggestions that the NOD mouse may be an incomplete model of human disease.20 This completely unbiased discovery of the GAD65 autoantigen in NOD mice demonstrates the utility of this chemical approach to elucidating molecular aspects of autoimmune disease.
Results and Discussion
Discovery of NOD Mouse IgG Ligand from a Small Molecule Library
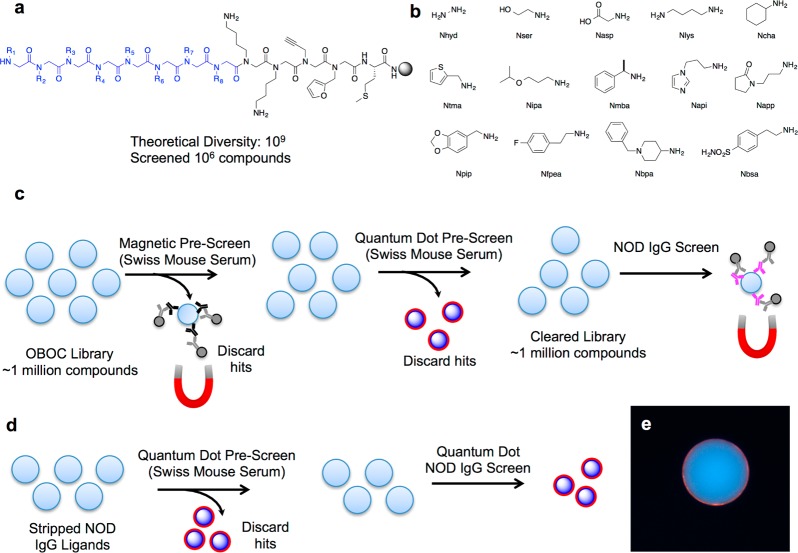
A peptoid library23,24 was subjected to differential screening4 between case and control samples in an attempt to discover ligands for IgG antibodies present in the NOD mouse. Split-and-pool solid-phase synthesis methodology25 was employed to create a diverse one-bead one-compound (OBOC) library of peptoids (Figure 1) comprised of ∼1 000 000 unique compounds. The invariant linker, shown in black in Figure 1a, was common to all library members. A methionine unit in the linker facilitated cleavage from the resin with CNBr postscreening. Furfuryl and propargyl groups were included to act as functional handles for future modifications, if desired. The aminobutyl groups (Nlys) served to enhance ligand solubility in aqueous solutions. The variable positions, shown in blue in Figure 1a, were synthesized by bromoacetylating the terminal amine on all beads followed by splitting the pool of bromoacetylated beads into 14 aliquots and displacement of bromide with one of the amines shown in Figure 1b. This process was repeated until eight monomeric units had been installed.
Figure 1.
Library design and screening strategy for ligands to autoimmune IgG. (a) Peptoid library scaffold with the invariant region in black and the variable positions in blue. (b) Primary amines used to install the variable region R groups via amination. The four-letter codes indicated are for the peptoid monomer derived from the primary amine. Using 14 amines, a library with a theoretical diversity of 109 was synthesized, and 1 000 000 compounds were screened. (c) Primary screen schematic. Ligands on beads that bound to healthy control antibodies were removed via magnetic pull-down after hybridizing with anti-IgG conjugated to magnetic DynaBeads. This pull-down of uninteresting ligands to control antibodies was repeated. A third control screen was performed, this time visualizing hits using red quantum dots. The library, having been completely denuded of ligands that bind to normal serum IgG, was incubated with NOD mouse serum, and hits were removed by magnetic pull down. (d) NOD mouse “hits” from the magnetic pull-down were stripped of all bound protein with organic solvents and rescreened using a red quantum dot-conjugated secondary antibody. Hits were removed from the library, and the bound ligand was ascertained by tandem mass spectrometry. (e) Representative photomicrograph of a “hit” bead displaying a characteristic red halo after incubation with fluorescent secondary antibody.
To identify ligands from the OBOC library that bind to NOD mouse IgG antibodies, the library was first cleansed of compounds that bind to antibodies abundant in the serum of control mice. A pooled sample (100 μg mL–1 total protein) of serum obtained from eight week-old Swiss mice was incubated with the beads. After washing, IgG antibody-binding beads were removed by incubation with IgG-coated iron oxide particles, and the magnetized TentaGel beads were removed using a strong magnet. This step was repeated again to attempt to remove uninteresting ligands from the library to the greatest extent possible. Indeed, because we have found that the magnetic “pullout” protocol is not 100% efficient (Mendes et al. in preparation), a third incubation with control serum was conducted, but in this case, IgG-binding beads were visualized by incubating the beads with red quantum dots (QDot655), permitting visual inspection of the hit beads followed by manual removal using a micropipette.26 These prescreening steps resulted in the removal of a few thousand beads.
The remainder of the library was screened against a single NOD mouse serum, which we called NOD18. This sample was obtained from a mouse 8 weeks of age that was still in the prodromal phase of the disease (hyperglycemia was not evident until 14 weeks of age). The serum (50 μg mL–1 total protein) was preincubated with insulin to block binding of anti-insulin autoantibodies to the library since we were interested in the identification of potentially novel species and anti-insulin antibodies are well-known to be present in NOD mice.21 After incubation with 50 μg mL–1 NOD18 serum, the library was incubated with anti-IgG-coated magnetic particles, and beads bearing hit ligands were removed after exposure to a strong magnet. The hits were stripped of bound proteins by incubating with aqueous acetonitrile. The stripped beads were then resubmitted to the screening conditions shown in Figure 1d using quantum dot-conjugated anti-IgG for visualization of hits. Beads that displayed a red halo, such as the one depicted in Figure 1e, were collected, stripped again, and released from the beads using CNBr. The sequence of each hit was determined by tandem MALDI-TOF mass spectrometry (MS). The MS data showed that four compounds were identified as ligands (Supporting Information Figure S2). One of the four compounds was isolated twice from the screen (compound 1, Figure 2a). Since false positives are quite common in these bead screens but “repeat hits” are generally bona fide IgG ligands,27 we chose to focus initially on the further characterization of this peptoid.
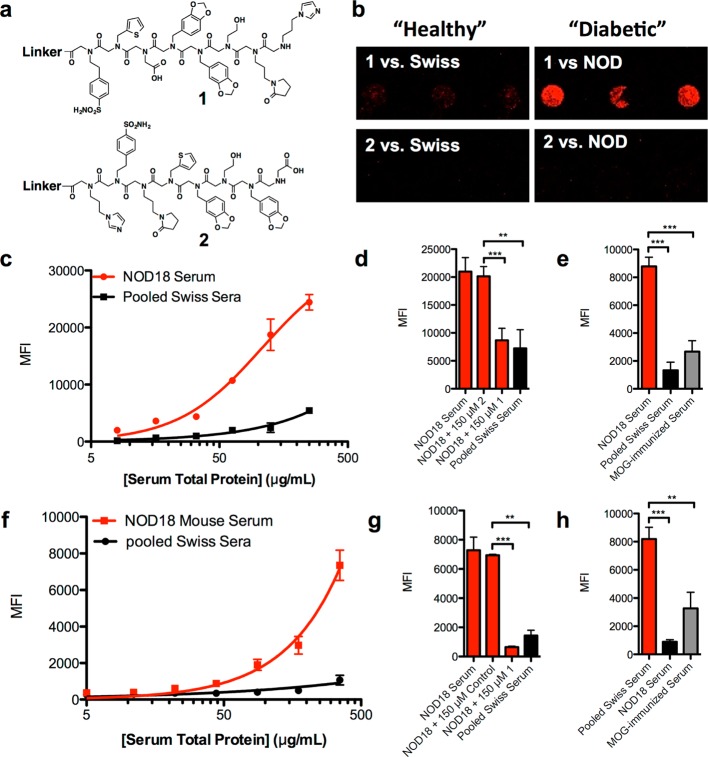
Figure 2.
Binding characterization of repeat hit 1. (a) Chemical structure of resynthesized hit 1 and control compound 2. (b) Representative fluorescence micrograph of a slide containing immobilized 1 and 2 after incubation with 125 μg mL–1 Swiss control mouse serum or autoimmune NOD mouse serum followed by fluorescent anti-IgG hybridization. (c) Binding titration curves generated using microarray analysis on glass slides containing 1 with varying concentrations of control or disease mouse serum. (d) Competitive binding was performed by preincubating 1 or 2 with disease mouse serum prior to addition to the microarray slide. (e) Binding of IgG to 1 on microarray slides using serum from Swiss, NOD, and MOG-immunized C57BL/6 mice at 125 μg mL–1 serum. (f) Binding titrations using the flow cytometry-based assay. 1 was immobilized onto 10 μm TentaGel microspheres, and the beads were incubated with case or control serum. (g) Competitive binding of 500 μg mL–1 serum preblocked with 250 μM 1 or 2 before being added to TentaGel beads containing immobilized 1. (h) IgG binding using 500 μg mL–1 serum taken from the three different mouse strains. Sera were incubated with 1 immobilized on 10 μm TentaGel beads to assess binding of IgG from other disease models. In all cases, data are reported as the mean ± s.d. from three experiments. Statistical significance was determined using an unpaired t test: **P < 0.01; ***P < 0.001; ns = not significant.
Binding Characterization of 1 on Microarray
To validate compound 1 as a NOD IgG ligand, it was resynthesized and purified using HPLC and spotted onto a chemically modified glass slide. Peptoid 2 (Figure 2a), with the same residues as hit 1, but in a scrambled order, was also synthesized and used as a control. The complete structures, including invariant linker structure, can be found in Supporting Information Figures S3–S5. After blocking the slide with StartingBlock, NOD18 serum, or pooled Swiss mouse sera, both drawn when the mice were 8 weeks old, was hybridized to the slide in the presence of 0.5% bovine serum albumin (BSA). After washing away unbound proteins, the slide was probed with an Alexa Fluor 647-conjugated antimouse secondary antibody. As shown in Figure 2b, 1 bound significantly more IgG antibodies from NOD18 than pooled Swiss mouse serum. The scrambled control 2 failed to capture serum antibodies from either mouse strain. The titration curve shown in Figure 2c displays the mean fluorescence intensity (MFI) of each spot over a range of serum concentrations. At higher concentrations of serum, the differential binding between NOD and Swiss mouse serum increased with a maximum differential of about 8-fold realized at ∼125 μg mL–1.
It is sometimes the case that serum samples contain antibodies that deposit on surfaces in an irreversible fashion, possibly reflecting the presence of very large immune complexes or mis-folded antibodies. Therefore, it is important to carry out competition experiments to determine how much of the observed signal can be competed by a soluble version of the “capture agent”. NOD18 serum was coincubated with a high concentration (150 μM) of soluble 1 or 2 before hybridization to the slide. Figure 2d shows that soluble 1 reduced the amount of IgG retained by immobilized 1 by about 60%. The remaining signal was similar to that observed when the slide was incubated with Swiss control mouse serum. The remainder of the signal must therefore correspond to some type of nonspecific binding of IgG’s to the surface. This reduction in binding was not observed when soluble 2 was incubated with the serum. We conclude that about 60% of the signal on the array represents selective binding of serum IgG’s to peptoid 1.
To assess the selectivity of 1 as a ligand for NOD-specific antibodies, serum from C57BL/6 mice that were immunized with a peptide fragment of the myelin oligodendrocyte glycoprotein (MOG(35–55)) was analyzed. This is a popular model for multiple sclerosis.28 Figure 2e shows that 1 bound much lower levels of IgG from MOG(35–55)-immunized animals than from NOD mice.
Similar experiments were carried out on a different, more convenient, analytical platform comprised of internally dye-encoded 10 μm TentaGel beads29 to which C-terminal Cys derivatives of peptoids 1 and 2 had been coupled to the surface after activation with bromoacetic acid. In this case, the beads were incubated with serum and, after washing, Alex Fluor 647-conjugated secondary antibody. After another wash, the beads were passed through a flow cytometer, and both the identities of the ligand on each bead (via the color code) and the amount of bound antibody were determined. As shown in Figure 2f–h, selective binding of NOD antibodies to peptoid 1 was observed on this analytical platform as well.
Peptoid 1 Facilitates Tracking NOD Antibodies over Time
The flow assay was then employed to measure relative levels of the antibodies that bind to peptoid 1 in several animals over several weeks. Blood was drawn biweekly from 20 five week old NOD, Swiss, and nonobese resistant (NOR) mice for six months. The latter are a backcrossed NOD strain that express diabetes-related IgG antibodies but infrequently develop insulitis.30 Each serum sample was diluted to 500 μg mL–1 and incubated with 1 linked to 10 μm TentaGel beads to track the immune response over time using the flow cytometer-based assay.29 The mean fluorescence intensity (MFI) from all of the control Swiss mouse samples was calculated (879 ± 545 fluorescence units). By adding three standard deviations to this number, a threshold of 2514 fluorescence intensity units was established, above which a sample was called positive. The full plots containing antibody titers for all mice at all blood draws can be found in Supporting Information Figure S7. Table 1, which summarizes these data and lists a mouse as diabetic if the level of antibodies binding to 1 exceeded the threshold at any time during its life, indicates that about one-third of NOD and NOR mice and 10% of Swiss mice test positive.
Table 1. Incidence of IgG Antibody Binding to Peptoid 1 Immobilized on TentaGel Beads Determined Using a Flow Cytometry-Based Assaya,b (60 Mice Were Tested: Swiss 1–20, NOD 1–20, and NOR 1–20).
| mouse ID | Swiss | NOD | NOR |
|---|---|---|---|
| 1 | |||
| 2 | + | + | |
| 3 | + | ||
| 4 | |||
| 5 | + | + | |
| 6 | + | + | |
| 7 | |||
| 8 | + | ||
| 9 | + | ||
| 10 | + | ||
| 11 | |||
| 12 | + | ||
| 13 | + | ||
| 14 | |||
| 15 | |||
| 16 | + | + | |
| 17 | + | ||
| 18 | |||
| 19 | |||
| 20 | + | + |
IgG binding conditions: 500 μg mL–1 serum total protein in 50 μL PBS containing 0.5% BSA, 0.05% Tween 20 was incubated with the beads containing immobilized 1 for 2 h at RT.
“+” indicates that the MFI was higher than the three-sigma value of 2514 fluorescence units for at least one blood draw during the animal’s lifetime.
Peptoid 1 Is a Ligand for Anti-GAD65 Autoantibodies
From the data shown in Table 1, peptoid 1 would obviously be of limited utility as a stand-alone biomarker with low sensitivity and imperfect specificity. However, we were mainly interested in using it as a tool to purify antibodies that bind to it by affinity chromatography. Toward this end, 1 was immobilized on sepharose, and a pooled sample of sera that exhibited high reactivity toward 1 was passed over the column. After washing, the bound proteins were eluted using a high salt buffer. The presence of IgG antibody was confirmed by SDS-PAGE analysis of the eluted protein. Titration of 1 immobilized on TentaGel beads with the original serum and the affinity-purified material suggested that the antibody had been enriched at least 10-fold. The wash fractions contained little of the 1-binding antibodies (Figure 3a).
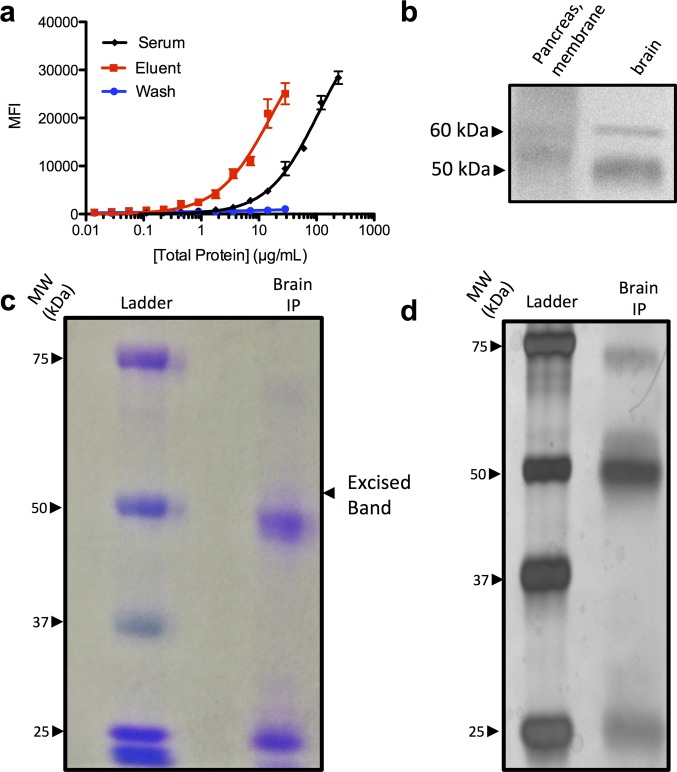
Figure 3.
Native antigen identification using enriched autoantibodies. (a) Binding titration of serum and affinity-purified eluent containing enriched peptoid-binding proteins. Antibody populations were probed against 1 immobilized onto 10 μm TentaGel microspheres. (b) Western blot analysis of SDS-PAGE separated pancreas (membrane fraction) and whole brain lysate. After transfer to nitrocellulose, the membrane was probed with enriched IgG, and bands were visualized via chemiluminescence. (c) Coomassie stained SDS-PAGE gel of brain proteins after the lysate was immunoprecipitated with enriched IgG autoantibodies. The band at ∼60 kDa was excised and submitted for LC-MS and proteomic analysis. (d) Silver stained SDS-PAGE gel of immunoprecipitated brain fraction.
Pancreas and brain tissue lysates were separated via nonreducing SDS-PAGE. After the proteins were blotted onto a nitrocellulose membrane, they were renatured in 0.1 M tris buffer containing 0.2% tween to attempt to refold any conformational epitopes that were lost during SDS treatment.31 The blot was blocked with 5% fat-free milk, and the affinity-purified IgG antibodies were hybridized to the membrane overnight. After washing, the membrane was incubated with an HRP-conjugated secondary antibody followed by development with chemiluminescent substrates. Figure 3b shows that faint bands corresponding to immune complexes at ∼50 kDa and ∼60 kDa were observed. The bands observed in the brain lysate were much stronger than those from the pancreas lysate. This tissue specificity was not entirely surprising given that T1DM antigens in humans are present in the brain and pancreas. Moreover, in mice, these antigens are sometimes better represented in the brain than in the pancreas due to an age-related reduction in antigen expression levels in the pancreas.32,33
To isolate and identify this putative autoantigen, affinity-purified IgG was incubated with nondenatured brain lysate overnight to allow the formation of immune complexes. The immune complexes were captured on a Protein A and Protein G column, which was washed extensively with buffer. Laemmli loading buffer containing 5% β-mercaptoethanol was added directly to the Protein A/Protein G beads, heated to 95 °C for 5 min, and the supernatant was separated by SDS-PAGE. Figure 3c shows a faint, 60 kDa band visualized after Coomassie staining, which correlated well to the band position on the Western blot in Figure 3b. This band was observed somewhat more clearly in a silver stained gel shown in Figure 3d. The Coomassie-stained band indicated in Figure 3c was excised and digested with trypsin for proteomic analysis. The trypsin digested fragments were separated by LC-MS analysis, and ∼50 protein hits were identified, of which 10 were specific to the excised band and not present in a neighboring control band. Table S1 lists the protein hits that are unique to the excised band and are of the expected molecular weight. Of the protein hits listed in Table S1, the 65 kDa isoform of glutamic acid decarboxylase (GAD65) was the only T1DM-related protein that is a T cell antigen, and thus appeared to be the most likely candidate for the native antigen to the IgG antibody population recognized by peptoid 1, despite the fact that previous efforts had failed to identify GAD65 as a humoral autoantigen in NOD mice.18−22
GAD65 Is a Specific Competitor of NOD and NOR Antibody Binding to 1
To determine if GAD65 binds to the antibodies that recognize peptoid 1, competition experiments were performed in which the ability of soluble GAD65, or other control proteins, to block binding of serum antibodies to immobilized 1 was assessed. NOR9 mouse serum drawn at 15 weeks of age was chosen due to the robust serum antibody binding to 1 observed in this sample. A total of 500 μg mL–1 of NOR9 serum was preincubated with soluble insulin, GAD65, or a control ligand 3 (Figure 4a). The blocked serum was incubated with TentaGel beads displaying 1 for 2 h at RT. After washing, binding was monitored by hybridizing Alexa Fluor 647-conjugated secondary antibody and quantifying fluorescence on a flow cytometer. Figure 4b shows that the murine isoform of GAD65 competed ∼50% of serum IgG binding to 1. A similar reduction in binding was observed when 500 nM 1 was used as the soluble competitor. Neither the control peptoid 3 nor insulin, when used as the soluble competitor, blocked binding of serum antibodies to immobilized 1. Interestingly, neither did human GAD65 (vide infra). To ensure that GAD65 did not compete antibody binding to antigen surrogates nonspecifically, we tested the ability of GAD65 to compete binding of serum IgG antibody to another, unrelated NOD mouse antigen surrogate. We chose 3, which is a small molecule found to bind specifically to NOD mouse IgG antibodies in a separate screen of a peptoid-like oligomer library (T.M.D. in preparation). We assessed binding of serum IgG to 3 in the absence of a competitor, or in the presence of 500 nM insulin or GAD65. Figure 4c shows that binding of serum IgG to 3 was unaffected in the presence of any of the competitors, indicating that GAD65 is a specific competitor of serum antibody binding to 1. These results are consistent with the proteomic data, which implicated murine GAD65 as the native antigen of the IgG antibodies that bind to 1.
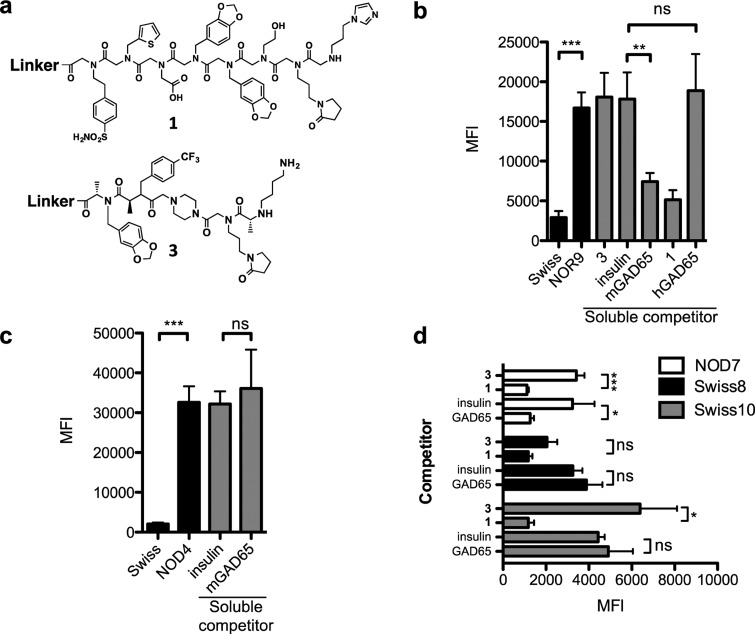
Figure 4.
Competitive binding to 1 using GAD65 as a specific competitor. (a) Chemical structures of 1 and 3. (b) Serum from pooled Swiss or NOR9 serum (500 μg mL–1) was incubated with 1 immobilized onto 10 μm TentaGel beads in a blocking buffer with no soluble competitor (black bars). Binding was monitored by flow cytometry after incubating the beads with Alexa Fluor 647-conjugated secondary antibody. Competitive binding (gray bars) to 1 was performed by preincubating 500 μg mL–1 NOR9 serum with 500 nM of either soluble 1, 3, insulin, murine GAD65 containing a GST fusion tag (mGAD65), or human GAD65 containing a GST fusion tag (hGAD65). (c) Binding to TentaGel beads containing 3 was performed by diluting pooled Swiss or NOD4 serum to 500 μg mL–1 in a blocking buffer (black bars). Competition binding to 3 (gray bars) was performed in the presence of 500 nM insulin or mGAD65. (d) Sera from two false positive samples, Swiss8 and Swiss10, and one true positive sample, NOD7, were diluted to 500 μg mL–1 and incubated in the presence of 500 nM 3, insulin, and murine GAD65 lacking a GST fusion tag (GAD65) for 5 min before incubating with immobilized 1 for 2 h. Beads were probed with Alexa Fluor 647-conjugated secondary antibody, and binding was quantified by flow cytometry. In all cases, data are reported as the mean ± s.d. from three experiments. Statistical significance was determined using an unpaired t test: *P < 0.05; **P < 0.01; ***P < 0.001; ns = not significant.
The same experiment was conducted with the Swiss mouse serum samples that registered as false positives in analyses using immobilized 1 (Swiss8 and Swiss10, Table 1). The question is whether these signals represent off-target binding of 1 to non-GAD65 antibodies. If so, we would anticipate that while most of the signal seen from these samples is competed by soluble 1, it would not be competed by soluble GAD65. As shown in Figure 4d, this was indeed the case. Thus, we conclude that Swiss mice 8 and 10 possess antibodies that bind to peptoid 1 with reasonable affinity, but that these antibodies do not recognize GAD65.
Immobilized GAD65 Supports a Blood Test for Anti-GAD65 Antibodies
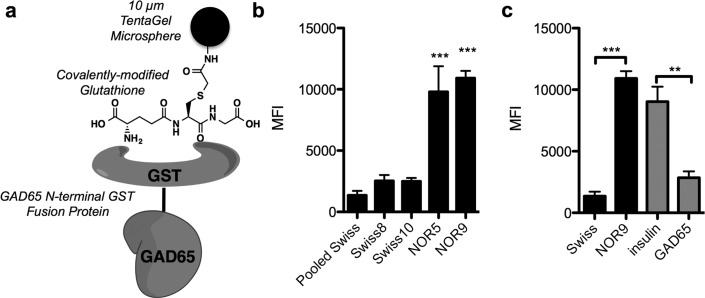
Given that the humoral response to GAD65 in humans is present in ∼70–80% of T1DM patients,34−37 it was of interest to determine if this is also the case in the mouse model. Initial attempts to do so by immobilizing murine GAD65 onto TentaGel beads via amide bond formation involving the protein lysines failed (Supporting Information Figure S11). We therefore linked glutathione to 10 μm TentaGel beads activated with bromoacetic acid via thioalkylation chemistry and then incubated them with a GAD65-GST fusion protein in hopes of avoiding destroying important epitopes in GAD65 during covalent cross-linking. An illustration of the 10 μm bead architecture is shown in Figure 5a. Efficient immobilization was confirmed by probing the beads with a monoclonal antibody to murine GAD65 (Supporting Information Figure S12). The GAD65-GST-displaying beads were incubated with 500 μg mL–1 NOR5 and NOR9 serum followed by hybridization with an Alexa Fluor 647-conjugated secondary antibody. IgG antibody binding was monitored using the flow cytometer-based assay. Figure 5b shows that GAD65-GST retained more than 7-fold more IgG antibody from the NOR5 and NOR9 serum samples than from pooled Swiss mouse serum. As expected, serum antibodies from neither NOR5 nor NOR9 elicited binding to immobilized GST protein lacking the GAD65 fusion (Supporting Information Figure S13). Notably, little signal above the background defined by the Swiss mouse pooled sample was observed when serum samples from the Swiss8 and Swiss10 mice, which gave false positive results using immobilized 1, were incubated with the GAD65-GST-displaying beads. Finally, when the serum was preblocked with soluble GAD65, most of the IgG binding to immobilized GAD65-GST was lost, while soluble insulin had only a minor effect. We conclude that this assay faithfully monitors the levels of anti-GAD65 antibodies in mouse serum.
Figure 5.
Immobilized GAD65-GST fusion protein as an anti-GAD65 IgG capture agent. (a) Schematic representation of the fusion protein-bead architecture. Beads were covalently modified with glutathione, and GST-GAD65 was immobilized onto the glutathione-bearing beads via the GST tag. (b) Binding of serum IgG antibodies from different mice to beads that were labeled with GAD65. Binding was performed using 500 μg mL–1 diluted into PBS containing 0.5% BSA and 0.05% Tween 20 and quantified by hybridizing with fluorescent secondary antibody. Fluorescence on the beads was measured using a flow cytometer. Significance was assessed for each serum sample relative to the pooled Swiss mouse sample. (c) Competition binding in which serum from NOR5 was incubated in the absence (black bars) or presence of 500 nM insulin or 500 nM GAD65 (without a GST tag, gray bars). Data reported are an average of three replicates ± s.d. Statistical significance was determined using an unpaired t test: **P < 0.01; ***P < 0.001; ns = not significant.
Given these encouraging results, all of the Swiss and NOD mouse sera were analyzed. The experiment was limited to serum samples collected up to 8 weeks of age, since this is the time frame when NOD mice first begin to express detectable levels of insulin autoantibodies,30 and we are interested in detecting very early immune responses. Serum was diluted to 500 μg mL–1 and combined with ∼200 10 μm TentaGel beads displaying the GAD65-GST fusion protein. The mixture was incubated for 2 h at RT, washed, and incubated with an Alexa Fluor 647-conjugated secondary antibody, and binding to the beads was monitored using a flow cytometer. Table 2 summarizes the data collected at a single serum concentration of 500 μg mL–1 total protein, again using the Swiss mouse average plus three standard deviations as the cutoff for calling a sample positive. A full plot of the antibody titers can be found in Supporting Information Figure S13. This experiment revealed that anti-GAD65 antibodies are readily detectable in 80% of the NOD mice prior to 8 weeks, whereas none of the Swiss mice exhibited levels above the threshold.
Table 2. Incidence of NOD and Swiss IgG Antibody Binding to GAD65 Immobilized on TentaGel Beads Determined Using a Flow Cytometry-Based Assaya,b (40 Mice Were Tested: Swiss 1–20 and NOD 1–20).
| mouse ID | Swiss | NOD |
|---|---|---|
| 1 | + | |
| 2 | + | |
| 3 | + | |
| 4 | + | |
| 5 | + | |
| 6 | + | |
| 7 | ||
| 8 | ||
| 9 | + | |
| 10 | + | |
| 11 | ||
| 12 | + | |
| 13 | + | |
| 14 | + | |
| 15 | + | |
| 16 | + | |
| 17 | ||
| 18 | + | |
| 19 | + | |
| 20 | + |
IgG binding conditions: 500 μg mL–1 serum total protein in 50 μL of PBS containing 0.5% BSA, 0.05% Tween 20 was incubated with the beads containing immobilized GAD65-GST fusion protein for 2 h at RT.
“+” indicates that the MFI was higher than the three-sigma value of 20 000 fluorescence units for either one of two serum samples collected prior to age 8.
Estimating the Polyclonal Anti-GAD Population That Binds 1
From the data shown above, it is clear that GAD65-GST is much better than peptoid 1 as a capture agent in Luminex-like assays, with superior diagnostic sensitivity and specificity. The lower specificity of 1 reflects its propensity to bind to off-target antibodies, as evidenced by the fact that the IgG’s captured by 1 from the sera of Swiss mice 8 and 10 can be blocked by soluble 1, but not soluble GAD65. However, the reason for the lower diagnostic sensitivity (38% vs 80% for peptoid 1 and GAD65-GST, respectively) is less clear. One explanation would be that 1 binds only some of the antibodies in the polyclonal spectrum that recognize GAD65. Indeed, although we refer to 1 as an antigen surrogate, it is more likely an epitope surrogate given its relatively small size. If there are autoantibodies that recognize different epitopes on GAD65, then it is unlikely that peptoid 1 could engage them all.
To test this idea, a depletion experiment was performed. NOD mouse serum was passed over TentaGel beads displaying immobilized 1 or, as a control, beads displaying immobilized 3. The flow through fractions were applied to beads displaying GAD65-GST, and after washing away unbound protein, the beads were hybridized with fluorescence secondary antibody and the amount of bound antibody was quantified using a flow cytometer. As shown in Figure 6, the sample passed over peptoid 3 exhibited a small decrease in GAD65 binding, presumably due to some amount of nonspecific antibody binding to immobilized 3. A significantly larger decline in antibody binding resulted from depleting peptoid 1-binding antibodies from the serum. Specifically, the signal from the 1-depleted serum sample was approximately 60% of that observed with the mock-depleted sample. This result supports the idea that peptoid 1 binds only about 40% of the polyclonal spectrum of anti-GAD65 autoantibodies.
Figure 6.
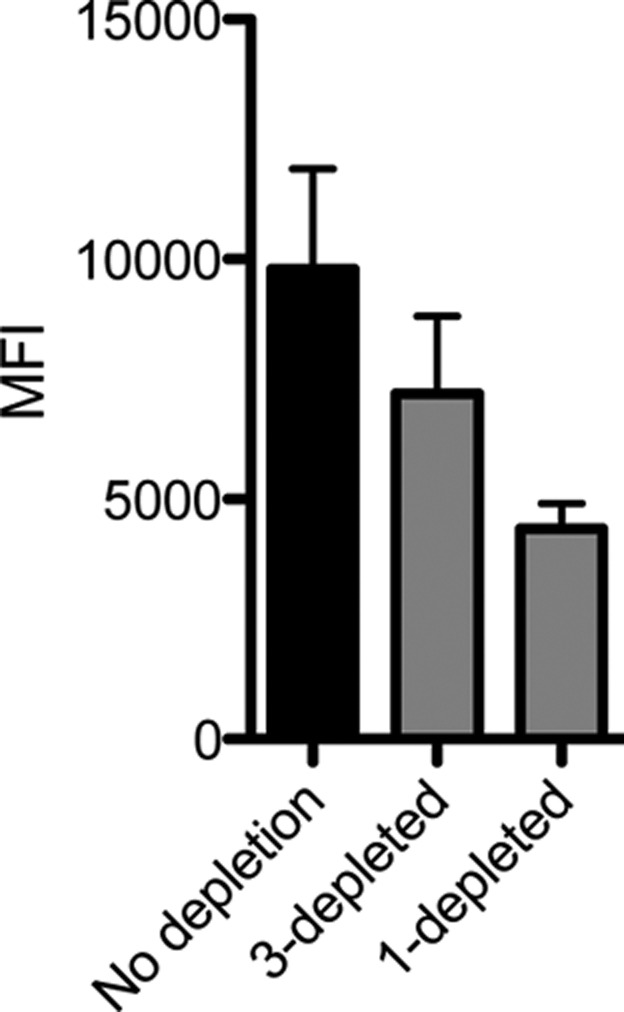
Estimation of anti-GAD polyclonal antibody fraction that recognizes 1. NOR9 serum was depleted of antibodies that bound 1 by passing 100 μL of 500 μg mL–1 serum over a TentaGel bead column displaying immobilized 1. As a control, serum was “mock depleted” by passing 500 μg mL–1 serum over a TentaGel column containing 3. A total of 50 μL of the resulting serum was incubated with GAD65-immobilized TentaGel beads, probed with Alexa Fluor 647 antimouse IgG, and analyzed by flow cytometry. Data reported are an average of three replicates ± s.d.
Discussion
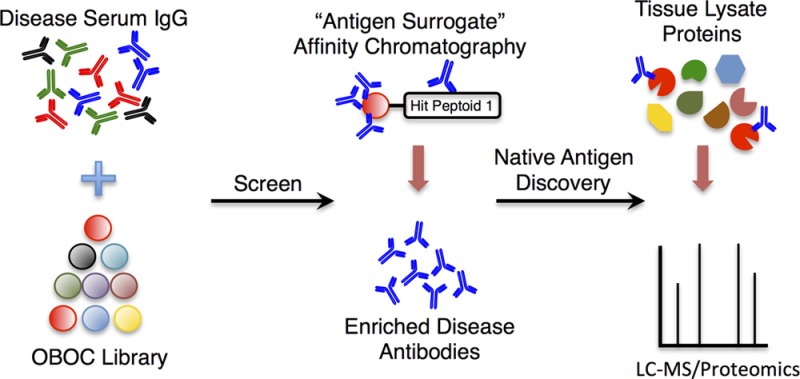
The discovery of autoantigens recognized by autoantibodies and T cells is a critical, but challenging, goal in understanding the etiology of autoimmune diseases. With respect to the antibody response, almost all efforts along these lines utilize some type of parallel or sequential screening strategy where serum from case and control samples are exposed to some collection of candidate antigens. Those that retain more antibody from the case than the control sample are characterized further. We have begun to explore a related, but different, approach in which a large library of unnatural synthetic molecules is substituted for the panel of candidate native antigens. The goal of such an exercise is to identify “antigen surrogates” that bind to autoantibodies with sufficient affinity and selectivity to retain them from serum. These synthetic surrogates, when mounted on a suitable analytical platform, could potentially be of diagnostic utility in monitoring autoantibody levels. However, this will likely require some degree of optimization of the primary screening hit to increase the affinity and selectivity for the target antibodies. Alternatively, these molecules might be useful as affinity reagents with which to purify the autoantibodies. These IgG’s, in turn, could then be used to hunt through appropriate tissue lysates for the native antigen.
In this study, we explored the latter issue in the context of the NOD mouse model for T1DM. A library of about 1 000 000 peptoids was first denuded of ligands to antibodies present in healthy mice, then screened against serum from a NOD mouse. Of the hits, a single peptoid was isolated twice, which was somewhat surprising since the library was not designed to be redundant. False positives are common in this kind of screen, but our recent work, carried out after this screen was initiated, has shown that hits isolated more than once are almost invariably bona fide ligands.27 Thus, we took advantage of this fortuitous occurrence and focused on the repeat hit peptoid 1 (Figure 2a).
When immobilized on a chemically modified glass slide or a TentaGel bead, 1 captured far more IgG antibody from some NOD mouse serum samples than most Swiss mouse (control) samples, suggesting that the antibodies it binds are indeed linked to T1DM. As a stand-alone diagnostic reagent, 1 leaves much to be desired. It binds above-background levels of antibodies from only about a third of the NOD and NOR mouse serum samples and cross-reacts with antibodies in 10% of the control Swiss mouse samples (Table 1).
However, as we demonstrate here, peptoid 1 proved useful as an affinity reagent with which to enrich the NOD-associated antibodies present in some mice (Figure 3). By probing tissue lysates by Western blotting using these enriched antibodies, an approximately 60 kDa band of interest was seen in brain tissue and, at lower levels, in a pancreatic membrane fraction (Figure 3B). Although we observed two bands in the immunoblot, one at 50 kDa was presumed to be the IgG heavy chain. An immunoprecipitation of brain lysate followed by proteomic analysis of the higher molecular weight band revealed two tryptic peptides from GAD65, suggesting that this protein might be the native autoantigen recognized by the antibodies that bind to peptoid 1. This explained the relative band intensities for various tissue lysates in the Western blot analysis (Supporting Information Figure S14). The lack of a band in pancreas whole tissue lysate and the presence of a faint band in the membrane fraction of pancreas lysates is reasonable given that GAD65 is palmitoylated to localize it to the membrane of secretory vesicles.32,38,39 The lower intensity of the band derived from the pancreas compared to that from brain lysate is consistent with a previous observation that GAD65 expression diminishes in the pancreas as the mice age.32,33
That GAD65 is the bona fide antigen recognized by 1-binding antibodies was supported by the observation that preincubation with murine GAD65, but not insulin, was able to block association of most of the serum antibodies that bind to 1 in NOD mice but did not interfere with binding of serum antibodies to a different synthetic molecule (Figure 4). Furthermore, when murine GAD65-GST fusion protein was immobilized on small glutathione-modified TentaGel beads, it bound above background levels of antibodies from about 80% of the NOD and NOR mice, but none of the control Swiss mice. Importantly, these interactions were blocked by excess soluble murine GAD65. We conclude that most of the NOR and NOD mice indeed have anti-GAD65 antibodies, in line with a similar prevalence of these antibodies in human T1DM patients.34−36
While it has long been known that GAD65 is a T cell antigen in NOD mice,19 the existence of anti-GAD65 autoantibodies in mice has been controversial.40,41 Initially, GAD65 antibodies were presumed to exist in NOD mice based on the fact that GAD65 enzymatic activity coprecipitated with serum antibodies in these animals.42 Moreover, binding of serum IgG’s to GAD65 adsorbed onto plastic ELISA plates was also observed,43 but this interaction was later deemed nonspecific since it could not be competed with the free protein.18,20−22 Binding of IgG from NOD mice to radiolabeled GAD65 has also been observed, but again, binding could not be competed with free GAD65.21 Thus, the current consensus in the field is that NOD mice do not have anti-GAD65 autoantibodies, which has resulted in speculation that the NOD mouse model does not accurately reflect human T1DM.20 Interestingly, careful reading of these papers reveals that the soluble protein competitor used in these experiments was rat, porcine, or human GAD65. We observed that, despite their high homology, mouse GAD65 competes with binding of serum antibodies to peptoid 1, but human GAD65 does not (Figure 4b). Thus, we suggest that some of the confusion was the result of the faulty assumption that GAD65 from organisms other than the mouse would bind to NOD autoantibodies, which is not the case for human GAD65 at least. However, it should also be noted that the murine and human GAD65 proteins used in our experiments were expressed in insect cells and wheat germ lysates, respectively, so we cannot rule out that some difference in post-translational modifications could also explain the striking difference in binding to NOD autoantibodies.
In any case, whatever the reason for these discrepancies in the literature, we believe that the data reported here settle this long-running controversy. More importantly, it validates the application of the antigen surrogate technology as an effective approach to eventual characterization of native autoantigens, by focusing on the use of screening hits as affinity reagents with which to purify reasonable quantities of interesting antibodies. These antibodies can then be used to hunt for the native autoantigen. This indirect, chemical biology-based approach to autoantigen identification is completely unbiased. While we identified a murine T1DM autoantigen, GAD65, whose existence had been examined inconclusively, there was nothing in the workflow employed that required any knowledge of candidate autoantigens.
Finally, a more specialized, but nonetheless important, point to emerge from this work is the effective use of the GAD65-GST fusion protein bound to 10 μm glutathione-modified TentaGel beads as an analytical platform for measurement of anti-GAD65 autoantibody levels. ELISA-like assays in which T1DM autoantigens are immobilized, either covalently or noncovalently, on most common analytical platforms have notoriously poor performance, probably due to obscuring important epitopes in the proteins. This has led to the widespread use of radioimmunoassays, which are difficult and tedious to carry out. While some recent advances have been achieved using electrochemiluminescence (ECL) detection44,45 and other more esoteric platforms,46 we suggest that the TentaGel bead/flow cytometer-based assay29 described here will be of significant utility, particularly if it proves to be useful for other known antigens. We demonstrated previously that the beads can be color-coded by covalent immobilization of specific ratios of Pacific Blue and Pacific Orange dyes. This allows for multiplexed analysis of dozens of antigens simultaneously, much as can be achieved with the conceptually similar Luminex platform but with greater sensitivity due to the much lower level of nonspecific IgG binding to the polyethylene glycol-coated TentaGel beads than the Luminex latex microspheres.
Materials and Methods
General
Organic reagent-grade solvents and chemicals were purchased from Sigma-Aldrich, Acros, or Fisher Scientific. These chemicals were used without further purification. Unless otherwise indicated, all steps involving water used deionized water that was additionally filtered through a Barnstead Nanopure water filtration system (Thermo Scientific). Knorr Amide MBHA resin, Fmoc-protected amino acids, and peptide coupling reagents HBTU and HOBt were purchased from EMD Millipore. Primary amines used in peptoid synthesis were purchased from Sigma-Aldrich, Tetra-Chem Industries, Acros Organics, or Oakwood Products.
The protocols employed for creating and screening the combinatorial libraries employed in this study are provided below. All other procedures used are described in detail in the Supporting Information.
OBOC Library Synthesis
Library synthesis was performed using standard solid-phase peptoid synthesis protocols.23,24 Tentagel S macrobeads (1 g, 90 μm, ∼2 860 000 beads, 0.28 mmol/g, Rapp-Polymere GmbH, Germany) were swelled in anhydrous DMF for 2 h before use. N-Fmoc-l-methionine (0.53 g, 1.4 mmol) was coupled to the beads overnight at RT using O-(benzotriazol-1-yl)-N,N,N,N-tetramethyluronium hexafluorophosphate (HBTU, 0.53 g, 1.4 mmol), hydroxybenzotriazole (HOBt, 0.21 g, 1.4 mmol), and N,N-diisopropylethylamine (DIPEA, 0.49 mL, 2.8 mmol). After washing in DMF (3 × 10 mL), Fmoc was deprotected using 20% piperidine. The common linker was synthesized by bromoacetylating the beads using 5 mL of 2 M bromoacetic acid (BAA) followed by 5 mL of 2.5 M N,N′-diisopropylcarbodiimide (DIC). The mixture was incubated at 37 °C for 10 min. Completion of each bromoacid coupling step was monitored by chloranil test. A clear negative result after a 5 min test indicated the amine was sufficiently acylated by the corresponding bromoacid. After washing (3 × 10 mL), amination was performed by adding a 1 M solution of the primary amine diluted in anhydrous DMF and incubating for 1 h. Purple beads following a chloranil test indicated that the amination had occurred in high yield. The final two monomeric units within the invariant linker were installed by diluting N-Boc-1,4 diaminobutane in anhydrous DMF and incubating with the activated resin at 37 °C for 1 h. Following the addition of the common linker, the beads were acetylated with bromoacetic acid using DIC activation. The resin was washed (3 × 10 mL) and split into 14 equal portions, and each primary amine from Figure 1b was added to the dried resin in each of the 14 aliquots. Each amination reaction was incubated for 1 h at 37 °C, or until a positive chloranil test resulted. The beads were then thoroughly washed and pooled together, wherein bromoacetylation was performed again. This split and pool procedure was continued until the peptoid chain was completed. After washing the resin in DCM (5 × 10 mL), the side chain protecting groups were removed by treating the pooled resin with 10 mL of 95% trifluoroacetic acid (TFA), 2.5% triisopropylsilane (TIPS), and 2.5% water with gentle shaking for 2 h at RT. The deprotected library was quenched with 10% DIPEA in DMF for 5 min and washed with DMF (5 × 10 mL). The library was prepared for screening by washing 10 times in water, followed by an overnight equilibration in water. The library was washed once more with water before washing with PBS (3 × 10 mL) and equilibrated in tris-buffered saline containing 0.05% Tween 20 (TBS-T) for 1 h. The beads were blocked with 100% StartingBlock (Thermo Scientific) for 1 h at RT.
Library Screening
A pooled sample of Swiss mouse serum derived from 10 mice at 8 weeks of age was diluted to a concentration of 100 μg mL–1. A total of 1.5 mL of the diluted serum sample was added to 300 mg of preblocked library beads and incubated overnight at 4 °C. The library was washed with TBS-T (5 × 1.5 mL). A 1:100 dilution of sheep antimouse IgG-conjugated to magnetic DynaBeads (Life Technologies) in StartingBlock containing 0.05% tween (herein referred to as screening buffer) was incubated with the library for 1 h, and hits were isolated using magnetic pull-down. The magnetic pull-down technique went as follows. The suspension of library beads and secondary antibody-conjugated DynaBeads was diluted to 40 mL with TBS-T in a 50 mL conical vial. Next, eight N52-grade cubic neodymium rare earth magnets (∼64 mm3 each) were stacked, wrapped in parafilm, and tied to string. The magnets were lowered into the suspension of library beads and agitated very gently for 5 min. The beads that were not influenced by the magnet were allowed to settle to the bottom of the tube. The magnets were removed from the OBOC library solution and set aside. The supernatant from the library solution was swiftly collected to ensure all hit beads that were detached from the magnet during magnet removal were collected before settling to the bottom. The parafilm was removed from the magnets, and hit beads were washed into a separate vial. This magnetic pull-down was repeated twice with the entire library. Since hit beads removed in this step represented hits to healthy Swiss mouse IgG, they were discarded, and the nonhit (e.g., nonmagnetized) beads were carried on for further screening. A total of 1 mL of the pooled Swiss mouse serum, diluted to 100 μg mL–1 in screening buffer, was added to the nonhit library beads and incubated overnight at 4 °C. After the beads were washed with TBS-T (3 × 1.5 mL) and incubated with secondary antibody-coated Dynabeads in screening buffer for 1 h at RT, the magnetic pull-down was repeated once again, and the hit beads were discarded. The remainder of the library was once again incubated with 100 μg mL–1 Swiss mouse control serum in screening buffer. After incubating overnight at 4 °C, the library beads were washed with TBS-T (3 × 1.5 mL). In the final round of Swiss mouse serum screening, Qdot 655-conjugated goat antimouse IgG secondary antibody (Life Technologies) was diluted 1:200 in screening buffer and incubated for 1 h at RT. The beads were washed with TBS-T (5 × 1.5 mL), and beads displaying a red halo under a fluorescence microscope using a DAPI emission filter were removed using a micropipette. To identify hit ligands that bound to NOD18 mouse IgG antibodies, NOD18 serum collected at 8 weeks of age was diluted to 50 μg mL–1 in StartingBlock containing 1 mM porcine insulin (Sigma-Aldrich) and added to the denuded library. The library was incubated overnight at 4 °C, followed by washing several times in TBS-T (5 × 2 mL). The library was incubated with secondary antibody-conjugated Dynabeads for 1 h at RT in screening buffer. Hit beads were removed using the magnetic pull-down method described above, with careful attention not to lose any of the isolated hits. In this round of screening, the hit beads were saved, stripped of binding proteins by incubating in 0.25% trypsin-EDTA (Life Technologies). They were washed in water (2 × 1 mL) and 50:50 acetonitrile/water at 37 °C for 1 h (2 × 1 mL). The beads were re-equilibrated in water (10 × 2 mL) with one extensive overnight water wash. The beads were prepared for screening by washing in TBS-T for 1 h followed by StartingBlock for 1 h. A control screen was performed by adding 50 μg mL–1 of pooled Swiss mouse serum to the stripped hit beads in screening buffer. The mixture was incubated overnight at 4 °C and washed with TBS-T (3 × 2 mL). The beads were hybridized with a 1:200 dilution of QDot 655-conjugated secondary antibody in screening buffer. After washing with TBS-T (4 × 2 mL), beads that captured IgG antibodies were distinguished visually by the appearance of a red halo under a DAPI filter on an inverted fluorescent microscope. “Hits” were manually removed using a micropipette and discarded. The remainder of the library was added to 50 μg mL–1 8 week old NOD18 mouse serum containing 1 mM porcine insulin (Sigma-Aldrich) and incubated overnight at 4 °C. The library was washed in TBS-T (4 × 2 mL), and a 1:200 dilution of QDot 655-conjugated secondary antibody was hybridized to the beads and washed and “hits” were removed under a fluorescent microscope. The beads were stripped of bound protein as described above and cleaved from the resin in 50 mg mL–1 cyanogen bromide (CNBr) dissolved in 5:4:1 acetonitrile/water/acetic acid overnight. The solvent was removed by evaporation, and the sequence of the ligand was determined by tandem MALDI-TOF MS using α-cyano-hydroxycinnamic acid as the matrix.
Acknowledgments
We thank K. Lowe and B. Torres in the Scripps Florida Flow Cytometry Core for technical assistance. We also thank Graham West, Ricardo Flefil, and Yelenis Mari of the Scripps Florida proteomics core for performing LC-MS experiments and providing insightful feedback. We are grateful to C. Tapia, M. Tapia, and A. Aditya, who aided in mouse serum collection. This work was supported by a grant to T.K. from the NIH (DP3 DK094309).
Supporting Information Available
Supplementary methods, blood sugar tracking, antibody binding profiles, detailed proteomic analysis, Western blots and SDS-PAGE gels, and chemical structures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): T.K. is a shareholder and member of the Scientific Advisory Board of Opko Health, Inc., which is working to commercialize this technology for the diagnosis of human diseases.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Balboni I.; Chan S. M.; Kattah M.; Tenenbaum J. D.; Butte A. J.; Utz P. J. (2006) Multiplexed protein array platforms for analysis of autoimmune diseases. Annu. Rev. Immunol. 24, 391–418. [DOI] [PubMed] [Google Scholar]
- Kanter J. L.; Narayana S.; Ho P. P.; Catz I.; Warren K. G.; Sobel R. A.; Steinman L.; Robinson W. H. (2006) Lipid microarrays identify key mediators of autoimmune brain inflammation. Nature Med. 12, 138–143. [DOI] [PubMed] [Google Scholar]
- Wang X.; Yu J.-Q.; Sreekumar A.; Varambally S.; Shen R.; Giachero D.; Mehra R.; Montie J. E.; Pienta K. J.; Sanda M. G.; Kantoff P. W.; Rubin M. A.; Wei J. T.; Ghosh D.; Chinnaiyan A. M. (2005) Autoantibody signatures in prostate cancer. N. Engl. J. Med. 355, 16–27. [DOI] [PubMed] [Google Scholar]
- Raveendra B.; Hao W.; Baccala R.; Reddy M. M.; Schilke J.; Bennett J. L.; Theofiliopolous A. N.; Kodadek T. (2013) Discovery of peptoid ligands for anti-Aquaporin 4 antibodies. Chem. Biol. 20, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. M.; Wilson R.; Wilson J.; Connell S.; Gocke A.; Hynan L.; German D.; Kodadek T. (2011) Identification of candidate IgG biomarkers for Alzheimer’s Disease via combinatorial library screening. Cell 144, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittelfelder R.; Riemer A. B.; Jensen-Jarolim E. (2009) Mimotope vaccination--from allergy to cancer. Expert Opin. Biol. Ther. 9, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth G. S. (1986) Type I diabetes mellitus. A chronic autoimmune disease. N. Eng. J. Med. 314, 1360–1368. [DOI] [PubMed] [Google Scholar]
- Herold K. C.; Vignali D. A.; Cooke A.; Bluestone J. A. (2013) Type 1 diabetes: translating mechanistic observations into effective clinical outcomes. Nature Rev. 13, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekkeskov S.; Aanstoot H. J.; Christgau S.; Reetz A.; Solimena M.; Cascalho M.; Folli F.; Richter-Olesen H.; De Camilli P. (1990) Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesizing enzyme glutamic acid decarboxylase. Nature 347, 151–156. [DOI] [PubMed] [Google Scholar]
- Kawasaki E.; Eisenbarth G. S.; Wasmeier C.; Hutton J. C. (1996) Autoantibodies to protein tyrosine phosphatase-like proteins in type I diabetes. Overlapping specificities to phogrin and ICA512/IA-2. Diabetes 45, 1344–1349. [DOI] [PubMed] [Google Scholar]
- Lu J.; Li Q.; Xie H.; Chen Z. J.; Borovitskaya A. E.; Maclaren N. K.; Notkins A. L.; Lan M. S. (1996) Identification of a second transmembrane protein tyrosine phosphatase, IA-2beta, as an autoantigen in insulin-dependent diabetes mellitus: precursor of the 37-kDa tryptic fragment. Proc. Natl. Acad. Sci. U. S. A. 93, 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini N.; Larigan J. D.; Genovese S.; Appella E.; Sinigaglia F.; Rogge L. (1995) The 37/40-kilodalton autoantigen in insulin-dependent diabetes mellitus is the putative tyrosine phosphatase IA-2. Proc. Natl. Acad. Sci. U. S. A. 92, 9412–9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacio E.; Lampasona V.; Genovese S.; Ferrari M.; Bosi E. (1995) Identification of protein tyrosine phosphatase-like IA2 (islet cell antigen 512) as the insulin-dependent diabetes-related 37/40K autoantigen and a target of islet-cell antibodies. J. Immunol. 155, 5419–5426. [PubMed] [Google Scholar]
- Lan M. S.; Wasserfall C.; Maclaren N. K.; Notkins A. L. (1996) IA-2, a transmembrane protein of the protein tyrosine phosphatase family, is a major autoantigen in insulin-depedent diabetes mellitus. Proc. Natl. Acad. Sci. U.S.A. 93, 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzlau J. M.; Juhl K.; Yu L.; Moua O.; Sarkar S. A.; Gottlieb P.; Rewers M.; Eisenbarth G. S.; Jensen J.; Davidson H. W.; Hutton J. C. (2007) The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc. Natl. Acad. Sci. U. S. A. 104, 17040–17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. S.; Bluestone J. A. (2005) The NOD mouse: a model of immune dysregulation. Annu. Rev. Immunol. 23, 447–485. [DOI] [PubMed] [Google Scholar]
- Simon R. J.; Kania R. S.; Zuckermann R. N.; Huebner V. D.; Jewell D. A.; Banville S.; Ng S.; Wang L.; Rosenberg S.; Marlowe C. K.; et al. (1992) Peptoids: a modular approach to drug discovery. Proc. Natl. Acad. Sci. U. S. A. 89, 9367–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velloso L. A.; Eizirik D. L.; Karlsson F. A.; Kampe O. (1994) Absence of autoantibodies against glutamate decarboxylase (GAD) in the non-obese diabetic (NOD) mouse and low expression of the enzyme in mouse islets. Clin. Exp. Imunol. 96, 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieg S.; Seissler J.; Herberg L.; Northemann W.; Scherbaum W. A. (1994) GAD65 is recognized by T-cells, but not by antibodies from NOD-mice. Autoimmunity 17, 189–194. [DOI] [PubMed] [Google Scholar]
- Mackay I. R.; Bone A.; Tuomi T.; Elliott R.; Mandel T.; Karopoulos C.; Rowley M. J. (1996) Lack of autoimmune serological reactions in rodent models of insulin dependent diabetes mellitus. J. Autoimmun. 9, 705–711. [DOI] [PubMed] [Google Scholar]
- Bonifacio E.; Atkinson M.; Eisenbarth G.; Serreze D.; Kay T. W.; Lee-Chan E.; Singh B. (2001) International Workshop on Lessons From Animal Models for Human Type 1 Diabetes: identification of insulin but not glutamic acid decarboxylase or IA-2 as specific autoantigens of humoral autoimmunity in nonobese diabetic mice. Diabetes 50, 2451–2458. [DOI] [PubMed] [Google Scholar]
- Yu L.; Robles D. T.; Abiru N.; Kaur P.; Rewers M.; Kelemen K.; Eisenbarth G. S. (2000) Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc. Natl. Acad. Sci. U. S. A. 97, 1701–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figliozzi G. M.; Goldsmith R.; Ng S. C.; Banville S. C.; Zuckermann R. N. (1996) Synthesis of N-substituted glycine peptoid libraries. Methods Enzymol. 267, 437–447. [DOI] [PubMed] [Google Scholar]
- Alluri P. G.; Reddy M. M.; Bachhawat-Sikder K.; Olivos H. J.; Kodadek T. (2003) Isolation of protein ligands from large peptoid libraries. J. Am. Chem. Soc. 125, 13995–14004. [DOI] [PubMed] [Google Scholar]
- Lam K. S.; Salmon S. E.; Hersh E. M.; Hruby V. J.; Kazmierski W. M.; Knapp R. J. (1991) A new type of synthetic peptide library for identifying ligand-binding activity. Nature 354, 82–84. [DOI] [PubMed] [Google Scholar]
- Olivos H. J.; Baccawat-Sikder K.; Kodadek T. (2003) Quantum dots as a visual aid for screening bead-bound combinatorial libraries. ChemBiochem 4, 1242–1245. [DOI] [PubMed] [Google Scholar]
- Doran T. M.; Gao Y.; Mendes K.; Dean S.; Simanski S.; Kodadek T. (2014) The utility of redundant combinatorial libraries in distinguishing high and low quality screening hits. ACS Comb. Sci. 16, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racke M. K. (2001) Experimental autoimmune encephalomyelitis (EAE), Current Protocols in Neuroscience (Crawley J. N., et al. , Eds.), Chapter 9, Unit 9.7, John Wiley & Sons, New York. [DOI] [PubMed] [Google Scholar]
- Doran T. M.; Kodadek T. (2013) A Liquid Array Platform for the Multiplexed Analysis of Synthetic Molecule-Protein Interactions. ACS Chem. Biol. 9, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abiru N.; Yu L.; Miao D.; Maniatis A. K.; Liu E.; Moriyama H.; Eisenbarth G. S. (2001) Transient insulin autoantibody expression independent of development of diabetes: comparison of NOD and NOR strains. J. Autoimmun. 17, 1–6. [DOI] [PubMed] [Google Scholar]
- Zeng F. Y.; Oka J. A.; Weigel P. H. (1996) Renaturation and ligand blotting of the major subunit of the rat asialoglycoprotein receptor after denaturing polyacrylamide gel electrophoresis. Glycobiology 6, 247–255. [DOI] [PubMed] [Google Scholar]
- Kim J.; Richter W.; Aanstoot H. J.; Shi Y.; Fu Q.; Rajotte R.; Warnock G.; Baekkeskov S. (1993) Differential expression of GAD65 and GAD67 in human, rat, and mouse pancreatic islets. Diabetes 42, 1799–1808. [DOI] [PubMed] [Google Scholar]
- Pleau J. M.; Esling A.; Bach J. F.; Dardenne M. (1996) Gene expression of pancreatic glutamic acid decarboxylase in the nonobese diabetic mouse. Biochem. Biophys. Res. Commun. 220, 399–404. [DOI] [PubMed] [Google Scholar]
- Petersen J. S.; Hejnaes K. R.; Moody A.; Karlsen A. E.; Marshall M. O.; Hoier-Madsen M.; Boel E.; Michelsen B. K.; Dyrberg T. (1994) Detection of GAD65 antibodies in diabetes and other autoimmune diseases using a simple radioligand assay. Diabetes 43, 459–467. [DOI] [PubMed] [Google Scholar]
- Bonifacio E.; Lampasona V.; Bernasconi L.; Ziegler A. G. (2000) Maturation of the humoral autoimmune response to epitopes of GAD in preclinical childhood type 1 diabetes. Diabetes 49, 202–208. [DOI] [PubMed] [Google Scholar]
- Velloso L. A.; Kampe O.; Hallberg A.; Christmanson L.; Betsholtz C.; Karlsson F. A. (1993) Demonstration of GAD-65 as the main immunogenic isoform of glutamate decarboxylase in type 1 diabetes and determination of autoantibodies using a radioligand produced by eukaryotic expression. J. Clin. Invest. 91, 2084–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.; Hagopian W. A.; Kockum I.; Li L. S.; Sanjeevi C. B.; Lowe R. M.; Schaefer J. B.; Zarghami M.; Day H. L.; Landin-Olsson M.; Palmer J. P.; Janer-Villanueva M.; Hood L.; Sundkvist G.; Lernmark A.; Breslow N.; Dahlquist G.; Blohme G. (2002) Genetic effects on age-dependent onset and islet cell autoantibody markers in type 1 diabetes. Diabetes 51, 1346–1355(Diabetes Incidence in Sweden Study, G., and Swedish Childhood Diabetes Study, G). [DOI] [PubMed] [Google Scholar]
- Kanaani J.; Patterson G.; Schaufele F.; Lippincott-Schwartz J.; Baekkeskov S. (2008) A palmitoylation cycle dynamically regulates partitioning of the GABA-synthesizing enzyme GAD65 between ER-Golgi and post-Golgi membranes. J. Cell Sci. 121, 437–449. [DOI] [PubMed] [Google Scholar]
- Christgau S.; Aanstoot H. J.; Schierbeck H.; Begley K.; Tullin S.; Hejnaes K.; Baekkeskov S. (1992) Membrane anchoring of the autoantigen GAD65 to microvesicles in pancreatic beta-cells by palmitoylation in the NH2-terminal domain. J. Cell Biol. 118, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson M. A.; Maclaren N. K. (1988) Autoantibodies in Nonobese Diabetic Mice Immunoprecipitate 64,000-Mr Islet Antigen. Diabetes 37, 1587–1590. [DOI] [PubMed] [Google Scholar]
- Elias D.; Markovits D.; Reshef T.; Zee R. v. d.; Cohen I. R. (1990) Induction and therapy of autoimmune diabetes in the non-obese diabetic (NOD/Lt) mouse by a 65-kDa heat shock protein. Proc. Natl. Acad. Sci. U.S.A. 87, 1576–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizpurua H. J. D.; French M. B.; Chosich N.; Harrison L. C. (1994) Natural History of Humoral Immunity to Glutamic Acid Decarboxylase in Non-Obese Diabetic (NOD) Mice. J. Autoimmun. 7, 643–653. [DOI] [PubMed] [Google Scholar]
- Tisch R.; Yang X.-D.; Singer S. M.; Liblau R. S.; Fugger L.; McDevitt H. O. (1993) Immune response to glutamic acid decarboxylase correlates with insulitis in non-obese diabetic mice. Nature 366, 72–75. [DOI] [PubMed] [Google Scholar]
- Miao D.; Guyer M. S.; Dong F.; Jiang L.; Steck A. K.; Rewers M.; Eisenbarth G. S.; Yu L. (2013) GAD65 autoantibodies detected by electrochemiluminescence assay identify high risk for type I diabetes. Diabetes 62, 4174–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.; Dong F.; Miao D.; Fouts A. R.; Wenzlau J. M.; Steck A. K. (2013) Proinsulin/Insulin autoantibodies measured with electrochemiluminescent assay are the earliest indicator of prediabetic islet autoimmunity. Diabetes Care 36, 2266–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Kumar R. B.; Dai H.; Feldman B. J. (2014) A plasmonic chip for biomarker discovery and diagnosis of type 1 diabetes. Nature Med. 20, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.