Abstract
Mass spectrometry (MS) has been a core technology for high sensitive and high-throughput analysis of the enriched glycoproteome in aspects of quantitative assays as well as qualitative profiling of glycoproteins. Because it has been widely recognized that aberrant glycosylation in a glycoprotein may involve in progression of a certain disease, the development of efficient analysis tool for the aberrant glycoproteins is very important for deep understanding about pathological function of the glycoprotein and new biomarker development. This review first describes the protein glycosylation-targeting enrichment technologies mainly employing solid-phase extraction methods such as hydrizide-capturing, lectin-specific capturing, and affinity separation techniques based on porous graphitized carbon, hydrophilic interaction chromatography, or immobilized boronic acid. Second, MS-based quantitative analysis strategies coupled with the protein glycosylation-targeting enrichment technologies, by using a label-free MS, stable isotope-labeling, or targeted multiple reaction monitoring (MRM) MS, are summarized with recent published studies. © 2014 The Authors. Mass Spectrometry Reviews Published by Wiley Periodicals, Inc. Rapid Commun. Mass Spec Rev 34:148–165, 2015.
Keywords: protein glycosylation, quantitative mass spectrometry, hydrazide, lectin, affinity enrichment, stable isotope labeling, multiple reaction monitoring
I. INTRODUCTION
Protein glycosylation has been reported to play important roles in adhesion, metastasis, and signaling through cell-to-cell interactions. Protein glycosylation in human contains several different types, including N-linked glycosylation, O-linked glycosylation, and C-glycosylation. N-linked glycosylation occurs on the asparagine in the sequence of Asn-X-Ser/Thr (and scarcely Cys) with X being any amino acid with the exception of proline. N-linked glycans have commonly core pentasaccharide moiety, and are classified by the type and position of monosaccharide residues added to the core pentasaccharide (Morelle et al., 2006; Rakus & Mahal, 2011). O-linked glycosylation is located on serine or threonine residues by addition of N-acetylgalactosamine (GalNAc), mannose, fucose, glucose, or N-acetylglucosamine (GlcNAc) (Ohtsubo & Marth, 2006). Mucin-type O-glycosylation, known to be involved in cell adhesion, invasion, and immune response is a typical glycosylation occurring in a hydroxyl group on Ser/Thr residues (Hollingsworth & Swanson, 2004). Except for these N-linked and O-linked glycosylation, various types of glycosylation such as C-glycosylation on tryptophan residue and further modification on glycan moiety by phosphorylation have also been reported (Wei & Li, 2009; Barnes et al., 2011).
Glycan moiety linked to a glycosylation site on a glycoprotein generally consists of numerous glycoforms having different glycan structures. Because of this glycan microheterogeneity in glycoprotein, glycoprotein shows especially high structural complexity and each protein glycoforms consisting of the glycoprotein is present with substoichiometric level far lower than total protein abundance of the glycoprotein. Furthermore, partial occupancy of glycan moiety on a glycosylation site of a given glycoprotein was also observed (Liu et al., 2010; Wada, 2012). These structural features in glycoprotein make difficult to efficiently separate, identify, and quantify the glycoprotein present in a complex biological sample like blood.
It has been reported that aberrant glycosylation in a glycoprotein can be related to the occurrence and progression of certain diseases. Abnormal glycosylation patterns, such as increased glycan size and extra branching of glycan chains with over-sialylation and -fucosylation, are closely associated with disease progression (Mizuochi et al., 1983; Pierce & Arango, 1986; Dennis et al., 1987; Orntoft & Vestergaard, 1999; Comunale et al., 2010). Analysis of altered cancer-related glycoprotein expression may facilitate discovery of potential biomarkers, as well as discovery of novel targets of therapeutics. Therefore, protein glycosylation analysis has become an important target in proteomic research field and has great potential for clinical applications like development of biomarkers. Many protein biomarkers that are known to be cancer biomarkers are also glycoproteins (Peracaula et al., 2003; Ohyama et al., 2004; Gomaa et al., 2009). Especially glycoproteins aberrantly glycosylated in the malignant cells can be secreted from the cells via common secretory pathways or shed from the cell membrane as a result of enhanced protease activity and reach the bloodstream, reflecting abnormal states of the malignant cells. Therefore, serological glycoprotein aberrantly glycosylated can be an attractive target for biomarker development.
Since the progression of certain diseases may involve aberrant glycosylation in a glycoprotein, the ability to analyze quantitatively aberrant protein glycoforms having a specific glycan structure provides a tool for monitoring differences between individual cases in the abundance of aberrant glycoforms of target proteins related to the diseases. So, efficient identification of these glycoproteins and quantification of the levels thereof between healthy and malignant individuals are useful for understanding the pathological mechanism of the malignant cells, and finally for developing specific disease biomarkers. A typical example involving the benefit obtainable from the analysis of aberrant protein glycoforms is alpha-fetoprotein (AFP), known as a potent biomarker for HCC (Gomaa et al., 2009). AFP-L3, a glycoform glycosylated aberrantly by fucosylation that shows increased affinity to fucose-specific lectins like lens culinaris agglutinin (LCA), has been reported to show improved specificity as a biomarker for hepatocellular carcinoma (HCC) (Kumada et al., 1999; Khien et al., 2001; Miyaaki et al., 2007).
Mass spectrometry (MS) is a core technology in the field of proteomics, capable of profiling a number of proteins from various complex biological media with high-throughput performance and accurate digitalized informatics. To conduct efficient mass analysis of glycoproteins playing important roles in adhesion, metastasis, and signaling through cell-to-cell interactions, many separation tools for enrichment of glycoproteins and analytical tools for mass analysis of the separated glycoproteins were developed. Especially through collaboration studies organized by Human Proteome Organization (HUPO) for mass analysis of serological glycoprotein purified and enriched from blood sample, the performance and reproducibility of MS-based strategy in structural identification and quantitative analysis of the purified glycoproteins was evaluated (Wada et al., 2007; Wada et al., 2010a). Over mass analysis focusing a purified target glycoprotein, MS has also received great attention for quantitative analysis of complex glycoproteome samples (Kirmiz et al., 2007; Hongsachart et al., 2009; Comunale et al., 2009; Jung, Cho, & Regnier, 2009; Ahn et al., 2012d). Nonetheless, there are still many obstacles to realize efficient analysis for complex glycoproteome present with wide ranges at concentration in complex biological medium, especially because of difficulties in identifying and quantifying altered glycoforms due to the high complex nature of the glycan structure, low abundance coming from substoichiometric occupation of altered glycoforms of a glycoprotein due to glycan microheterogeneity, and severe masking effects due to highly abundant serological proteins in the cases of blood sample.
To overcome these problems, a variety of sample preparation approaches targeting protein glycosylation have been developed to increase sample separation efficiency, decrease sample complexity, and enrich low abundant glycoproteins of interest ( Fig. 1). Combined strategies systematically integrating separation techniques were tried for glycoprotein- or glycopeptide-targeting sample preparation, prior to subsequent MS-based analysis of the prepared samples. The enrichment techniques based on solid-phase extraction rather than gel-based techniques are generally advantageous, since the solid-phase-based techniques are far simple for subsequent sample manipulations like enzyme digestion and therefore feasible to construct high-throughput MS-based analytical systems. In general, glycoproteins samples separated are subjected to enzymatic digestion to afford low molecular-weight peptide mixture or glycan mixture, and can further be purified and separated by on-line or off-line technique prior to mass analysis. Mass spectrometry of the prepared peptides can be realized by label-free quantification methods or by quantitative methods with stable isotope-labeling or stable isotope-coded internal standard spiking to the prepared sample. Since great number of studies have been reported involving sample separation technologies and quantitative mass analysis technologies in the proteomic field, the scope of this review will be limited to the recent studies about glycoprotein/glycopeptide-targeting separation methods based on solid-phase extraction and high-throughput MS-based quantification methods coupled with the glycoprotein/glycopeptide-targeting separation methods.
FIGURE 1.
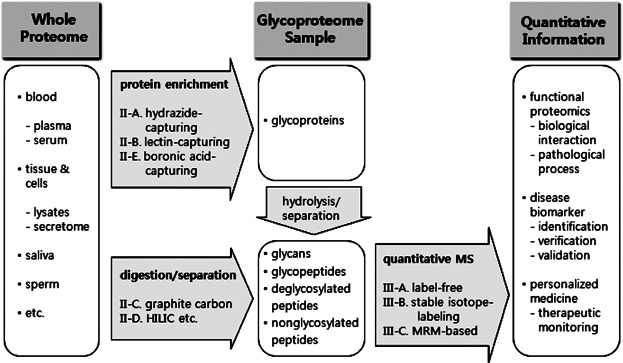
Glycoproteome-targeting separation and MS-based quantification approaches.
II. SEPARATION TECHNIQUES TARGETING PROTEIN GLYCOSYLATION
Hydrazide-Capturing Techniques
Hydrazide-capturing technique, a chemical reaction-based approach between bead-immobilized hydrazide group and dialdehyde group derived from glycan moiety of glycoprotein, has been developed to enrich glycoproteins or glycopeptides by selectively targeting cis-diols present in monosaccharides on glycan moieties ( Fig. 2). In the hydrazide chemistry, cis-diols on glycans are oxidized into aldehydes by sodium periodate, then covalently coupled with hydrazide immobilized to solid beads (Zhang et al., 2003; Liu et al., 2005; Zeng et al., 2010a). Bound glycoproteins are generally trypsin-digested in situ, and then nonglycosylated or unbound peptides are removed by washing. Covalently coupled glycopeptides are then released by PNGase F, and the resulting deglycopeptides are analyzed by mass spectrometry. In the process of the PNGases F-mediated enzyme action, the asparagine residue of the NX(S/T) consensus sequence of N-glycosites hydrazide-captured is converted into an aspartic acid containing COOH functionality at side chain, as shown in Figure 2b.
FIGURE 2.
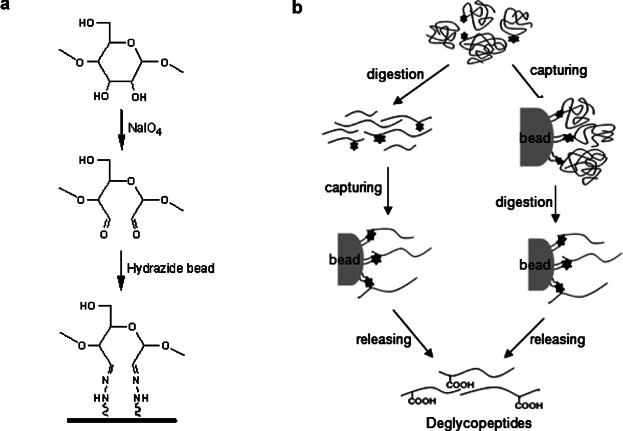
Graphical features in hydrazide-capturing method for glycoproteome enrichment.
The bound glycopeptides can also be released by acidic hydrolysis rather than enzymatic cleavage to analyze selectively sialylated glycopeptides and identify their glycosylation sites (Nilsson et al., 2009). In this approach, sialylated glycoproteins were selectively periodate-oxidized, captured on hydrazide beads, trypsinized and released by acid hydrolysis of sialic acid glycosidic bonds. The chemical reaction-based approach based on the hydrazide-capturing technique developed for N-glycoproteins has also been applied for the enrichment of O-GlcNAc modified proteins with appropriate modifications (Klement et al., 2010). O-linked glycopeptides, captured on hydrazide resin and on-resin digested, can be released by hydroxylamine to give hydroxylamine-modified glycopeptides. This enrichment strategy offers a fringe benefit in mass spectrometry analysis, because the presence of the open carbohydrate ring on the modified glycopeptides leads to characteristic fragmentation facilitating both glycopeptide identification and site assignment.
Some examples using magnetic beads to immobilize hydrazide probe were also reported (Berven et al., 2010; Wang et al., 2012). The use of magnetic beads can be helpful to automation of the hydrazide-capturing method through the use of a kind of magnetic particle processor. Glycopeptides rather than intact glycoproteins can also be enriched by the hydrazide-capturing method (Tian et al., 2007; Cao et al., 2009). Recently ultrasmall gold nanoparticles with core diameter of 1.2 nm were functionalized with hydrazide groups, and these conjugates were used for isolation/enrichment of N-glycosylated peptides (Tran et al., 2012). Hydrazide-functionalized gold nanoparticles showed excellent stability in biological samples and exhibited a large capacity for peptide capturing. Superparamagnetic silica particles were also utilized to immobilized hydrazide groups on the solid surface, and further these conjugates were evaluated as the solid support for solid-phase extraction of glycopeptides (Zou et al., 2008). The hydrazide-functionalized silica particles containing superparamagnetic iron oxide cores displayed a strong response to the external magnetic field, and this feature made possible to capture and release the particles easily for automated, high-throughput sample preparation of glycopeptides.
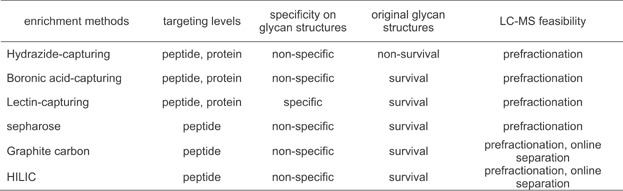
These hydrazide-capturing methods based on hydrazide chemistry are useful for capturing glycosylated molecules from highly complex sample and characterizing the captured molecules with high-throughput analytical performance. Nonetheless, since this hydrazide-based approach utilizes dialdehyde groups, generated from glycan of glycoproteins or glycopeptides by the oxidative cleavage, to capture glycan moieties, structural information for intact glycan structures on glycoproteins or glycopeptides is lost inevitably (Table 1). Also, these chemical reaction-based capturing are mainly selective to cis-diols on monosaccharides consisting of glycans, but is not promising for selective capturing of protein glycoforms having a specific glycan structure, produced by post-translational modification of glycoproteins in a biological process (i.e., aberrant protein glycoforms in cancer cells) (Table 1). So, specific capturing for protein glycoforms having a specific glycan structure originating by abnormal biological process is not achievable using only this hydrazide-capturing approach.
Table 1.
Comparison of enrichment tools for glycoproteome
 |
Lectin-Specific Capturing Techniques
Lectin-based capturing is one of powerful techniques to separate and enrich of a kind of interesting glycoproteins from complex biological mixture in proteomic field (Kaji et al., 2003; Drake et al., 2006). A variety of lectins have been widely used to capture protein glycoforms showing specific binding affinity to the used lectins. The most notable feature is its capability of enriching specifically a unique protein glycoforms having a specific glycan-structure active to a lectin used ( Fig. 3). Lectin-based separation of glycoproteomics was utilized to explore glyco-alteration and analyze cancer-related glycoprotein biomarkers (Dai et al., 2009; Ito et al., 2009). These lectin-based methods can be utilized complementary with other chemical reaction-based technologies using hydrazide and boronic acid to capture glycan moieties of glycoproteome (Sparbier et al., 2005; Pan et al., 2006). To get the best recovery for glycoproteome including lots kind of protein glycoforms from complex proteome sample, multi-lectin columns prepared by pre-mixing multiple lectins were developed (Heo et al., 2007; Kullolli, Hancock, & Hincapie, 2008; Zeng et al., 2011). Since the multi-lectin column consisting of multiple lectins mixed in advance before lectin fractionation of proteome sample shows binding activity simultaneously for various glycoforms of glycoproteins, it makes possible to collect lots kind of glycoproteins having a variety of glycan structures. But this approach using multi-lectin column makes impossible to extract selectively a protein glycoforms specifically active to each lectin used.
FIGURE 3.
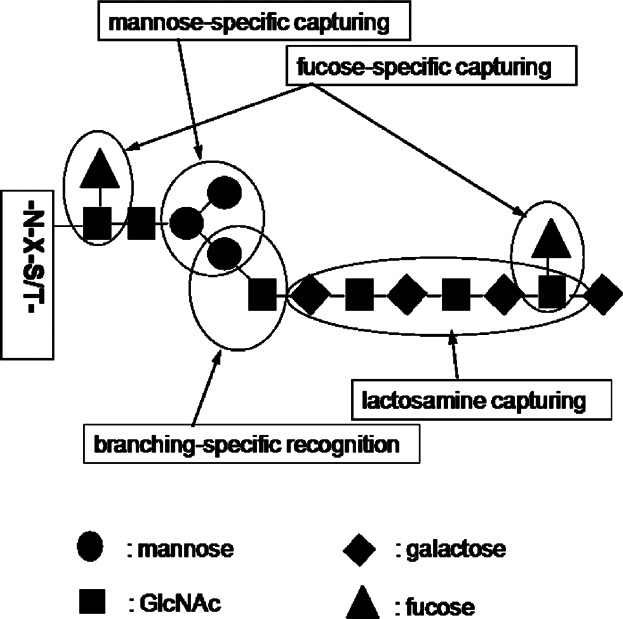
Graphical features in glycoproteome-capturing by lectins showing specific-binding affinity to glycan structures.
Since one of major characteristics in lectin fractionation is its binding ability to glycoproteins according to structural featuring on glycan moieties of the glycoproteins, single lectin capturing makes possible to fractionate separately protein glycoforms specific to the used each lectin from complex proteome sample, rather than to separate total glycoforms of glycoproteins from the proteome sample (Geng et al., 2001; Kim et al., 2008; Drake et al., 2011). This feature in lectin fractionation based on specificity to glycan structures is especially noteworthy as comparing with other methods like the chemical reaction-based approach (Table 1). Another feature in the lectin-capturing is the remove of a negative masking effect by protein glycoforms inactive to a lectin used. Because a lectin with binding affinity for a specific glycan structure, in general, captures lectin-specific protein glycoforms populating in a substoichiometrically low portion of the total amounts of glycoproteins, most highly abundant protein glycoforms that do not show affinity to the used lectin can be removed before conducting subsequent mass spectrometry (Jung, Cho, & Regnier, 2009; Ahn et al., 2012c). In addition to using a single lectin or a multi-lectin column premixed with several lectins, the multiplex lectin-channel monitoring (LCM) approach, based on a multiplex parallel lectin-capturing to fractionate separately protein glycoforms active to each multiple lectins used, was also tried (Jung, Cho, & Regnier, 2009; Ahn et al., 2012a). So, each lectins showing narrow specificity for a specific glycan structure is expected to be good selectors for enriching protein glycoforms of interest.
Lectin affinity chromatography was also evaluated as a powerful method to enrich glycoproteins in biofluid and cell/tissue lysates, especially membrane glycoproteins (Wei, Dulberger, & Li, 2010). Isolation and characterization of the hydrophobic fraction of the membrane proteome sample have been greatly limited. However, the use of detergents in lectin-specific capturing for membrane glycoproteins can minimize nonspecific binding and facilitate the elution of hydrophobic glycoproteins. Instead of using immobilized lectin, the using free lectin solution was also tried for affinity entrapment of oligosaccharides and glycopeptides (Yodoshi et al., 2011). In this approach, oligosaccharide–lectin conjugates with ammonium sulfate were formed and separated through the salting out using ethanol precipitation from nonconjugated protein glycoforms inactive to the used lectins. The glycopeptide–lectin conjugates were also specifically trapped on a centrifugal ultra-filtration membrane with cut-off of 10 kDa for subsequent glycosylation analyses by LC-MS. Lectin-specific capturing approach for glycoproteome was further integrated with microarray technology for construction of a semi-automated high-throughput glycoprotein biomarker discovery platform (Comunale et al., 2009; Choi et al., 2011).
Affinity Separation Techniques Using Porous Graphite Carbon
With recent advances in proteomics, a variety of approaches to collect glycopeptides and glycans from digested mixture of the glycoproteome sample have been developed. Glycoproteins fractionated by various techniques discussed above, such as the hydrazide-capturing techniques and the lectin-specific capturing techniques, can usually be hydrolyzed to give smaller molecules by a specific enzyme for analytical efficiency in subsequent MS. N-linked glycans is routinely detach from protein moiety of glycoproteins by endoglycosidases like PNGases F (Maley et al., 1989). Chemical elimination technique is also employed to release O-linked glycans from glycoproteins (Wells et al., 2002; Miura et al., 2010). In these glycan moiety-targeting approaches, the resulting deglycosylated proteins, or their hydrolyzed peptides are generally separated from the glycans before subsequent mass analyses. Thereby the complexity of the enriched glycans sample is dramatically decreased. This glycan-targeting approach is very powerful method for structural identification of the glycans of purified glycoproteins and quantitative analysis of the glycosylation patterns of the glycans, as evaluated by HUPO collaboration studies (Wada et al., 2007; Wada et al., 2010a).
Porous graphite carbon is attractive to capture and separate hydrophilic glycans that are released from glycoproteins but not retained by reverse phase chromatography (Davies et al., 1992; Wada et al., 2007; Grass et al., 2011; Lam et al., 2011). Glycans separated can be analyzed by mass spectrometry, in their free glycans or in their derivative forms labeled by various tagging techniques, such as permethylation (Ciucanu & Kerek, 1984; Costello, Contado-Miller, & Cipollo, 2007), peracylation (Shetty & Holloway, 1994), derivatization with amine tag (Kotani & Takasaki, 1998; Gil, Kim, & Kim, 2008; Nakano et al., 2009; Walker et al., 2011), or active methylene tag (Ahn & Yoo, 1998; Markely et al., 2010), for high sensitive detection or improved chromatographic separation of the glycans to be analyzed. Unfortunately this glycan-targeting approach has disadvantage such that information for glycoprotein or glycosylation site generating a specific glycan component of interest disappears inevitably during glycan-preparing process via deglycosylation reaction by enzymatic or chemical method (Maley et al., 1989; Wells et al., 2002; Miura et al., 2010). As a supplementary approach for the glycan-targeting method, the analysis of the deglycosylated protein parts of glycoproteins that drop glycan moieties via deglycosylation, is useful for glycoprotein identification and glycosite (lately, deglycosylated) identification through profiling experiment by LC/tandem MS.
The graphite carbon is also useful to the analysis of a complex mixture of glycopeptides. Glycopeptides originated by enzymatic hydrolysis using various endoproteinases like trypsin from total proteome or fractionated glycoproteome can be targeted instead of glycan moieties (Alley, Mechref, & Novotny, 2009). Glycopeptides are also routinely separated by porous graphite carbon and chromatographic methods at peptide level before tandem MS-based analysis. This glycopeptide-targeting approach has advantage such that information for glycosylation sites and glycan structures linked to each glycosylation sites can be concurrently obtainable by tandem mass analysis of each glycopeptides although exact structural identification of glycopeptides is much more challenging because of their high structural complexity due to glycan microheterogeneity, complex fragmentation patterns in tandem mass analysis, and poor chromatographic separation due to linkage isomerism in glycosidic bond. A micro fluidic chip packed with porous graphitized carbon was used for chromatographic separation of complex glycopeptides and identification of N-linked and O-linked glycopeptides via MS and MS/MS analyses (Alley, Mechref, & Novotny, 2009; Froehlich et al., 2011; Nwosu et al., 2011; Hua et al., 2012). These approaches present a platform to simultaneously characterize N- and O-glycosites in the same mixture with extensive site heterogeneity and to allow isomer-sensitive glycoprotein analysis.
Sometimes because of the size of glycopeptides, they are not often amenable to tandem MS. To overcome this problem, glycoproteins were digested with multiple proteases including pronase to produce glycopeptides that are of suitable size for tandem MS analysis, which leaded to maximize glycan and peptide information via using optimized conditions for tandem MS of graphite carbon-enriched sample (Froehlich et al., 2011; Nwosu et al., 2011). A simple and economical technique using ZipTip microcolumn packed with a 1:1 ratio of graphite carbon to activated charcoal (w/w) (termed as GA-ZipTip microcolumn) was also introduced to efficiently isolate and enrich N-glycopeptides from digested mixtures of individual glycoproteins (Xin et al., 2012).
Affinity Separation Techniques Using HILIC
Hydrophilic interaction liquid chromatography (HILIC) was introduced as promising enrichment and separation method for glycopeptides and glycans originated from glycoproteins. HILIC is characterized by using a hydrophilic stationary phase, based on functionality of cationic exchange (Alpert, 1990; Lindner et al., 1996), anionic exchange (Alpert, 2008), zwitterionic interaction (Naidong, 2003), and sepharose (Wada, Tajiri, & Yoshida, 2004), and by using a relatively hydrophobic organic mobile phase. Due to the impact of the hydrophilic carbohydrate moiety, glycopeptides were more strongly retained on the HILIC column and separated from the remaining nonglycosylated peptides present in the digest. Thereby each glycans or glycopeptides were also chromatographically resolved and analyzed by mass spectrometry detectors. So, HILIC is performed usually with water miscible solvents and elution is achieved by a water gradient (Alpert, 1990; Naidong, 2003). The hydrophilicity of oligosaccharides of glycopeptides makes them ideal candidates for separation by HILIC (Lam et al., 2010; Gilar et al., 2011; Neue et al., 2011; Wan et al., 2011; Zhao et al., 2011b).
The combinations of the HILIC with other technologies, such as the hydrazide-capturing and lectin-specific fractionation for glycoproteins, have been attractive strategy for subsequent MS-based analysis of complex glycoproteome in the glycoproteomics field (Kaji et al., 2006; Palmisano et al., 2010; Parker et al., 2011). The HILIC method can also be used through combination with a specific enzymatic hydrolysis method. Glycopeptides enriched by the HILIC are generally treated with PNGase F before subsequent LC-MS/MS to identify glycosylation sites (Parker et al., 2011; Qu et al., 2011; Kuo et al., 2012). The HILIC approach can also be applied to identification of core fucosylated N-glycans and O-glycosylation site mapping of glycoproteins through partial deglycosylation performed by a specific enzyme reaction (Hagglund et al., 2004, 2007). In these studies, digestion of glycopeptides is conducted by using endo-beta-N-acetylglucosaminidases (Endo H) that cleave the glycosidic bond between the two GlcNAc residues in the conserved N-glycan core structure. Thereby, single GlcNAc residues with putative fucosyl side moieties are leaved onto the peptide chains. These partial deglycosylated peptides can be analyzed by mass spectrometry to afford unambiguous assignment of their glycosylation sites. The HILIC method can also be used through combination with the graphite carbon. N-linked glycans enriched with graphitized carbon cartridge and more separated over a TSK-Gel Amide80 column under HILIC conditions were analyzed by LC-coupled Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) to determine the sugar composition, linkage pattern, and attachment sites of N-linked glycans (Wang, Emmett, & Marshall, 2010).
Affinity Separation Techniques Using Boronic Acid
Boronic acid has also been used to enrich glycoproteins or glycopeptides by selectively capturing glycan moieties. Boronic acid shows selective binding affinity to cis-diols present in monosaccharides on glycan moieties via formation of boronate heterocyclic diester. The glycoproteins or glycopeptides captured by the boronic acid and purified by a washing buffer is released from the boronic acid via the exchange with monosaccharides present in a eluting buffer at high concentration. In general, boronic acid immobilized on magnetic beads has been used to capture glycoproteins and glycopeptides through formation of the boronate diesters with vicinal diols on glycan moieties (Lee et al., 2005; Sparbier et al., 2005, 2007). On-plate versions using boronic acid-modified gold nanoparticles or gold-coated Si wafer were also introduced for enrichment of glycopeptides and subsequent matrix-assisted laser desorption/ionization (MALDI) MS (Tang et al., 2009; Xu et al., 2010). Boronate affinity monolithic columns were developed for selective enrichment of glycopeptides and glycoproteins and evaluated their capturing performances by capillary liquid chromatography (LC) (Chen et al., 2009; Lin et al., 2011; Yang et al., 2011).
These boronic acid-capturing methods are useful for capturing glycosylated molecules from highly complex sample with intact glycan structures and characterizing the captured glycans with high-throughput analytical performance. Nonetheless, the boronic acid-capturing are mainly selective to only cis-diols on monosaccharides consisting of glycans, but is not promising for selective capturing of protein glycoforms having a specific glycan structure (i.e., produced by aberrant protein glycosylation in cancer cells) (Table 1).
In general, glycans and glycopeptides enriched from complex proteome samples using various enrichment techniques addressed above can be analyzed by high-throughput tandem mass spectrometry. However, although peptide identification employing tandem MS data obtained is now routinely achievable, automated glycopeptide identification from these tandem MS data for glycans and glycopeptides remains a considerable challenge in the areas of glycomics and glycoproteomics. In the past decade, numerous software programs are released to identify glycan structure from tandem MS data for glycans such as OSCAR (Lapadula et al., 2005), GLYCH (Tang, Mechref, & Novotny, 2005), and GlycosidIQ (Joshi et al., 2004). Data obtained from tandem MS analysis of glycopeptides are much more challenging to handle because of the high complexity and size of the molecules and the highly complex fragmentation patterns in tandem analysis. Several software programs were released to identify glycopeptides using high-throughput tandem MS data obtained from protein cocktails or complex proteome samples, such as Branch-and-Bound (Peltoniemi, Joenväärä, & Renkonen, 2009), GlycoMiner (Ozohanics et al., 2008), GP Finder (Nwosu et al., 2011), and Medical N-glycosite library (Joenväärä et al., 2008). Therefore, notable progress in development of software program for glycopeptides identification has been realized. Nonetheless, the development of an innovative software program capable of monitoring quantitative variation of glycopeptides as well as identifying glycopeptide compositions and/or structures is required to better understand the functions and bioactivities of glycoproteins.
MASS SPECTROMETRY-BASED QUANTIFICATION METHODS
Glycoproteome samples can be generally purified and enriched in a state of glycoproteins, glycopeptides, and glycans by using various separation technologies discussed above including the hydrazide-capturing and the lectin-specific fractionation, and affinity separation methods. Each enriched samples are primarily analyzed qualitatively for structure identification of glycans or proteome profiling by tandem mass spectrometry. To obtain quantitative information for these enriched samples, a variety of analytical approaches depending on features of the each enriched samples are employed. For example, quantitative analysis in glycomics using glycan or glycopeptide samples mainly focuses increase or decrease of specific glycoforms in the glycan profile attached to a glycoprotein or in the glycan pool of whole glycoproteins. In different, quantitative analysis for glycoprotein samples in glycoproteomics largely focuses measurement of the concentration of specific glycoproteins, absolutely or relatively to other molecules, as well as quantitative comparison of the levels of specific glycoproteins between individuals or sample groups to be compared. Many MS-based analytical techniques to obtain quantitative information for these enriched samples have been employed such as label-free MS method, stable isotope-labeling method, and multiple reaction monitoring (MRM)-based quantification method with stable isotope standard (SIS). And these analytical techniques have been utilized independently or cooperatively in quantitative glycomic and glycoproteomic fields based on high-throughput mass spectrometry.
Quantitative Analysis by Label-Free MS
Quantitative analysis for glycans or glycopeptides obtained using various separation technologies discussed above and proper enzymatic or chemical proteolysis have been conducted efficiently by label-free MS technique to monitor changes in the glycan patterns attached to a glycoprotein or in the glycan pool of whole glycoproteins (Table 2). The label-free quantitative analysis of glycans has been applied for analysis of complex proteome samples like serum (An et al., 2003; Kirmiz et al., 2007). The dramatic reduction in sample complexity coming from glycan-targeting enrichment can also make the direct mass analysis of the purified glycans and glycopeptides enable to be accomplished using MALDI FT-ICR MS showing high resolving power (>100,000 full width at half height) and mass accuracy (<5 ppm). This glycan-targeting approach using MALDI MS was applied to development of breast cancer biomarker. To improve mass sensitivity of glycans to be analyzed, permethylation method for glycan moieties was also utilized in a study for breast cancer diagnosis (Kyselova et al., 2008). Although these benefits in the glycan-targeting approach, this approach has disadvantage such that information for glycoprotein or glycosylation site generating a specific glycan component of interest disappears inevitably during glycan-preparing process via deglycosylation reaction by chemical or enzymatic method. Since glycan sample prepared from a complex proteome sample is still highly complex, further separation for the glycans using HILIC can be conducted before mass spectrometric analysis to N-linked glycosylation profiling of pancreatic cancer serum (Zhao et al., 2007; Mann et al., 2012).
Table 2.
Summary of typical methods for glycoproteome-targeting quantitative analysis using label-free MS
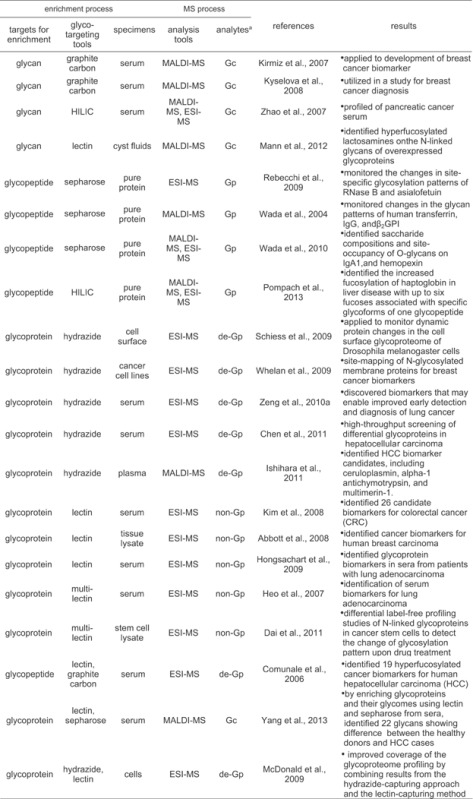 |
aGc, Gp, de-Gp, and non-Gp represent glycans, glycopeptides, deglycopeptides, and nonglycopeptides released from glycoprotein and detected by MS, respectively.
As a label-free quantitative method targeting glycopeptides, a model study using a single pure glycoprotein and a glycoprotein mixture was introduced (Rebecchi et al., 2009). In this study, through the label-free analysis for N-linked glycopeptides enriched by sepharose, the changes in glycosylation patterns were monitored in a glycosylation site-specific manner on a single glycoprotein and a glycoprotein mixture. Also, Usefulness of label-free quantitative analysis of glycans released from purified glycoproteins was evaluated by coupling with sepharose-based enrichment (Wada, Tajiri, & Yoshida, 2004; Wada, Tajiri, & Ohshima, 2010b). In the study, label-free analysis for N-, O-linked glycopeptides released from glycoproteins purified from serum by polyclonal antibody was tried to identify saccharide compositions and site-occupancy of N-, O-glycans on the glycoproteins. From the signal intensity of glycopeptide ions in the mass spectra and tandem mass spectra from electron transfer dissociation, the variability in glycan modifications among individuals was evaluated and the applicability of this label-free analysis method to detection of disease-related alterations was introduced. Recently, a study to examine site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma (HCC) was introduced (Pompach et al., 2013). By two-dimensional separation composed of hydrophilic interaction and nano-reverse phase chromatography coupled to QTOF mass spectrometry of the enriched glycopeptides, it was found that fucosylation of haptoglobin in liver disease increased with up to six fucoses associated with specific glycoforms of one glycopeptides.
As a glycoprotein-targeting method, a chemical reaction-based approach based on hydrazide chemistry was utilized for label-free quantification of hydrazide-captured glycoproteins (Schiess et al., 2009). The MS-based strategy for the specific detection and quantification of cell surface proteome changes was tried based on the hydrazide-based capturing of cell surface glycoproteome and the label-free quantification of peptide patterns for the captured cell surface proteome by mass spectrometry. This cell surface capturing technology that selectively enriches glycopeptides exposed to the cell exterior was applied to monitor dynamic protein changes in the cell surface glycoproteome of Drosophila melanogaster cells. Hydrazide-based capturing approaches coupled label-free quantification mass spectrometry have also been tried for biomarker discovery targeting breast cancer (Whelan et al., 2009), lung cancer (Zeng et al., 2010a), and HCC (Chen et al., 2011; Ishihara et al., 2011).
Glycoproteomes fractionated by lectin-specific capturing have also been used for the label-free quantification of the lectin-specific glycoproteomes via mass profiling of tryptic digests of the fractionated glycoproteins. This lectin-capturing approach coupled with tandem MS was applied to serum proteome sample for serological biomarker discovery in colorectal cancer (Kim et al., 2008). This study indicates that lectin enrichment strategy targeting an abnormal change in glycan microheterogeneity associated with malignancy in cells can be an effective method for discovery of potential cancer biomarkers. A lectin-captured sample from tissue lysate proteome was also used to identify cancer biomarkers for human breast carcinoma (Abbott et al., 2008), lung adenocarcinoma (Hongsachart et al., 2009). With these lectin-capturing approaches using a specific single lectin, a multi-lectin column prepared by in advance mixing multiple lectins was also developed and utilized for identification of serum biomarkers for lung adenocarcinoma (Heo et al., 2007), breast cancer (Zeng et al., 2010b), and ovarian cancer (Abbott et al., 2010). Differential label-free profiling studies of N-linked glycoproteins in cancer stem cells were tried to detect the change of glycosylation pattern upon drug treatment (Dai et al., 2011; He et al., 2011).
The combination strategy using different separation techniques may be useful for better enrichment of glycoproteins or glycopeptides. In a study, the glycan enrichment by porous graphite carbon and the glycopeptides enrichment by lectin-capturing technique were used together for label-free quantitative analysis (Comunale et al., 2006). From this comparative data analysis, 19 hyperfucosylated glycoproteins were identified as HCC biomarker candidates. A Con A-magnetic particle conjugate-based method was also utilized to selectively isolate the glycoproteins and their glycomes from the healthy donor and HCC case sera (Yang et al., 2013). By MALDI-TOF/TOF-MS of glycans prepared by sepharose enrichment of tryptic digests derived from the lectin-capturing glycoproteins, 22 glycans showing difference between the healthy donors and HCC cases were identified. It was further confirmed the differences of the identified glycoproteins between the healthy donors and HCC cases were caused by the change of both protein expression and their glycosylation levels. The hydrazide-capturing technique was also compared with the lectin-specific capturing technique by using label-free tandem MS for lysates of cancer cells (McDonald et al., 2009). The combinational application of two methods, one that uses periodate to glycoproteins of intact cells and a hydrazide resin to capture the oxidized glycoproteins and another that targets glycoproteins with sialic acid residues using lectin affinity chromatography, in conjunction with liquid chromatography-tandem mass spectrometry is effective for plasma membrane glycoprotein identification. As a result, it was observed that combining results from the hydrazide-capturing approach and the lectin-capturing method substantially improves the coverage of the glycoproteome profiled.
Quantitative Analysis by Isotope Tag-Labeling Approach
In the past decade, one of the techniques widely adopted for quantitative proteomic studies is a stable isotope-labeling approach, including reductive amination using isotope-coded amine tag, permethylation using isotope-coded methyl group, and enzyme-mediated incorporation of isotope-coded reagent (Table 3). These isotope tag-labeling techniques have been applied for comparative analysis between proteomic samples to be compared such as disease and healthy samples. Glycans released from glycoproteome have been labeled through reductive amination method using various isotope-coded amine tags, and analyzed comparatively by mass spectrometric-based methods (Xia et al., 2009; Bowman & Zaia, 2010; Prien et al., 2010). Similarly hydrazide tag was also used for the relative quantification of N-linked glycans by ESI mass spectrometry (Walker et al., 2011). A novel strategy, quantification by isobaric labeling (QUIBL), using permethylation reaction by stable isotope-coded methyl iodide was introduced to facilitate comparative glycomics (Atwood et al., 2008). Permethylation of a glycan with 13CH3I or 12CH2DI generates a pair of isobaric derivatives, which have the same nominal mass. However, since each methylation site introduces a mass difference of 0.002922 Da, the total mass difference for the isobaric pair due to multiple methylation sites allows separation and quantification at a resolution of approximately 30,000 m/Delta m. QUIBL facilitates relative quantification over a linear dynamic range of two orders of magnitude and permits the relative quantification of isomeric glycans. As an enzyme-mediated incorporation method using an isotope-coded reagent, endoglycosidase-mediated incorporation of 18O into glycans was tried for relative quantification of N-linked glycans collected from sera of HCC patient (Zhang et al., 2011).
Table 3.
Summary of typical methods for glycoproteome-targeting quantitative analysis using stable isotope tag-labeling method
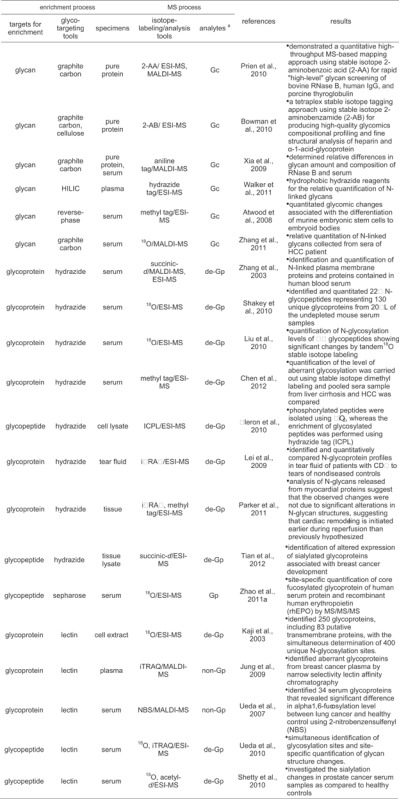 |
aGc, Gp, de-Gp, and non-Gp represent glycans, glycopeptides, deglycopeptides, and nonglycopeptides released from glycoprotein and detected by MS, respectively.
Besides glycan-targeting quantitative analysis, many studies targeting glycopeptides or glycoproteins have been tried for quantitative analysis of protein glycosylation. The stable isotope-labeling approach is also useful for comparative quantification of glycoproteins enriched by the hydrazide-capturing. The most notable feature in hydrazide-capturing method for glycoprotein enrichment is its capability of enriching globally overall protein glycoforms and its viability in stable isotope-labeling strategy for the hydrazide-captured glycoproteins to do quantitative mass analysis. Hydrazide bead-bound glycopeptides generated from glycoproteins captured by the hydrazide can be labeled isotopically by a stable isotope-coded tag for comparative quantification (Zhang et al., 2003). The isotope-labeling can also be accomplished by the enzyme-mediated incorporation technique during tryptic hydrolysis of the hydrazide-captured glycoproteins, or during enzymatic deglycosylation of glycopeptides generated by the tryptic hydrolysis. This enzyme-mediated 18O-labeling for hydrazide-captured glycoproteins was accomplished sequentially in the hydrolysis step of the captured glycoproteins and the deglycosylation step of bead-bound glycopeptides (Liu et al., 2010; Shakey, Bates, & Wu, 2010). And the resulting 18O3-isotope-labeled and deglycosylated peptides are generally analyzed quantitatively by tandem MS. A combined approach using hydrazide-based capturing, and enzymatic cleavage by endoglycosidase F3, and isotopically differential dimethylation was introduced for the mass spectrometric identification and quantification of aberrant N-glycosylation in HCC sera (Chen et al., 2012).
A novel post-digest labeling strategy using isotope-coded protein label (ICPL) was developed for quantitative characterization of phosphorylated and glycosylated proteins in prostate-related metastatic adenocarcinoma cell lines originating from two distinct metastasis sites (Fleron et al., 2010). In this study, phosphorylated peptides were isolated using TiO2, whereas the enrichment of glycosylated peptides was performed using hydrazide-based chemistry. An isobaric tagging reagent iTRAQ was also used for quantitative analysis of deglycosylated peptides collected from N-linked glycoproteins in tear fluid by hydrazide-based capturing and enzymatic deglycosylation by endoproteinase (Lei et al., 2009). A quantitative analysis of N-linked glycoproteins in cardiovascular disease, particularly ischemia, and reperfusion injury was conducted with hydrazide-based capturing, HILIC separation, and iTRAQ labeling techniques (Parker et al., 2011). The hydrazide-capturing technique as glycopeptides-targeting method was further applied the sialoglycoproteomic method to the profiling of breast cancer tissues and compared findings with the results from the total glycoproteomic analysis (Tian et al., 2012). From this study, altered expression of sialoglycoproteins, as well as the total glycoprotein changes associated with breast cancer was identified using isotope-labeled succinic anhydride. The glycopeptides-targeting approach was also used for tandem mass analysis of the core fucosylated and 18O-labeled glycopeptides separated by sepharose (Zhao et al., 2011a). The core fucosylated peptides were prepared through partial deglycosylation of enzyme-mediated 18O incorporated glycopeptides and separated by sepharose. In the core fucosylated peptides, the innermost GlcNAc and fucose groups are kept as glycopeptides through Endo F3-catalyzed partial deglycosylation. Thus the relative quantification of the core fucosylated and 18O-incorporated glycopeptides was achieved by tandem MS analysis.
The stable isotope-labeling for glycoproteins enriched by a specific lectin has also been tried for comparative quantification of protein glycoforms, showing a specific binding affinity to the used lectin. The most notable feature in lectin-specific enrichment for glycoproteins is its capability of enriching specifically a unique protein glycoforms having a specific glycan-structure active to a lectin used. Based on the lectin-specific capture of a set of glycopeptides in tryptic digests, and the following enzyme-mediated 18O incorporation into the N-glycosylation-specific site, a strategy for the large-scale identification and quantification of lectin-specific protein glycoforms from a complex biological sample was tried with LC-tandem MS (Kaji et al., 2003). The isobaric iTRAQ reagent was also useful for comparative quantification of protein glycoforms captured by multiple lectins showing different binding affinity to glycan structures (Jung, Cho, & Regnier, 2009). The use of lectin showing narrow selectivity can make it possible to characterize glycoprotein diversity and to recognize protein glycoforms associated with aberrant glycosylation. A tryptophan-specific labeling tag, 2-nitrobenzensulfenyl (NBS) was also used for comparative profiling of protein glycoforms lectin-captured from serum glycoproteome (Ueda et al., 2007). The NBS-labeled peptides were analyzed by MALDI-QIT-TOF mass spectrometric analysis for the lung cancer biomarker discovery.
As a glycopeptides-targeting method, a combined labeling approach using both the isobaric iTRAQ labeling and enzyme-mediated incorporation of 18O to lectin-captured protein glycoforms were introduced to simultaneous identify glycosylation sites and quantify changes of glycan structure in each specific glycosylation sites (Ueda et al., 2010). A lectin-directed tandem labeling (LTL) quantitative proteomics strategy was used for investigation of sialylation aberration in N-linked glycopeptides (Shetty et al., 2010). The sialylated glycopeptides enriched by SNA and labeled at its N-terminus by acetic anhydride-d reagents, were enzymatically deglycosylated in the presence of heavy water. The differentially labeled deglycopeptides were performed LC/MS/MS analysis to investigate the sialylation changes in prostate cancer serum samples as compared to healthy controls.
Targeted Multiple Reaction Monitoring (MRM)-Based Analysis
Targeted MRM-based MS has been widely used as a quantification technique of various target molecules because of its high specificity and high sensitivity in mass detection for the targets, and easy multiplexing for targets. The high specificity for a selected target in the LC-coupled MRM-based method comes from highly selective monitoring through sequential triplicate selections including separation of the target by liquid chromatography, precursor mass selection of the target peptide, and selective monitoring for fragment ions originated from the selected precursor target by tandem MS. LC-coupled MRM-based MS has been recognized to be a powerful tool for multiplexed quantitative monitoring of multiple target molecules in proteomics. A novel strategy, namely reverse glycoblotting technique, was introduced for the multiplexed quantitative mouse serum glycoproteomics based on a specific chemical ligation focusing sialic acids (Kurogochi et al., 2010). In this strategy, sialylated glycopeptides captured by hydrazide were released from the hydrazide beads through incorporation of a fluorescence tag, 2-aminopyridine, into the sialyl moieties on the captured glycopeptides. The released amine-tagged glycopeptides were profiled by tandem MS, and analyzed by MRM-based method. Because of unavailability of each isotope-coded reference peptides corresponding amine-tagged target glycopeptides for the MRM-based quantitative analysis, the study focused to compare quantitatively differences between diabetic model mouse and control mouse (Table 4).
Table 4.
Summary of typical methods for glycoproteome-targeting MRM-based quantitative analysis
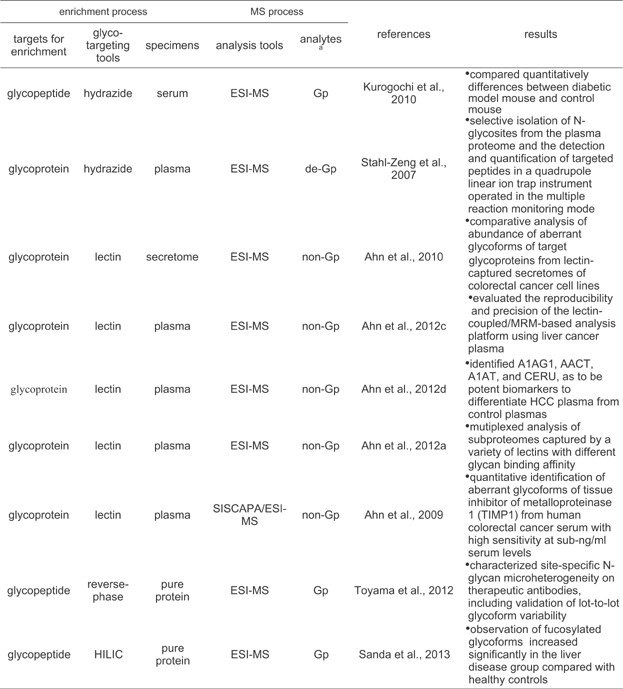 |
aGp, de-Gp, and non-Gp represent glycopeptides, deglycopeptides, and nonglycopeptides released from glycoprotein and detected by MS, respectively.
This LC-coupled MRM-based technique with stable isotope-coded standard has been utilized to analyze quantitatively glycoproteins of interest enriched from complex biological samples like plasma (Stahl-Zeng et al., 2007; Ahn et al., 2012a, c) and cell secretome (Ahn et al., 2010). This targeted quantification method using stable isotope-coded standard provides excellent precision and reproducibility in quantitative measurements of targeted peptides by virtue of using the internal standards of each target peptides (Table 4). The hydrazide-capturing technique to enrich glycoproteome from complex biological samples can be combined efficiently with the MRM-based quantification method with stable isotope standard. By virtue of capability of enriching globally overall protein glycoforms, the hydrazide-capturing technique can be an attractive tool for high-throughput quantitative mass analysis of complex glycoproteome. A quantitative analysis of N-glycosites of target glycoproteins of interest in plasma samples were conducted by targeted MRM-based method using tryptic peptide mixtures, prepared by tryptic digestion and sequential PNGases F-mediated deglycosylation of hydrazide beads-bound glycoproteins (Stahl-Zeng et al., 2007). In the process of the PNGases F-mediated enzyme action, the asparagine residue of the NX(S/T) consensus sequence of N-glycosites on the glycopeptides hydrazide bead-captured is converted into an aspartic acid ( Fig. 2). In this study the application of MRM-based method to N-glycosites isolated from plasma shows excellent limit of detection of peptides in the low ng/mL to sub-ng/mL range concentrations. In addition, it was confirmed that the use of stable isotope-labeled reference peptides makes the targeted analytes be accurately quantified over a dynamic range of near five orders of magnitude.
Lectin fractionation enabling to capture lectin-specific protein glycoforms is provides the basis for detecting and quantifying different types of protein glycoforms from complex biological samples. The targeted MRM-based method was used for measuring an abundance of aberrant glycoforms of target glycoproteins from lectin-captured secretomes of colorectal cancer cell lines (Ahn et al., 2010). The lectin-coupled/MRM-based quantification approach was confirmed to be a powerful tool for measuring the abundance of a specific glycoforms involving in aberrant protein glycosylation, rather than measuring total glycoforms of the target protein using human plasma sample (Ahn et al., 2012c, d). In these studies using aleuria aurantia lectin (AAL) showing a specific affinity to fucose on N-linked glycans, the lectin-coupled/MRM-based method was evaluated to give reproducibility and precision sufficient to distinguish a difference in the abundance of AAL-captured protein glycoforms between control and HCC plasmas, even without highly abundant protein depletion or antibody-based enrichment against target proteins. Since a variety of lectins are available, this method can be easily extended to the independent analysis of various glycan-specific subproteomes captured by a variety of lectins with different binding affinity for glycan structures. A multiplex LCM consisting of multiplex parallel lectin-fractionation and MRM-based analysis was tried to quantitatively monitor the diversity of serological protein glycosylation in a human serum sample (Ahn et al., 2012a).
Many biomarker candidates identified from studies in glycoproteomic field present at low abundant concentration levels in complex proteome samples. Further the concentration level of the aberrant glycoforms of a biomarker candidate may be far low compared with that of total glycoforms of the candidate. For enriching the target peptide as a surrogate of the aberrant protein glycoforms of the biomarker, and to decrease the sample complexity, a target peptide enrichment protocol, using a stable isotope standard and capture by anti-peptide antibody (SISCAPA) technique, is efficient since the enriched antigen (target peptide) is directly subjected to mass analysis without further sample treatment (Anderson et al., 2004). A quantitative identification of aberrant glycoforms of tissue inhibitor of metalloproteinase 1 (TIMP1) was accomplished from human colorectal cancer serum by the systematically combined strategy using a lectin-capturing and SISCAPA/MRM technique with high sensitivity at sub-ng/mL serum levels (Ahn et al., 2009). TIMP1 in colorectal cancer serum was further confirmed by 15T MALDI FT-ICR MS with high resolving power (>400,000 full width at half-height) and mass accuracy (<0.5 ppm) (Ahn et al., 2012b).
Recently, a new method combining MRM MS technique with energy-resolved structural analysis, termed “energy-resolved oxonium ion monitoring,” was introduced (Toyama et al., 2012). Monitoring the yields of oligosaccharide-derived fragment ions (oxonium ions) over a wide range of collision-induced dissociation (CID) energy applied to a glycopeptide precursor exhibits a glycan structure-unique fragmentation pattern. In the analysis of purified immunoglobulin glycopeptides, the energy-resolved oxonium ion profile was shown to clearly distinguish between isomeric glycopeptides. Therefore, both quantification of glycopeptides and assignment of their glycan structures were achieved by a simple analysis procedure. This method was further applied for characterizing site-specific N-glycan microheterogeneity on therapeutic antibodies, including validation of lot-to-lot glycoform variability. The data suggest that energy-resolved oxonium ion monitoring could fulfill the regulatory requirement on the routine quality control analysis of forthcoming biosimilar therapeutics. Similarly an LC-MS-MRM workflow was used to analyze the disease-related haptoglobin glycoforms in liver cirrhosis and HCC (Sanda et al., 2013). In this study, LC-MS-MRM analysis for samples prepared by treatment of a tryptic digest of haptoglobin with exoglycosidase was conducted differentially though monitoring oligosaccharide-derived fragment ions (oxonium ions) and a glycopeptide-specific ion. Because of unavailability of each isotope-coded reference glycopeptides for the MRM-based quantitative analysis, this exoglycosidase-assisted LC-MS-MRM workflow focused to compare quantitatively differences between disease groups.
PERSPECTIVES
Because of the high complexity and deep dynamic range in protein constituents in a clinical proteome sample as well as the microheterogeneity in a glycoprotein due to the diversity of glycan structure and the linkage complexity in glycosidic binding of the glycoprotein, there is still great challenge for comprehensive analysis of glycoprotein. Nonetheless by virtue of momentous efforts in analytical proteomic fields, mass spectrometry is now recognized as a core technology for glycoproteomics research. Mass spectrometry showing excellent capability in high-throughput analytical performance affords accurate digitalized informatics through profiling efficiently large number of glycoproteins from various complex biological media. Over profiling qualitatively large number of target proteins, mass spectrometry has also become central roles in quantitative glycoproteomics through the development of efficient separation tools for glycoproteome and powerful techniques for quantification of the separated glycoproteome, and the systematic integration of the developed technologies for glycoprotein separation and quantification. As shown many reports that aberrant protein glycosylation is involved in abnormal pathological function of cells like in cancer, lectin specificity-based glycoprotein enrichment approach, capable of targeting protein glycoforms having glycan structures active specifically to a used lectin rather than targeting total glycoforms of the glycoprotein, has been received particularly great attention in the clinical proteomic field. In addition, MRM-based quantification approach showing notable advantage in targeted proteomics is also expected to play central roles in high-throughput validation studies for tremendous number of biomarker candidates newly discovered but requiring more validation studies.
Nonetheless there are still many obstacles to overcome for the MS-based quantification approach to be utilized regularly in clinical proteomic study and biomarker development. Many successful studies to identify glycosylation patterns of glycans or glycopeptides released from a glycoprotein and monitor changes in the glycosylation patterns have been conducted mainly through analysis of single glycoprotein pre-purified by affinity-based enrichment using antibody. Therefore, the development of more powerful tool is required enable to monitor changes in glycan patterns directly from complex proteome samples even without using individual antibody. Thus, the combination of the global enrichment technique for glycopeptides (using hydrazide-capturing, porous graphite carbon, or HILIC) and the aberrant glycoform-specific enrichment tool like lectin-capturing may be effective. This integrated strategy should be coupled practically with a high-sensitive tandem mass analysis technique for high-throughput identifying of aberrant protein glycoform and monitoring its quantitative change from complex samples. Another to be solved for the MS-based quantification tool to be utilized regularly in clinical proteomic study and biomarker development is the development of a verification tool for target glycoforms identified as aberrantly glycosylated but requiring further verification against large number of cohorts. For this, systematical integration of an enrichment technique for the aberrantly glycosylated target identified and a high-throughput MS-based assay system may be an attractive approach. This high-throughput MS-based assay platform focusing aberrant glycosylation will play important role in verification study for biomarker candidate involved in aberrant protein glycosylation.
Acknowledgments
The research was supported by the Converging Research Center Program through the Ministry of Science, ICT and Future Planning, Korea (Grant Number 2013K000426).
REFERENCES
- Abbott KL, Aoki K, Lim JM, Porterfield M, Johnson R, O'Regan RM, Wells L, Tiemeyer M, Pierce M. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res. 2008;7:1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott KL, Lim JM, Wells L, Benigno BB, McDonald JF, Pierce M. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics. 2010;10:470–481. doi: 10.1002/pmic.200900537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn YH, Yoo JS. Malononitrile as a new derivatizing reagent for high-sensitivity analysis of oligosaccharides by electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 1998;12:2011–2015. doi: 10.1002/(SICI)1097-0231(19981230)12:24<2011::AID-RCM429>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Lee JY, Lee JY, Kim YS, Ko JH, Yoo JS. Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J Proteome Res. 2009;8:4216–4224. doi: 10.1021/pr900269s. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Kim YS, Ji ES, Lee JY, Jung JA, Ko JH, Yoo JS. Comparative quantitation of aberrant glycoforms by lectin-based glycoprotein enrichment coupled with multiple-reaction monitoring mass spectrometry. Anal Chem. 2010;82:4441–4447. doi: 10.1021/ac1001965. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Ji ES, Shin PM, Kim KH, Kim YS, Ko JH, Yoo JS. A multiplex lectin-channel monitoring method for human serum glycoproteins by quantitative mass spectrometry. Analyst. 2012a;137:691–703. doi: 10.1039/c1an15775b. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Kim KH, Shin PM, Ji ES, Kim H, Yoo JS. Identification of low-abundance cancer biomarker candidate TIMP1 from serum with lectin fractionation and peptide affinity enrichment by ultrahigh-resolution mass spectrometry. Anal Chem. 2012b;84:1425–1431. doi: 10.1021/ac2024987. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Shin PM, Ji ES, Kim H, Yoo JS. A lectin-coupled, multiple reaction monitoring-based quantitative analysis of human plasma glycoproteins by mass spectrometry. Anal Bioanal Chem. 2012c;402:2101–2112. doi: 10.1007/s00216-011-5646-3. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Shin PM, Oh NR, Park GW, Kim H, Yoo JS. A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J Proteomics. 2012d;75:5507–5515. doi: 10.1016/j.jprot.2012.06.027. [DOI] [PubMed] [Google Scholar]
- Alley WR, Jr, Mechref Y, Novotny MV. Use of activated graphitized carbon chips for liquid chromatography/mass spectrometric and tandem mass spectrometric analysis of tryptic glycopeptides. Rapid Commun Mass Spectrom. 2009;23:495–505. doi: 10.1002/rcm.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert AJ. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J Chromatogr. 1990;499:177–196. doi: 10.1016/s0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- Alpert AJ. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 2008;80:62–76. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
- An HJ, Peavy TR, Hedrick JL, Lebrilla CB. Determination of N-glycosylation sites and site heterogeneity in glycoproteins. Anal Chem. 2003;75:5628–5637. doi: 10.1021/ac034414x. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- Atwood JA, III, Cheng L, Alvarez-Manilla G, Warren NL, York WS, Orlando R. Quantitation by isobaric labeling: Applications to glycomics. J Proteome Res. 2008;7:367–374. doi: 10.1021/pr070476i. [DOI] [PubMed] [Google Scholar]
- Barnes J, Lim JM, Godard A, Blanchard F, Wells L, Steet R. Extensive mannose phosphorylation on leukemia inhibitory factor (LIF) controls its extracellular levels by multiple mechanisms. J Biol Chem. 2011;286:24855–24864. doi: 10.1074/jbc.M111.221432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berven FS, Ahmad R, Clauser KR, Carr SA. Optimizing performance of glycopeptide capture for plasma proteomics. J Proteome Res. 2010;9:1706–1715. doi: 10.1021/pr900845m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman MJ, Zaia J. Comparative glycomics using a tetraplex stable-isotope coded tag. Anal Chem. 2010;82:3023–3031. doi: 10.1021/ac100108w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Shen C, Wang H, Shen H, Chen Y, Nie A, Yan G, Lu H, Liu Y, Yang P. Identification of N-glycosylation sites on secreted proteins of human hepatocellular carcinoma cells with a complementary proteomics approach. J Proteome Res. 2009;8:662–672. doi: 10.1021/pr800826u. [DOI] [PubMed] [Google Scholar]
- Chen M, Lu Y, Ma Q, Guo L, Feng YQ. Boronate affinity monolith for highly selective enrichment of glycopeptides and glycoproteins. Analyst. 2009;134:2158–2164. doi: 10.1039/b909581k. [DOI] [PubMed] [Google Scholar]
- Chen R, Tan Y, Wang M, Wang F, Yao Z, Dong L, Ye M, Wang H, Zou H. Development of glycoprotein capture-based label-free method for the high-throughput screening of differential glycoproteins in hepatocellular carcinoma. Mol Cell Proteomics. 2011;10:M110.006445. doi: 10.1074/mcp.M110.006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Wang F, Tan Y, Sun Z, Song C, Ye M, Wang H, Zou H. Development of a combined chemical and enzymatic approach for the mass spectrometric identification and quantification of aberrant N-glycosylation. J Proteomics. 2012;75:1666–1674. doi: 10.1016/j.jprot.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Choi E, Loo D, Dennis JW, O'Leary CA, Hill MM. High-throughput lectin magnetic bead array-coupled tandem mass spectrometry for glycoprotein biomarker discovery. Electrophoresis. 2011;32:3564–3575. doi: 10.1002/elps.201100341. [DOI] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, Evans AA, Hann HW, Block TM, Mehta AS. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, Block T, Mehta A. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: Implications for a biomarker of hepatocellular carcinoma. PLoS ONE. 2010;5:e12419. doi: 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello CE, Contado-Miller JM, Cipollo JF. A glycomics platform for the analysis of permethylated oligosaccharide alditols. J Am Soc Mass Spectrom. 2007;18:1799–1812. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Zhou J, Qiu SJ, Liu YK, Fan J. Lectin-based glycoproteomics to explore and analyze hepatocellular carcinoma-related glycoprotein markers. Electrophoresis. 2009;30:2957–2966. doi: 10.1002/elps.200900064. [DOI] [PubMed] [Google Scholar]
- Dai L, Liu Y, He J, Flack CG, Talsma CE, Crowley JG, Muraszko KM, Fan X, Lubman DM. Differential profiling studies of N-linked glycoproteins in glioblastoma cancer stem cells upon treatment with γ-secretase inhibitor. Proteomics. 2011;11:4021–4028. doi: 10.1002/pmic.201100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M, Smith KD, Harbin AM, Hounsell EF. High-performance liquid chromatography of oligosaccharide alditols and glycopeptides on a graphitized carbon column. J Chromatogr. 1992;609:125–131. doi: 10.1016/0021-9673(92)80155-n. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Laferte S, Waghorne C, Breitman ML, Kerbel RS. Beta 1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science. 1987;236:582–585. doi: 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- Drake RR, Schwegler EE, Malik G, Diaz J, Block T, Mehta A, Semmes OJ. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- Drake PM, Schilling B, Niles RK, Braten M, Johansen E, Liu H, Lerch M, Sorensen DJ, Li B, Allen S, Hall SC, Witkowska HE, Regnier FE, Gibson BW, Fisher SJ. A lectin affinity workflow targeting glycosite-specific, cancer-related carbohydrate structures in trypsin-digested human plasma. Anal Biochem. 2011;408:71–85. doi: 10.1016/j.ab.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleron M, Greffe Y, Musmeci D, Massart AC, Hennequiere V, Mazzucchelli G, Waltregny D, De Pauw-Gillet MC, Castronovo V, De Pauw E, Turtoi A. Novel post-digest isotope coded protein labeling method for phospho- and glycoproteome analysis. J Proteomics. 2010;73:1986–2005. doi: 10.1016/j.jprot.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Froehlich JW, Barboza M, Chu C, Lerno LA, Jr, Clowers BH, Zivkovic AM, German JB, Lebrilla CB. Nano-LC-MS/MS of glycopeptides produced by nonspecific proteolysis enables rapid and extensive site-specific glycosylation determination. Anal Chem. 2011;83:5541–5547. doi: 10.1021/ac2003888. [DOI] [PubMed] [Google Scholar]
- Geng M, Zhang X, Bina M, Regnier F. Proteomics of glycoproteins based on affinity selection of glycopeptides from tryptic digests. J Chromatogr B Biomed Sci Appl. 2001;752:293–306. doi: 10.1016/s0378-4347(00)00550-8. [DOI] [PubMed] [Google Scholar]
- Gil GC, Kim YG, Kim BG. A relative and absolute quantification of neutral N-linked oligosaccharides using modification with carboxymethyl trimethylammonium hydrazide and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem. 2008;379:45–59. doi: 10.1016/j.ab.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Gilar M, Yu YQ, Ahn J, Xie H, Han H, Ying W, Qian X. Characterization of glycoprotein digests with hydrophilic interaction chromatography and mass spectrometry. Anal Biochem. 2011;417:80–88. doi: 10.1016/j.ab.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Gomaa AI, Khan SA, Leen EL, Waked I, Taylor-Robinson SD. Diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2009;15:1301–1314. doi: 10.3748/wjg.15.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass J, Pabst M, Chang M, Wozny M, Altmann F. Analysis of recombinant human follicle-stimulating hormone (FSH) by mass spectrometric approaches. Anal Bioanal Chem. 2011;400:2427–2438. doi: 10.1007/s00216-011-4923-5. [DOI] [PubMed] [Google Scholar]
- Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- Hagglund P, Matthiesen R, Elortza F, Hojrup P, Roepstorff P, Jensen ON, Bunkenborg J. An enzymatic deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins. J Proteome Res. 2007;6:3021–3031. doi: 10.1021/pr0700605. [DOI] [PubMed] [Google Scholar]
- He J, Liu Y, Zhu TS, Xie X, Costello MA, Talsma CE, Flack CG, Crowley JG, Dimeco F, Vescovi AL, Fan X, Lubman DM. Glycoproteomic analysis of glioblastoma stem cell differentiation. J Proteome Res. 2011;10:330–338. doi: 10.1021/pr101158p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SH, Lee SJ, Ryoo HM, Park JY, Cho JY. Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics. 2007;7:4292–4302. doi: 10.1002/pmic.200700433. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MA, Swanson BJ. Mucins in cancer: Protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- Hongsachart P, Huang-Liu R, Sinchaikul S, Pan FM, Phutrakul S, Chuang YM, Yu CJ, Chen ST. Glycoproteomic analysis of WGA-bound glycoprotein biomarkers in sera from patients with lung adenocarcinoma. Electrophoresis. 2009;30:1206–1220. doi: 10.1002/elps.200800405. [DOI] [PubMed] [Google Scholar]
- Hua S, Nwosu CC, Strum JS, Seipert RR, An HJ, Zivkovic AM, German JB, Lebrilla CB. Site-specific protein glycosylation analysis with glycan isomer differentiation. Anal Bioanal Chem. 2012;403:1291–1302. doi: 10.1007/s00216-011-5109-x. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Fukuda I, Morita A, Takinami Y, Okamoto H, Nishimura S, Numata Y. Development of quantitative plasma N-glycoproteomics using label-free 2-D LC-MALDI MS and its applicability for biomarker discovery in hepatocellular carcinoma. J Proteomics. 2011;74:2159–2168. doi: 10.1016/j.jprot.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Ito H, Kuno A, Sawaki H, Sogabe M, Ozaki H, Tanaka Y, Mizokami M, Shoda J, Angata T, Sato T, Hirabayashi J, Ikehara Y, Narimatsu H. Strategy for glycoproteomics: Identification of glyco-alteration using multiple glycan profiling tools. J Proteome Res. 2009;8:1358–1367. doi: 10.1021/pr800735j. [DOI] [PubMed] [Google Scholar]
- Joenväärä S, Ritamo I, Peltoniemi H, Renkonen R. N-glycoproteomics. An automated workflow approach. Glycobiology. 2008;18:339–349. doi: 10.1093/glycob/cwn013. [DOI] [PubMed] [Google Scholar]
- Joshi HJ, Harrison MJ, Schulz BL, Cooper CA, Packer NH, Karlsson NG. Development of a mass fingerprinting tool for automated interpretation of oligosaccharide fragmentation data. Proteomics. 2004;4:1650–1664. doi: 10.1002/pmic.200300784. [DOI] [PubMed] [Google Scholar]
- Jung K, Cho W, Regnier FE. Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J Proteome Res. 2009;8:643–650. doi: 10.1021/pr8007495. [DOI] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Kaji H, Yamauchi Y, Takahashi N, Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat Protoc. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- Khien VV, Mao HV, Chinh TT, Ha PT, Bang MH, Lac BV, Hop TV, Tuan NA, Don LV, Taketa K, Satomura S. Clinical evaluation of lentil lectin-reactive alpha-fetoprotein-L3 in histology-proven hepatocellular carcinoma. Int J Biol Markers. 2001;16:105–111. doi: 10.1177/172460080101600204. [DOI] [PubMed] [Google Scholar]
- Kim YS, Son OL, Lee JY, Kim SH, Oh S, Lee YS, Kim CH, Yoo JS, Lee JH, Miyoshi E, Taniguchi N, Hanash SM, Yoo HS, Ko JH. Lectin precipitation using phytohemagglutinin-L(4) coupled to avidin-agarose for serological biomarker discovery in colorectal cancer. Proteomics. 2008;8:3229–3235. doi: 10.1002/pmic.200800034. [DOI] [PubMed] [Google Scholar]
- Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S, Lebrilla CB, Miyamoto S. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- Klement E, Lipinszki Z, Kupihár Z, Udvardy A, Medzihradszky KF. Enrichment of O-GlcNAc modified proteins by the periodate oxidation-hydrazide resin capture approach. J Proteome Res. 2010;9:2200–2206. doi: 10.1021/pr900984h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani N, Takasaki S. Analysis of 2-aminobenzamide-labeled oligosaccharides by high-pH anion-exchange chromatography with fluorometric detection. Anal Biochem. 1998;264:66–73. doi: 10.1006/abio.1998.2820. [DOI] [PubMed] [Google Scholar]
- Kullolli M, Hancock WS, Hincapie M. Preparation of a high-performance multi-lectin affinity chromatography (HP-M-LAC) adsorbent for the analysis of human plasma glycoproteins. J Sep Sci. 2008;31:2733–2739. doi: 10.1002/jssc.200800233. [DOI] [PubMed] [Google Scholar]
- Kumada T, Nakano S, Takeda I, Kiriyama S, Sone Y, Hayashi K, Katoh H, Endoh T, Sassa T, Satomura S. Clinical utility of Lens culinaris agglutinin-reactive alphafetoprotein in small hepatocellular carcinoma: Special reference to imaging diagnosis. J Hepatol. 1999;30:125–130. doi: 10.1016/s0168-8278(99)80016-6. [DOI] [PubMed] [Google Scholar]
- Kuo CW, Wu IL, Hsiao HH, Khoo KH. Rapid glycopeptide enrichment and N-glycosylation site mapping strategies based on amine-functionalized magnetic nanoparticles. Anal Bioanal Chem. 2012;402:2765–2776. doi: 10.1007/s00216-012-5724-1. [DOI] [PubMed] [Google Scholar]
- Kurogochi M, Matsushista T, Amano M, Furukawa J, Shinohara Y, Aoshima M, Nishimura S. Sialic acid-focused quantitative mouse serum glycoproteomics by multiple reaction monitoring assay. Mol Cell Proteomics. 2010;9:2354–2368. doi: 10.1074/mcp.M110.000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH, Novotny MV. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin Chem. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- Lam MP, Siu SO, Lau E, Mao X, Sun HZ, Chiu PC, Yeung WS, Cox DM, Chu IK. Online coupling of reverse-phase and hydrophilic interaction liquid chromatography for protein and glycoprotein characterization. Anal Bioanal Chem. 2010;398:791–804. doi: 10.1007/s00216-010-3991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MP, Lau E, Siu SO, Ng DC, Kong RP, Chiu PC, Yeung WS, Lo C, Chu IK. Online combination of reversed-phase/reversed-phase and porous graphitic carbon liquid chromatography for multicomponent separation of proteomics and glycoproteomics samples. Electrophoresis. 2011;32:2930–2940. doi: 10.1002/elps.201100092. [DOI] [PubMed] [Google Scholar]
- Lapadula AJ, Hatcher PJ, Hanneman AJ, Ashline DJ, Zhang H, Reinhold VN. Congruent strategies for carbohydrate sequencing. 3. OSCAR: An algorithm for assigning oligosaccharide topology from MSn data. Anal Chem. 2005;77:6271–6279. doi: 10.1021/ac050726j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim Y, Ha MY, Lee EK, Choo J. Immobilization of aminophenylboronic acid on magnetic beads for the direct determination of glycoproteins by matrix assisted laser desorption ionization mass spectrometry. J Am Soc Mass Spectrom. 2005;16:1456–1460. doi: 10.1016/j.jasms.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lei Z, Beuerman RW, Chew AP, Koh SK, Cafaro TA, Urrets-Zavalia EA, Urrets-Zavalia JA, Li SF, Serra HM. Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J Proteome Res. 2009;8:1992–2003. doi: 10.1021/pr800962q. [DOI] [PubMed] [Google Scholar]
- Lin ZA, Pang JL, Lin Y, Huang H, Cai ZW, Zhang L, Chen GN. Preparation and evaluation of a phenylboronate affinity monolith for selective capture of glycoproteins by capillary liquid chromatography. Analyst. 2011;136:3281–3288. doi: 10.1039/c1an15180k. [DOI] [PubMed] [Google Scholar]
- Lindner H, Sarg B, Meraner C, Helliger W. Separation of acetylated core histones by hydrophilic-interaction liquid chromatography. J Chromatogr A. 1996;743:137–144. doi: 10.1016/0021-9673(96)00131-8. [DOI] [PubMed] [Google Scholar]
- Liu T, Qian WJ, Gritsenko MA, Camp DG, II, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cao J, He Y, Qiao L, Xu C, Lu H, Yang P. Tandem 18O stable isotope labeling for quantification of N-glycoproteome. J Proteome Res. 2010;9:227–236. doi: 10.1021/pr900528j. [DOI] [PubMed] [Google Scholar]
- Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- Mann BF, Goetz JA, House MG, Schmidt CM, Novotny MV. Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins. Mol Cell Proteomics. 2012;11:M111.015792. doi: 10.1074/mcp.M111.015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markely LR, Ong BT, Hoi KM, Teo G, Lu MY, Wang DI. A high-throughput method for quantification of glycoprotein sialylation. Anal Biochem. 2010;407:128–133. doi: 10.1016/j.ab.2010.07.029. [DOI] [PubMed] [Google Scholar]
- McDonald CA, Yang JY, Marathe V, Yen TY, Macher BA. Combining results from lectin affinity chromatography and glycocapture approaches substantially improves the coverage of the glycoproteome. Mol Cell Proteomics. 2009;8:287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y, Kato K, Takegawa Y, Kurogochi M, Furukawa J, Shinohara Y, Nagahori N, Amano M, Hinou H, Nishimura S. Glycoblotting-assisted O-glycomics: Ammonium carbamate allows for highly efficient o-glycan release from glycoproteins. Anal Chem. 2010;82:10021–10029. doi: 10.1021/ac101599p. [DOI] [PubMed] [Google Scholar]
- Miyaaki H, Nakashima O, Kurogi M, Eguchi K, Kojiro M. Lens culinaris agglutinin-reactive alpha-fetoprotein and protein induced by vitamin K absence II are potential indicators of a poor prognosis: A histopathological study of surgically resected hepatocellular carcinoma. J Gastroenterol. 2007;42:962–968. doi: 10.1007/s00535-007-2117-x. [DOI] [PubMed] [Google Scholar]
- Mizuochi T, Nishimura R, Derappe C, Taniguchi T, Hamamoto T, Mochizuki M, Kobata A. Structures of the asparagine-linked sugar chains of human chorionic gonadotropin produced in choriocarcinoma: Appearance of triantennary sugar chains and unique biantennary sugar chains. J Biol Chem. 1983;258:14126–14129. [PubMed] [Google Scholar]
- Morelle W, Canis K, Chirat F, Faid V, Michalski JC. The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics. 2006;6:3993–4015. doi: 10.1002/pmic.200600129. [DOI] [PubMed] [Google Scholar]
- Naidong W. Bioanalytical liquid chromatography tandem mass spectrometry methods on underivatized silica columns with aqueous/organic mobile phases. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;796:209–224. doi: 10.1016/j.jchromb.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Nakano M, Higo D, Arai E, Nakagawa T, Kakehi K, Taniguchi N, Kondo A. Capillary electrophoresis-electrospray ionization mass spectrometry for rapid and sensitive N-glycan analysis of glycoproteins as 9-fluorenylmethyl derivatives. Glycobiology. 2009;19:135–143. doi: 10.1093/glycob/cwn115. [DOI] [PubMed] [Google Scholar]
- Neue K, Mormann M, Peter-Katalinić J, Pohlentz G. Elucidation of glycoprotein structures by unspecific proteolysis and direct nanoESI mass spectrometric analysis of ZIC-HILIC-enriched glycopeptides. J Proteome Res. 2011;10:2248–2260. doi: 10.1021/pr101082c. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Rüetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods. 2009;6:809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German BJ, Lebrilla CB. Simultaneous and extensive site-specific N- and O-glycosylation analysis in protein mixtures. J Proteome Res. 2011;10:2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Ohyama C, Hosono M, Nitta K, Oh-eda M, Yoshikawa K, Habuchi T, Arai Y, Fukuda M. Carbohydrate structure and differential binding of prostate specific antigen to Maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology. 2004;14:671–679. doi: 10.1093/glycob/cwh071. [DOI] [PubMed] [Google Scholar]
- Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20:362–371. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Ozohanics O, Krenyacz J, Ludányi K, Pollreisz F, Vékey K, Drahos L. GlycoMiner: A new software tool to elucidate glycopeptide composition. Rapid CommunMass Spectrom. 2008;22:3245–3254. doi: 10.1002/rcm.3731. [DOI] [PubMed] [Google Scholar]
- Palmisano G, Lendal SE, Engholm-Keller K, Leth-Larsen R, Parker BL, Larsen MR. Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat Protoc. 2010;5:1974–1982. doi: 10.1038/nprot.2010.167. [DOI] [PubMed] [Google Scholar]
- Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin J, Li JG, Zhu D, Pan C, Zhang J. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- Parker BL, Palmisano G, Edwards AV, White MY, Engholm-Keller K, Lee A, Scott NE, Kolarich D, Hambly BD, Packer NH, Larsen MR, Cordwell SJ. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Mol Cell Proteomics. 2011;10:M110.006833. doi: 10.1074/mcp.M110.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltoniemi H, Joenväärä S, Renkonen R. De novo glycan structure search with the CID MS/MS spectra of native N-glycopeptides. Glycobiology. 2009;19:707–714. doi: 10.1093/glycob/cwp034. [DOI] [PubMed] [Google Scholar]
- Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13:457–470. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- Pierce M, Arango J. Rous sarcoma virus-transformed baby hamster kidney cells express higher levels of asparagine-linked tri- and tetraantennary glycopeptides containing [GlcNAc-beta (1,6)Man-alpha (1,6)Man] and poly-N-acetyllactosamine sequences than baby hamster kidney cells. J Biol Chem. 1986;261:10772–10777. [PubMed] [Google Scholar]
- Pompach P, Brnakova Z, Sanda M, Wu J, Edwards N, Goldman R. Site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol Cell Proteomics. 2013;12:1281–1293. doi: 10.1074/mcp.M112.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prien JM, Prater BD, Qin Q, Cockrill SL. Mass spectrometric-based stable isotopic 2-aminobenzoic acid glycan mapping for rapid glycan screening of biotherapeutics. Anal Chem. 2010;82:1498–1508. doi: 10.1021/ac902617t. [DOI] [PubMed] [Google Scholar]
- Qu Y, Xia S, Yuan H, Wu Q, Li M, Zou L, Zhang L, Liang Z, Zhang Y. Integrated sample pretreatment system for N-linked glycosylation site profiling with combination of hydrophilic interaction chromatography and PNGase F immobilized enzymatic reactor via a strong cation exchange precolumn. Anal Chem. 2011;83:7457–7463. doi: 10.1021/ac201665e. [DOI] [PubMed] [Google Scholar]
- Rakus JF, Mahal LK. New technologies for glycomic analysis: Toward a systematic understanding of the glycome. Annu Rev Anal Chem (Palo Alto Calif) 2011;4:367–392. doi: 10.1146/annurev-anchem-061010-113951. [DOI] [PubMed] [Google Scholar]
- Rebecchi KR, Wenke JL, Go EP, Desaire H. Label-free quantitation: A new glycoproteomics approach. J Am Soc Mass Spectrom. 2009;20:1048–1059. doi: 10.1016/j.jasms.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MS-MRM) analysis of site-specific glycoforms of haptoglobin in liver disease. Mol Cell Proteomics. 2013;12:1294–1305. doi: 10.1074/mcp.M112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiess R, Mueller LN, Schmidt A, Mueller M, Wollscheid B, Aebersold R. Analysis of cell surface proteome changes via label-free, quantitative mass spectrometry. Mol Cell Proteomics. 2009;8:624–638. doi: 10.1074/mcp.M800172-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakey Q, Bates B, Wu J. An approach to quantifying N-linked glycoproteins by enzyme-catalyzed 18O3-labeling of solid-phase enriched glycopeptides. Anal Chem. 2010;82:7722–7728. doi: 10.1021/ac101564t. [DOI] [PubMed] [Google Scholar]
- Shetty HU, Holloway HW. Assay of myo-inositol in cerebrospinal fluid and plasma by chemical ionization mass spectrometry of the hexaacetate derivative. Biol Mass Spectrom. 1994;23:440–444. doi: 10.1002/bms.1200230710. [DOI] [PubMed] [Google Scholar]
- Shetty V, Nickens Z, Shah P, Sinnathamby G, Semmes OJ, Philip R. Investigation of sialylation aberration in N-linked glycopeptides by lectin and tandem labeling (LTL) quantitative proteomics. Anal Chem. 2010;82:9201–9210. doi: 10.1021/ac101486d. [DOI] [PubMed] [Google Scholar]
- Sparbier K, Koch S, Kessler I, Wenzel T, Kostrzewa M. Selective isolation of glycoproteins and glycopeptides for MALDI-TOF MS detection supported by magnetic particles. J Biomol Tech. 2005;16:407–413. [PMC free article] [PubMed] [Google Scholar]
- Sparbier K, Asperger A, Resemann A, Kessler I, Koch S, Wenzel T, Stein G, Vorwerg L, Suckau D, Kostrzewa M. Analysis of glycoproteins in human serum by means of glycospecific magnetic bead separation and LC-MALDI-TOF/TOF analysis with automated glycopeptide detection. J Biomol Tech. 2007;18:252–258. [PMC free article] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- Tang H, Mechref Y, Novotny MV. Automated interpretation of MS/MS spectra of oligosaccharides. Bioinformatics. 2005;21:i431–439. doi: 10.1093/bioinformatics/bti1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Liu Y, Qi D, Yao G, Deng C, Zhang X. On-plate-selective enrichment of glycopeptides using boronic acid-modified gold nanoparticles for direct MALDI-QIT-TOF MS analysis. Proteomics. 2009;9:5046–5055. doi: 10.1002/pmic.200900033. [DOI] [PubMed] [Google Scholar]
- Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Esteva FJ, Song J, Zhang H. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Mol Cell Proteomics. 2012;11:M111.011403. doi: 10.1074/mcp.M111.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama A, Nakagawa H, Matsuda K, Sato TA, Nakamura Y, Ueda K. Quantitative structural characterization of local N-glycan microheterogeneity in therapeutic antibodies by energy-resolved oxonium ion monitoring. Anal Chem. 2012;84:9655–9662. doi: 10.1021/ac3023372. [DOI] [PubMed] [Google Scholar]
- Tran TH, Park S, Lee H, Park S, Kim B, Kim OH, Oh BC, Lee D, Lee H. Ultrasmall gold nanoparticles for highly specific isolation/enrichment of N-linked glycosylated peptides. Analyst. 2012;137:991–998. doi: 10.1039/c1an15810d. [DOI] [PubMed] [Google Scholar]
- Ueda K, Katagiri T, Shimada T, Irie S, Sato TA, Nakamura Y, Daigo Y. Comparative profiling of serum glycoproteome by sequential purification of glycoproteins and 2-nitrobenzensulfenyl (NBS) stable isotope labeling: A new approach for the novel biomarker discovery for cancer. J Proteome Res. 2007;6:3475–3483. doi: 10.1021/pr070103h. [DOI] [PubMed] [Google Scholar]
- Ueda K, Takami S, Saichi N, Daigo Y, Ishikawa N, Kohno N, Katsumata M, Yamane A, Ota M, Sato TA, Nakamura Y, Nakagawa H. Development of serum glycoproteomic profiling technique; simultaneous identification of glycosylation sites and site-specific quantification of glycan structure changes. Mol Cell Proteomics. 2010;9:1819–1828. doi: 10.1074/mcp.2010/000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y. Label-free analysis of O-glycosylation site-occupancy based on the signal intensity of glycopeptide/peptide ions. Mass Spectrom. 2012;1:A0008. doi: 10.5702/massspectrometry.A0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinic J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17:411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- Wada Y, Dell A, Haslam SM, Tissot B, Canis K, Azadi P, Bäckström M, Costello CE, Hansson GC, Hiki Y, Ishihara M, Ito H, Kakehi K, Karlsson N, Hayes CE, Kato K, Kawasaki N, Khoo KH, Kobayashi K, Kolarich D, Kondo A, Lebrilla C, Nakano M, Narimatsu H, Novak J, Novotny MV, Ohno E, Packer NH, Palaima E, Renfrow MB, Tajiri M, Thomsson KA, Yagi H, Yu SY, Taniguchi N. Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1. Mol Cell Proteomics. 2010a;9:719–727. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Tajiri M, Ohshima S. Quantitation of saccharide compositions of O-glycans by mass spectrometry of glycopeptides and its application to rheumatoid arthritis. J Proteome Res. 2010b;9:1367–1373. doi: 10.1021/pr900913k. [DOI] [PubMed] [Google Scholar]
- Walker SH, Budhathoki-Uprety J, Novak BM, Muddiman DC. Stable-isotope labeled hydrophobic hydrazide reagents for the relative quantification of N-linked glycans by electrospray ionization mass spectrometry. Anal Chem. 2011;83:6738–6745. doi: 10.1021/ac201376q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yan J, Yu L, Sheng Q, Zhang X, Xue X, Li X, Liang X. Zirconia layer coated mesoporous silica microspheres as HILIC SPE materials for selective glycopeptide enrichment. Analyst. 2011;136:4422–4430. doi: 10.1039/c1an15554g. [DOI] [PubMed] [Google Scholar]
- Wang X, Emmett MR, Marshall AG. Liquid chromatography electrospray ionization Fourier transform ion cyclotron resonance mass spectrometric characterization of N-linked glycans and glycopeptides. Anal Chem. 2010;82:6542–6548. doi: 10.1021/ac1008833. [DOI] [PubMed] [Google Scholar]
- Wang L, Aryal UK, Dai Z, Mason AC, Monroe ME, Tian ZX, Zhou JY, Su D, Weitz KK, Liu T, Camp DG, II, Smith RD, Baker SE, Qian WJ. Mapping N-linked glycosylation sites in the secretome and whole cells of Aspergillus niger using hydrazide chemistry and mass spectrometry. J Proteome Res. 2012;11:143–156. doi: 10.1021/pr200916k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Li L. Comparative glycoproteomics: Approaches and applications. Brief Funct Genomic Proteomic. 2009;8:104–113. doi: 10.1093/bfgp/eln053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Dulberger C, Li L. Characterization of murine brain membrane glycoproteins by detergent assisted lectin affinity chromatography. Anal Chem. 2010;82:6329–6333. [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Whelan SA, Lu M, He J, Yan W, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J Proteome Res. 2009;8:4151–4160. doi: 10.1021/pr900322g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Feasley CL, Sachdev GP, Smith DF, Cummings RD. Glycan reductive isotope labeling for quantitative glycomics. Anal Biochem. 2009;387:162–170. doi: 10.1016/j.ab.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Zhang H, Liu H, Li Z. Equal ratio of graphite carbon to activated charcoal for enrichment of N-glycopeptides prior to matrix-assisted laser desorption/ionization time-of-flight mass spectrometric identification. Rapid Commun Mass Spectrom. 2012;26:269–274. doi: 10.1002/rcm.5327. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhang L, Lu H, Yang P. On-plate enrichment of glycopeptides by using boronic acid functionalized gold-coated Si wafer. Proteomics. 2010;10:1079–1086. doi: 10.1002/pmic.200900097. [DOI] [PubMed] [Google Scholar]
- Yang F, Lin Z, He X, Chen L, Zhang Y. Synthesis and application of a macroporous boronate affinity monolithic column using a metal-organic gel as a porogenic template for the specific capture of glycoproteins. J Chromatogr A. 2011;1218:9194–9201. doi: 10.1016/j.chroma.2011.10.049. [DOI] [PubMed] [Google Scholar]
- Yang G, Cui T, Wang Y, Sun S, Ma T, Wang T, Chen Q, Li Z. Selective isolation and analysis of glycoprotein fractions and their glycomes from hepatocellular carcinoma sera. Proteomics. 2013;13:1481–1498. doi: 10.1002/pmic.201200259. [DOI] [PubMed] [Google Scholar]
- Yodoshi M, Oyama T, Masaki K, Kakehi K, Hayakawa T, Suzuki S. Affinity entrapment of oligosaccharides and glycopeptides using free lectin solution. Anal Sci. 2011;27:395–400. doi: 10.2116/analsci.27.395. [DOI] [PubMed] [Google Scholar]
- Zeng X, Hood BL, Sun M, Conrads TP, Day RS, Weissfeld JL, Siegfried JM, Bigbee WL. Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J Proteome Res. 2010a;9:6440–6449. doi: 10.1021/pr100696n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Hincapie M, Haab BB, Hanash S, Pitteri SJ, Kluck S, Hogan JM, Kennedy J, Hancock WS. The development of an integrated platform to identify breast cancer glycoproteome changes in human serum. J Chromatogr A. 2010b;1217:3307–3315. doi: 10.1016/j.chroma.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, Wang H, Hancock WS. A proteomics platform combining depletion, multi-lectin affinity chromatography (M-LAC), and isoelectric focusing to study the breast cancer proteome. Anal Chem. 2011;83:4845–4854. doi: 10.1021/ac2002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang H, Tang H, Yang P. Endoglycosidase-mediated incorporation of 18O into glycans for relative glycan quantitation. Anal Chem. 2011;83:4975–4981. doi: 10.1021/ac200753e. [DOI] [PubMed] [Google Scholar]
- Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jia W, Wang J, Ying W, Zhang Y, Qian X. Fragmentation and site-specific quantification of core fucosylated glycoprotein by multiple reaction monitoring-mass spectrometry. Anal Chem. 2011a;83:8802–8809. doi: 10.1021/ac201676a. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yu L, Guo Z, Li X, Liang X. Reversed-phase depletion coupled with hydrophilic affinity enrichment for the selective isolation of N-linked glycopeptides by using Click OEG-CD matrix. Anal Bioanal Chem. 2011b;399:3359–3365. doi: 10.1007/s00216-011-4652-9. [DOI] [PubMed] [Google Scholar]
- Zou Z, Ibisate M, Zhou Y, Aebersold R, Xia Y, Zhang H. Synthesis and evaluation of superparamagnetic silica particles for extraction of glycopeptides in the microtiter plate format. Anal Chem. 2008;80:1228–1234. doi: 10.1021/ac701950h. [DOI] [PubMed] [Google Scholar]


