Abstract
Current treatment options for epilepsy are inadequate as too many patients suffer from uncontrolled seizures and from negative side effects of treatment. Along with major challenges in treatment, scientific understanding of epilepsy is also incomplete, with key questions in epilepsy research remaining unanswered. The major benefit of optogenetic and designer receptor technology is the unprecedented and much needed specificity they provide, allowing spatial, temporal, and cell-type selective modulation of neuronal circuits. Equipped with such tools, it is now possible to begin to address some of the fundamental unanswered questions in epilepsy, to dissect epileptic neuronal circuits, and to develop new intervention strategies. Such specificity of intervention also has the potential for direct therapeutic benefits, allowing healthy tissue and network functions to continue unaffected. In this Perspective, we discuss promising uses of these technologies for the study of seizures and epilepsy, as well as potential use of these strategies for clinical therapies.
The need for new approaches to treating epilepsy is clear. There are 65 million people worldwide with epilepsy and 150,000 new cases of epilepsy diagnosed every year in the US alone 1. Traditional pharmacological approaches to epilepsy lack temporal, regional, and cell-type specificity, and, therefore not surprisingly, often have negative side effects, including nausea, tremor, fatigue, low blood counts, abnormal liver function, cognitive impairment, bone loss, mood changes, and teratogenic effects 2. Not only can such a ‘hammer’ approach produce a variety of unwanted and sometimes debilitating side effects, but, critically, current pharmacological approaches also fail to adequately control seizures for many patients. It is estimated that one third of epilepsy patients will develop drug-refractory epilepsy 3. For some of these patients, surgical removal of brain tissue may be an option. Clearly, this is a non-reversible treatment strategy that lacks temporal and cell-type specificity, removing an entire area or areas believed to be the seizure focus, and is therefore also not without major negative side effects 4. In part due to these considerations and referral practices, for those for whom such surgery is an option, patients wait on average 10–20 years before attempting such treatment 5. For those patients undergoing such surgery, approximately 60–70% will initially experience freedom from generalized seizures 6. Unfortunately, the percent of seizure free patients after surgery decreases with time, and is less than 50% ten years after surgery 5. Furthermore, such surgical approaches (with the potential exclusion of some palliative approaches, such as corpus callosotomy) are only an option for a limited number of patients, requiring a well-defined seizure focus that can be removed without devastating consequences. It is therefore not an option for patients with bilateral temporal lobe epilepsy, patients with a seizure focus near eloquent cortex, or patients with seizures without a clear focus. New technological advances that allow manipulation of neuronal populations with unprecedented specificity, including optogenetics and designer receptors with designer drugs, may pave the way for improved interventions for epilepsy.
Optogenetics makes use of light sensitive proteins called opsins. Opsins include excitatory channels, inhibitory channels and pumps, and G-protein coupled receptors (Box 1). A key feature of the optogenetic toolbox is that these proteins can be expressed in select cell types, and light can be delivered to select areas, allowing control of spatially and genetically defined neuronal populations. Expression of these exogenous proteins essentially requires gene therapy and is approached in much the same way as selective expression of any introduced protein, generally resting on the use of viral vectors or transgenic animals (or both), although other methods such as electroporation are also used 7. It is possible to preferentially target some cell populations through the use of different viral serotypes 8,9, enhancers 10, or cell-type specific promoters. As placing the opsin directly under a specific promoter may produce weak expression, systems such as the flip-excision (FLEX; also referred to as double-floxed inverse open reading frame -- DIO) Cre/loxP recombination system 11–13 are often used. In this and related scenarios, the opsin itself is placed under a strong (rather than cell-type specific) promoter, but expression is made to be Cre dependent. As only low levels of Cre expression is required, Cre can be placed under the weaker cell-type specific promoter (or, for example, attached to wheat germ agglutinin [WGA] to allow expression based on the projection profiles of neurons 14). Similar approaches can be used with the analogous Flp/Frt system, and a recently developed strategy combines Flp/Frt and Cre/loxP recombination systems to allow even greater selectivity in opsin expression through intersectional approaches 15.
Box 1. Optogenetics and DREADDs: Tools of specificity.
Selective-expression of light-sensitive proteins (opsins) and DREADDs allows direct and selective control of neuronal populations. There is a wide, and rapidly expanding, assortment of DREADDs and light-sensitive tools. This box provides a basic introduction and overview of some of these tools.
DREADDs
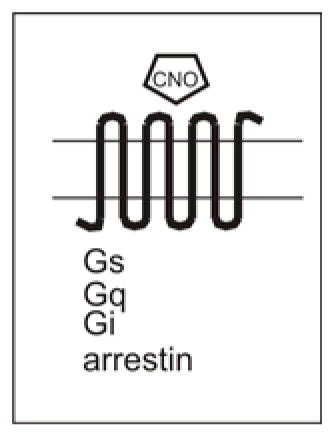
The crux of DREADDs is the idea of dual specificity – that is, both the ligand and the receptor are exclusive partners. The receptor is designed to be unresponsive to natively expressed ligands, but is instead activated solely by a designer drug (CNO -- clozapine-N-oxide), which in turn is inert at natively expressed receptors (for recent DREADD reviews, see references 56 and 61). In this regard, DREADDs mark an improvement over RASSL technology, whose ligands are typically active also at natively expressed receptors. However, metabolites of DREADD ligands may activate native receptors as well 62, a factor which should be considered when designing experiments or therapeutic interventions. DREADDs are G-protein coupled receptors (GPCRs) and come in a few flavors. DREADDs can couple to Gi (hM4Di), Gq (rM3Dq, hM3q), and Gs (rM3Ds), or partner primarily with arrestin (rM3Darr) 19–21. Embedded within each receptor’s name is the receptor subtype from which the receptor is primarily derived as well as the preferential signaling pathway, and the letter D to indicate that it is a DREADD. For example, hM3Dq is a DREADD (D) primarily derived from the human (h) M3 muscarinc receptor (M3) and couples preferentially with Gq (q).
Opsins
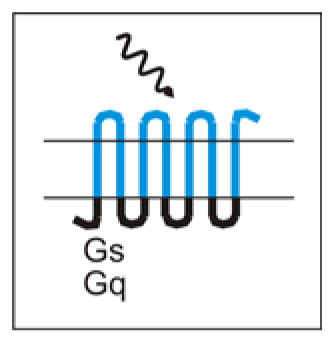
OptoXRs are GPCRs which, instead of being activated by designer drugs, are activated by light. OptoXRs have extracellular and membranous domains taken from bovine rhodopsin, and intracellular domains taken from either the hamster β2 adrenergic receptor (for Gs signaling) or the human α1 adrenergic receptor (for Gq signaling) 68.
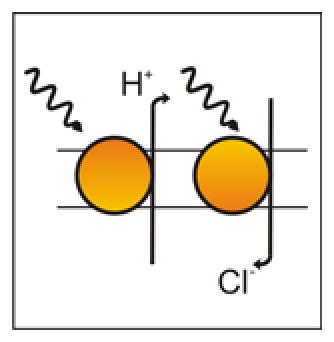
Inhibitory Pump Opsins include the chloride pumps halorhodopsin (HR) 69 and the red-shifted cruxhalorhodopsin (JAWS) 41, and the proton pump (Arch) 70. When activated by light, HR (including improved versions, such as eNpHR3.0 14) and JAWS actively pump chloride ions into the cell, hyperpolarizing it and inhibiting firing. In contrast, when activated by light, Arch actively pumps protons out of the cell. The end result is largely the same – hyperpolarization and inhibition.
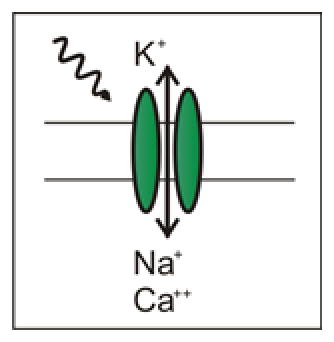
Excitatory Channel Opsins, when activated by light, allow cations to pass into the cell, depolarizing the cell. If sufficient current is induced, an action potential can be generated. Excitatory opsins include ChR2 (including the H134R variety) 71, C1V1 (which is red-shifted) 42, and SSFO (stabilized step-function opsins, which can be turned on with one wavelength of light, and off with another) 42.
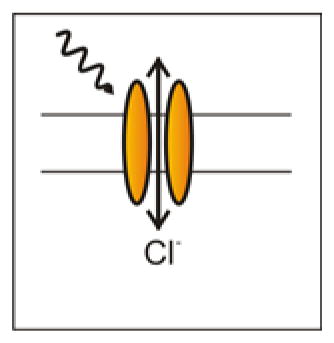
Inhibitory Channel Opsins are relatively new to the scene. Unlike the inhibitory chloride pumps, which actively pump chloride into the cell, iC1C2 and SwiChR (a step-function opsin version, which can be turned on with one wavelength of light, and off with another) are chloride channels, passing chloride as dictated by the electrochemical driving force 45. Because these opsins are channels, they change the cell’s membrane resistance, and can therefore inhibit neurons through a shunting mechanism unlike inhibitory pump opsins.

Additional light-sensitive technology include tools for activating intracellular signaling proteins, such as opto-SOS 72, and tools for regulating gene transcription, including LITEs (light-inducible transcriptional effectors)73 and VP-EL222 74. Several systems, including both the opto-SOS system and LITEs, make use of light-induced protein interactions. Specifically, the opto-SOS system makes use of the phytochrome B (Phy)-PIF interaction system, which can be linked to signaling proteins whose activity is controlled by recruitment. Phy-PIF can be turned ‘on’ (recruit proteins) with red light and ‘off’ with infrared light. In the opto-SOS system, PIF is fused to the catalytic domain of the SOS protein, allowing light driven activation of the signaling protein Ras 72. Similarly, LITEs rest on the light-induced interaction between two proteins: the light sensitive protein cryptochrome 2 (CRY2) and its interacting partner (CIB1). CRY2 is fused to a TALE binding domain (allowing targeting of endogenous genomic loci) and CIB1 is fused to an effector (e.g., VP64) to mediate transcriptional modulation 73. In contrast, VP-EL222 -- a version of EL222 modified for use in eukaryotic systems -- rest on light-induced disruption of binding. VP-EL222 contains a photosensory LOV doman and a helix-turn-helix (HTH) DNA-binding domain. Blue light disrupts LOV-domain to HTH-domain binding, allowing the HTH domain to bind to the DNA 74.
Importantly, when combined with seizure detection, optogenetics can be used in an on-demand fashion 16–18. Additionally, unlike electrical stimulation, opsins allow direct control over the direction of modulation – excitation or inhibition. Equipped with these tools, the experimenter, and potentially one day the physician, can modulate neuronal activity with temporal, spatial, cell-type, and direction of modulation specificity. Such improved specificity allows unparalleled dissection of neuronal circuits critically involved in seizures, provides an avenue for treatment options with fewer side effects, and may improve seizure control for patients in need of new treatment options.
DREADDs, or Designer Receptors Exclusively Activated by Designer Drugs (and closely related RASSLs, or Receptors Activated Solely by Synthetic Ligands) are receptors engineered to be solely activated by synthetic ligands (Box 1). Through just two point mutations, the human muscarinic receptor was engineered to be unresponsive to acetylcholine, and further engineering provided an array of DREADDs, coupling to Gi, Gq, or Gs, or partnering primarily with arrestin 19–21. Importantly, these receptors are not only unresponsive to natively expressed ligands, but the synthetic ligand (CNO -- clozapine-N-oxide) for DREADDs is also inert at natively expressed receptors. This restrictive pairing provides a means for selective modulation of cells expressing DREADDs through designer drug delivery. Just as opsins can be expressed in select areas and neuronal populations, selective expression of designer receptors can provide a means to manipulate neuronal populations with impressive specificity, albeit with less temporal specificity than that achieved with optogenetics.
Optogenetic and designer receptor technologies, and the ability to manipulate neuronal populations with exceptional specificity that they provide, have already made a significant impact on epilepsy research. In time, these tools may make a substantial impact on epilepsy interventions, both directly and indirectly, as discussed below.
Specificity is key to unlocking icto- and epileptogenesis
The recent expansion of optogenetic and designer receptor technologies, and the recent use of these technologies to study epilepsy, is ushering in an era of using selective modulation of neurons in functional circuits to address some fundamental questions in epilepsy including what are the key networks, cell types, and conditions involved in initiating, sustaining, propagating, and terminating seizures. These tools can be applied both to the study of epileptogenesis (the development of epilepsy, a condition of spontaneous recurrent seizures), and to ictogenesis (the generation of a seizure). Ictogenesis can be studied for both acutely provoked seizures (in epileptic or non-epileptic animals) and for spontaneous seizures that arise in an epileptic animal during the chronic phase of the disorder.
Optogenetic and DREADD-mediated inhibition of seizures has been demonstrated across a range of epilepsy models using a variety of approaches (Table 1). As discussed in more detail below, designer receptor and drug technology has been used to inhibit acute seizures (intracerebral pilocarpine and picrotoxin models) and chronic seizures (tetanus toxin model) 22 (Fig. 1). Similarly, seizure inhibition has been achieved using optogenetics in both acute models (including acute systemic lithium-pilocarpine in rat 23 and acute intrahippocampal bicuculline methiodide 24 or 4-Aminopyridine 25 in mouse) and chronic models (including stroke-induced thalamocortical epilepsy in rats 16, tetanus toxin model of focal neocortical epilepsy in rat 26, and intrahippocampal kainate model of temporal lobe epilepsy in mouse 18,27) (Fig. 2), in addition to inhibiting epileptiform activity in slices 24,28 and in silico 29. The broad and successful use of these tools indicates that their utility is not limited to a single approach, model, or type of epilepsy. However, these technologies have yet to be applied to genetic models of epilepsy, including epileptic encephalopathies -- an important next step. Using these techniques to study genetic epilepsies presents additional challenges, including multiple, progressing, or diffuse seizure focus/foci, but presents equal opportunities to advance understanding (e.g., identifying circuitry capable of inhibiting such seizures).
Table 1. In Vivo Optogenetic and DREADD Studies of Seizures and Epilepsy.
Optogenetic and DREADD technology has been applied for the induction of seizures as well as the inhibition of seizures across a range of seizure and epilepsy models. For simplicity, all versions of halorhodopsin are abbreviated HR, and all versions of channelrhodopsin as ChR2. Promoters listed are those responsible for providing cell-type specificity, and may not be the promoter driving the opsin/DREADD expression directly. NB: transgene expression from the Thy1 promoter results in expression in different subsets of neurons in different lines 75.
| Technique | Specific Tool | Method of Gene Delivery | Cell-type Specificity | Regional Specificity | Temporal Specificity | Model | Outcome | Ref. |
|---|---|---|---|---|---|---|---|---|
| Optogenetics | ChR2 | Transgenic rats | (Thy-1 promoter) | Light delivered to hippocampus | (acute) | - | Seizure Induction | 39 |
| Optogenetics | ChR2 | AAV | (CAG promoter) | Light and vector delivered to hippocampus | (acute) | - | Seizure Induction | 39 |
| DREADD | hM3Dq | Transgenic mice | CamKIIα promoter | - | (drug effects peaked at 45–50min) | - | Seizure Induction | 30 |
| Optogenetics | HR | AAV | CamKIIα promoter | Light and vector delivered to hippocampus | (acute) | Acute systemic lithium-pilocarpine | Seizure Inhibition | 23 |
| Optogenetics | HR | AAV | (hSyn promoter) | Light and vector delivered to hippocampus | (acute) | Acute intrahippocampal bicuculline methiodide | Seizure Inhibition | 24 |
| Optogenetics | ChR2 | Transgenic mice | (Thy-1 promoter) | Light delivered to hippocampus | (acute) | Acute intrahippocampal 4-AP | Seizure Inhibition | 25 |
| Optogenetics | HR | Lentivirus | CamKIIα promoter | Light and vector delivered to motor cortex | (not on- demand) | Chronic tetanus toxin model of focal cortical epilepsy | Seizure Inhibition | 26 |
| Optogenetics | HR | AAV | CamKIIα promoter | Light and vector delivered to thalamus | On-demand | Cortical stroke induced chronic thalamocortical epilepsy | Seizure Inhibition | 16 |
| Optogenetics | HR | Transgenic mice | CamKIIα promoter | Light delivered to hippocampus | On-demand | Intrahippocampal kainate model of chronic temporal lobe epilepsy | Seizure Inhibition | 18 |
| Optogenetics | ChR2 | Transgenic mice | Parvalbumin promoter | Light delivered to hippocampus | On-demand | Intrahippocampal kainate model of chronic temporal lobe epilepsy | Seizure Inhibition | 18,27 |
| Optogenetics | ChR2 | Transgenic mice | Parvalbumin promoter | Light delivered to the cerebellum | On-demand | Intrahippocampal kainate model of chronic temporal lobe epilepsy | Seizure Inhibition | 27 |
| Optogenetics | HR | Transgenic mice | Parvalbumin promoter | Light delivered to the cerebellum | On-demand | Intrahippocampal kainate model of chronic temporal lobe epilepsy | Seizure Inhibition | 27 |
| Optogenetics | ChR2 | Transgenic mice | Purkinje cell protein promoter | Light delivered to the cerebellum | On-demand | Intrahippocampal kainate model of chronic temporal lobe epilepsy | Seizure Inhibition | 27 |
| DREADD | hM4Di | AAV | CamKIIα promoter | Vector delivered to motor cortex | - | Acute intracortical pilocarpine | Seizure Inhibition | 22 |
| DREADD | hM4Di | AAV | CamKIIα promoter | Vector delivered to motor cortex | - | Acute intracortical picrotoxin | Seizure Inhibition | 22 |
| DREADD | hM4Di | AAV | CamKIIα promoter | Vector delivered to motor cortex | - | Tetanus toxin model of chronic focal cortical epilepsy | Seizure Inhibition | 22 |
Figure 1. DREADD attenuation of acute seizures.
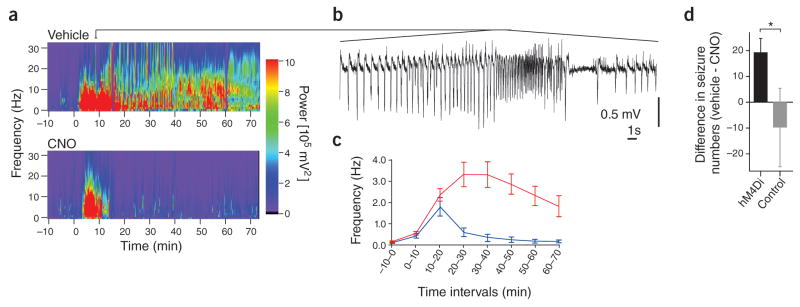
Intracortical injection of pilocarpine evoked acute seizures (Morlet-wavelet spectra of the EEG is shown in a; pilocarpine was administered at time 0; a segment of the EEG trace for a portion of the induced seizure shown in a is provided in panel b). The inhibitory DREADD hM4Di was expressed in cortical excitatory cells. Activation of these receptors by CNO delivery (via intraperitoneal injection) immediately after pilocarpine injection reduced seizure severity (an example is shown in the bottom of panel a), and reduced the frequency of induced spiking (c, red trace: no CNO delivery; blue trace: with CNO delivery). Activation of hM4Di receptors also reduced the number of severe behavioral seizures following intracortical injection of picrotoxin (d, black bar: animals expressing hM4Di; gray bar: animals not expressing DREADDs). Error bars represent s.e.m. Image modified from ref 22, in accordance with the creative commons license http://creativecommons.org/licenses/by/3.0/
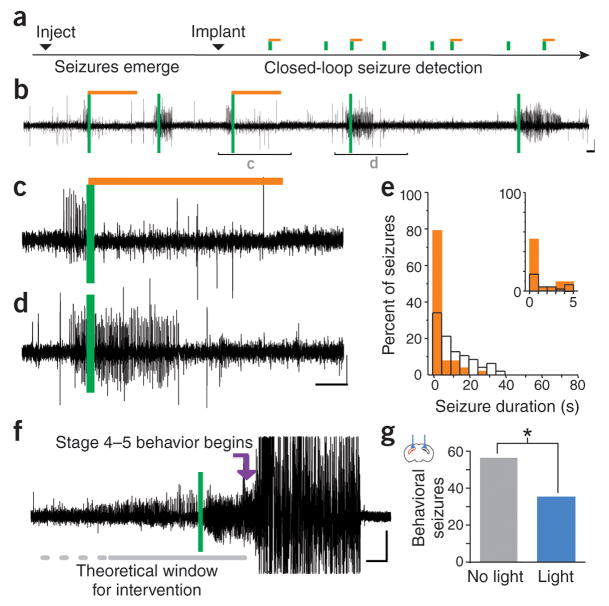
Figure 2. On-demand inhibition of spontaneous seizures.
In the unilateral intrahippocampal kainate model of temporal lobe epilepsy, on-demand optogenetic inhibition of excitatory cells during the chronic phase of the disorder inhibits spontaneous seizures (a–e). Example electrographic seizures are shown in b–d. Seizures were detected on-line (vertical green bars), activating amber light (589nm) randomly for 50% of events (light delivery denoted by horizontal amber bars; expanded examples in c and d). (e) Typical example distribution of post-detection seizure durations (5s bin size) during light (solid amber bars) and no-light internal control conditions (open black bars). Inset: first 5s bin expanded, 1s bin size. Additionally, inhibition of seizures, including a reduction in the number of severe behavioral seizures (an example of a severe seizure not receiving light intervention is shown in f), can be achieved through bilateral optogenetic activation of parvalbumin-expressing inhibitory interneurons (g). Scale bars 100 μV, 5 s. Red hippocampus in the inset indicates the side previously injected with kainate. Image modified from ref 18, in accordance with the creative commons license http://creativecommons.org/licenses/by/3.0/
While in many regards the use of these tools to study the epilepsies is in its infancy, substantial progress and benefits have been garnered by the studies mentioned above. For example, Katzel and colleagues examined the potential for DREADD technology to inhibit focal cortical seizures in rats 22 (Figure 1). Seizures were induced by local application of pilocarpine (a muscarinic receptor agonist), picrotoxin (a GABAA receptor blocker), or tetanus toxin (which blocks neurotransmitter release) into layer 5 of the motor cortex. The inhibitory Gi coupled receptor hM4Di was expressed in the affected area in excitatory cells through an adeno-associated virus (AAV) with a CamKIIα promoter. Intraperitoneal injection of the DREADD agonist CNO reduced electrographic and motor seizure activity. As mentioned above, CNO is inert at natively expressed receptors, and these findings therefore suggest that selective focal inhibition of excitatory cells through such a gene therapy approach can inhibit focal cortical seizures. This finding is supported by prior optogenetic work 26. However, as discussed more below, potential metabolites of CNO may be biologically active at native muscarinic receptors, potentially jeopardizing the specificity of intervention. In this regard, it is interesting that the effects of CNO delivery appeared to be greatest for seizures evoked by pilocarpine, a muscarinic receptor agonist. Critically, however, no significant reduction of seizure activity was seen with CNO delivery to animals not expressing the DREADD 22, indicating that off-target effects could not explain the seizure reduction observed in rats expressing the DREADD in these experiments. It is also worth noting that it is possible to actually induce seizures with DREADD technology by exciting, rather than inhibiting, excitatory cells. Specifically, a study expressing the Gq coupled DREADD (hM3Dq, Box 1) in CamKIIα-expressing cells (including CA1 pyramidal cells) found that CNO delivery depolarized and increased the firing rate of expressing neurons, and induced acute behavioral seizures in mice 30. While the finding that broadly increasing the excitation of excitatory neurons can result in seizures is not entirely surprising, this study does show that CNO delivery itself need not be anti-ictogenic, and that results will critically depend upon the type of modulation. Future use of such technology can expand on these findings, for example attempting to better parse the role of specific neuronal populations, including subpopulations of excitatory neurons in focal cortical seizures 31–33.
A benefit of optogenetic technologies, over DREADDs, is the improved temporal resolution possible. That is, through timed light delivery, optogenetics can provide great temporal specificity, in addition to regional and cell-type specificity. However, in order to harness this temporal specificity for interventions, it is necessary to know when to deliver light, a non-trivial requirement given the intermittent and rather sporadic nature of spontaneous seizures. An on-demand intervention approach, in which intervention is delivered in a responsive fashion only at the time of seizures, requires fast, on-line seizure detection. A flexible system for on-line seizure detection, in which the experimenter can select from a variety of algorithms and tune the detector to the specific EEG signature of a given animal has been developed and made widely available 17. However, the appearance of seizures on EEG (or any recording modality) can vary substantially between individual animals, even within the same model, and certainly across different types of epilepsy. Additionally, detecting seizures early may require knowledge of the seizure focus, which is often not available. Therefore, while this detection software is flexible, it may not work for all experimental needs. Optogenetic approaches can be used to study a variety of disorders, and a particular strength of the use of optogenetics for epilepsy research per se, especially when used in an on-demand manner, is the immediate read out available in the EEG. Despite the potential hurdles in the implementation of responsive systems, on-demand optogenetics has been successfully applied in models of thalamocortical 16 and temporal lobe epilepsies 18,27 (Figure 2).
One such study examining the use of on-demand optogenetics to inhibit spontaneous seizures used the intrahippocampal kainate model of temporal lobe epilepsy. In this model, kainate is injected unilaterally into the hippocampus, provoking acute seizures. Over a period of weeks, spontaneous seizures then emerge, typically arising ipsilateral and slightly posterior to the site of previous kainate injection 34,35. A variety of optogenetic approaches to inhibit seizures in the chronic phase were examined. First, the inhibitory opsin halorhodopsin (eNpHR3.0 14) was expressed in excitatory cells through a transgenic mouse approach. On-demand light delivery to the hippocampus broadly inhibited excitatory cells, and successfully inhibited seizures (Figure 2). These findings demonstrated that on-demand optogenetic approaches can be used to inhibit temporal lobe seizures.
Making use of the potential for these technologies to investigate the role of cell populations beyond excitatory principal cells in epilepsy, on-demand optogenetics was then employed to instead excite a subpopulation of inhibitory interneurons 18 (Figure 2). The excitatory opsin channelrhodopsin (Box 1) was expressed in parvalbumin-expressing interneurons and, despite directly targeting less than 5% of illuminated neurons 36, this approach also inhibited seizures. This identifies parvalbumin-expressing interneurons as a potential therapeutic target, and, importantly, improves understanding of mechanisms of ictogenesis. Specifically, it has been proposed that fast-spiking parvalbumin-expressing interneurons may be responsible for putting a break on seizures (a brake which fails when a seizure spreads), but it has also been proposed these neurons are instead (or potentially also) capable of inducing seizures by introducing aberrant synchrony 37. Optogenetics allows testing of such hypotheses and examining causality in a previously impossible manner.
Interestingly, seizure inhibition could be achieved whether parvalbumin-expressing interneurons were excited ipsilateral or contralateral to the site of previous kainate injection 18, indicating that optogenetically targeting a site physically remote from the initial site of insult can inhibit seizures 38. An even more striking example of remote control of seizures comes from a recent study in which on-demand optogenetic intervention targeting the cerebellum inhibited seizures recorded from the hippocampus 27. This study also reported a unique a reduction in seizure frequency (seen as an increase in time to next seizure), which was seen only with excitation of the midline cerebellum, and was therefore both location and direction of modulation sensitive. A final example illustrating that a site distant from the initial injury can be an effective target for intervention is a study using a very different model of epilepsy – cortical stroke induced epilepsy 16. In this study, on-demand optogenetic inhibition of thalamic (rather than cortical) neurons interrupted cortical stroke induced seizures, supporting the theory that the cortical strokes produced thalamocortical seizures 16. These studies illustrate how optogenetic technologies can be used to investigate critical brain regions, networks, and mechanisms of seizures and seizure suppression.
Clearly, however, the full power and potential for specificity of modulation, available through technologies such as optogenetics and designer receptors, is still to be harnessed. In this regard, it is worthwhile to consider a recent study which used optogenetic activation of hippocampal neurons to induce, rather than inhibit seizures 39. The excitatory opsin channelrhodopsin (ChR2) was expressed using either a transgenic rat line (Thy1.2-ChR2) or via injection of an AAV vector in wild-type Wistar rats (using a non-specific cytomegalovirus enhancer/chicken beta-actin ‘CAG’ promoter). Optogenetic activation produced acute electrographic and behavioral seizures, and was used to study network directional dynamics during light-evoked seizures 39. This study nicely illustrates the potential to use optogenetic techniques to study seizure network dynamics, but makes little use of the cell-type specificity achievable with an optogenetic approach. Through more selective targeting of cell populations, these technologies can be applied to better dissect the network, and determine under which conditions which networks and cell types within those networks can push the system to a seizure.
Moving forward, in addition to further tackling cell-type and circuit level questions, the consequences of seizure inhibition or induction can be examined. Optogenetic and designer receptor technologies can potentially even be used to create new models of epilepsy. Importantly, these technologies may also provide a means to address and understand the causes of common comorbidities of epilepsy, including cognitive and psychiatric comorbidities. Furthermore, treatment options ideally would not only repress seizures or treat comorbidities at the time of intervention, but also have an effect on the underlying disorder itself (e.g., disease modifying). There are new and developing tools of specificity which have yet to be applied to epilepsy, including optogenetic tools to alter gene expression or manipulate signaling molecules (Box 1), which may be particularly useful in this regard. There are also evolving technologies for improving selectivity of targeting 8,15,40, for reducing the invasive nature of optogenetics through red-shifted opsins 41–44, and for decreasing the amount of light required for sustained opsin activity through stabilized step function opsins 42,45. The use of tools of specificity such as optogenetics and designer receptors in the study of epilepsy is just beginning, and continued application of these and new technologies will allow researchers to make ever larger strides.
Basic epilepsy research that disentangles epileptic networks and illuminates the roles of specific cell types during seizures can lead to new treatment options for epilepsy using more traditional approaches such as pharmacology. For example, if a particular cell type was identified as a critical node for seizures, a more traditional pharmacological approach could potentially be developed to target this cell population. However, the challenge remains to develop such an approach that could selectively target this population without having effects on other cell populations as well. Similarly, a region outside the seizure focus identified by optogenetics to modulate the frequency of seizures 27 could be targeted clinically with electrical stimulation. Again, however, this approach would lack the specificity of the newer technologies of optogenetics and DREADDs. We propose therefore that optogenetic and designer receptor technology may not only be critical for research purposes to guide future treatments, but that the unparalleled specificity achievable with these technologies warrants direct consideration as therapeutic possibilities.
Selective intervention: a potential therapy of the future
As outlined above, the potential benefits of an optogenetic or DREADD intervention are clear and can be summarized in one word: specificity. With improved specificity of intervention, healthy networks can continue unaltered. When coupled with a responsive system and used in an on-demand fashion, normal physiological functioning, even in tissue critically engaged during seizures, may be able to continue unaltered. This specificity can reduce side-effects and potentially improve cognitive outcomes compared to traditional pharmacological and resective surgical approaches. Improved specificity may have further benefits, such as increasing seizure control and reaching new patients. For example, patients that are unresponsive to anti-epileptic drugs (AEDs) and not good candidates for traditional resective surgery may be reached by an optogenetic or DREADD approach.
While the need for new intervention strategies is clear, introducing optogenetic or DREADD technology to the clinic is certainly not without major challenges. Although no hurdle is necessarily insurmountable, none are trivial. Luckily, many are not unique to applying optogenetic and designer drug and receptor technology to epilepsy. For example, devices for on-demand electrical stimulation can inform device design for on-demand light delivery. Similarly, as these technologies rely on the expression of exogenous proteins, they fall under a larger umbrella of gene therapy approaches 46, and therefore benefit from gene therapy work broadly (for a recent review of gene therapy for Parkinson’s, including progress and remaining hurdles, see reference 47).
Obtaining safe & appropriate exogenous protein expression
Safety of mechanisms for exogenous gene expression is obviously a requirement. In this regard in particular, progress in gene therapy for neurological disorders more broadly is particularly encouraging. Specifically, it has been shown that viral vectors can be used without major adverse effects. Adeno-associated virus (AAV) use has been reported to be safe for at least 2 years after injection 48, and has been used safely in several phase I and II clinical trials 46,49, including a phase I clinical trial for neuronal ceroid lipofuscinosis (NCL -- a disorder for which seizures are often the first manifestation 50). While many trials utilize AAV vectors for gene delivery, they are certainly not the only viral vector being tested 46,49,51. For example, lentiviral vectors have also been used in a clinical trial without significant adverse effects.
Indeed, viral vectors are being explored with increasing knowledge, modification, and sophistication of use 8,46. While new vectors may require their own safety vetting, the expanding toolbox can provide unique solutions and opportunities. For example, it is now known that certain AAVs can cross the blood brain barrier (BBB) after intravenous injection 52, or preferentially target areas with a compromised BBB after seizures 53. Engineering vectors can also aid in achieving and maintaining cell-type selective expression, and further advances are being made on this front as well, including new vectors which allow targeting based on multiple features using Boolean logic 15. For example, certain neuropeptides such as somatostatin are transiently expressed in principal cells after seizures 54, complicating attempts to selectively target somatostatin-expressing interneurons, especially using Cre-based systems 7. Seizure-induced off-target expression could be resolved through an intersectional approach.
Safety will also need to be demonstrated for the expression of the exogenous protein itself, including stability of expression, lack of an immune response to expression of the non-native protein, and limited constitutive signaling or other effects. In this regard, it is important to note that the proteins are also being engineered to improve compatibility with mammalian systems, and opsin expression has been successful in non-human primates 55. Also, while earlier RASSLs had confounding constitutive signaling, newer DREADDs appear to have no or limited constitutive signaling 56. However, the concern of potentially disrupting normal signaling pathways or scaffolding proteins remains, and achieving appropriate dosing will be critical. Unlike in animal studies, where preliminary expression studies can be done and expression directly examined, human dosing requires either an iterative trial and error approach or new methods to assess viral vector delivery and gene expression in humans in vivo 47. While recordings of neuronal activity, for example, could aid in in vivo assessments, the delay between vector injection and stable protein expression still presents a challenge.
In brief, while animal studies and the increasing knowledge and engineering of vectors discussed above can aid in finding appropriate and safe methods, ultimately, translation to the human brain can present its own challenges, and unforeseen consequences are always possible. Therefore, while the technology may eventually be most beneficial for patients who are not currently surgical candidates, for safety reasons a first attempt is likely to occur in a situation in which the viral vector-infected tissue could be removed should the need arise. For example, a patient who was eligible for resective surgery (e.g., for unilateral temporal lobe epilepsy) could first try an optogenetic or DREADD approach. If this failed, or any safety concerns arose necessitating removal of the tissue, one could revert to surgical resection 51. Once safety has been demonstrated in such patients, use of optogenetics and DREADDs could be applied to a larger patient population.
Challenges unique to optogenetic approaches
For optogenetic approaches, light delivery is an additional unique consideration. It will be necessary to have a device which can safely deliver light, and deliver enough light to reach a critical volume of tissue. There is continual development of new and improved devices for light delivery 57,58, and red-shifted opsins 41–44 will help in the scaling-up of optogenetics from rodents (in which most of the research has currently been done) to humans (as red light is absorbed less by the tissue, and thus reaches a greater volume of cells).
Additionally, in order for on-demand or responsive optogenetics to be realized clinically, the device (or a companion device) will need to be able to predict or detect seizures in real time. Some modest success has been achieved with a device for seizure forecasting 59, and a device capable of responsive neurostimulation has received FDA approval 60. Therefore, a device for on-demand light delivery is well within the realm of feasibility. As discussed below, there are additional challenges to identify where and how to best detect seizures and intervene.
Challenges unique to designer receptor and drugs
Activation of DREADD proteins is more straightforward and less invasive than opsin activation. CNO (clozapine-N-oxide), the synthetic ligand capable of activating current DREADDs (Box 1), can cross the BBB and can even be delivered orally 61. As noted above, a benefit of DREADDs, over most RASSLs, is that this synthetic ligand is not active at native receptors, theoretically limiting off-target drug effects 56.
However, the pharmacokinetics of drugs, including CNO, is a potential area of concern for both experimental studies (as discussed earlier), as well as critically in a clinical settings. Importantly, it has been reported that CNO is readily metabolized in humans into clozapine 62, which is active at native receptors. However, in rodents at least, CNO delivery itself did not have large effects in animals not expressing DREADDs 22,30. Patient to patient variability in drug metabolism will also need to be considered.
No single form of epilepsy, no single intervention
An important final consideration in implementing designer receptor and optogenetic technologies in a clinical setting is individualization of care. While these technologies have shown promise in a range of epilepsy models, the specific approach taken will need to be tailored to the specific epilepsy, and the specific patient being treated. For example, approaches will likely need to target either the seizure focus (/foci) or a region capable of modulating this area 16,17,26,38. This will necessitate identification of the focus or understanding of the network underpinnings for the patient’s seizures. As mentioned previously, for genetic epilepsies this information is especially lacking. In cases where there are multiple foci or no clear foci, it may be necessary to have opsin/DREADD expression (and in the case of optogenetics, light delivery) in multiple locations. In cases where the cells to be targeted are too diffuse and widespread, a DREADD, rather than optogenetic, approach coupled with widespread vector delivery (perhaps via an AAV vector which crosses the BBB) may be better suited. For any approach, extensive testing in animal models will be necessary.
Exciting new advances are certainly not limited to optogenetic and designer receptor technology, and some of these areas may be combined with, or may indirectly advance or benefit from work implementing optogenetic and designer receptor and designer drug technology. For example, transplanted or induced pluripotent stem cells (iPS cells) 63,64 could be combined with DREADD or optogenetic technology to allow selective manipulation of these cells 65. Gene therapies can strive for the goal of specificity of intervention, for example increasing neuropeptide Y or potassium channel expression in a localized area and particular cell type 26,66,67. Considering the strides that have already been made, it may be reasonable or at least inspiring to dream big. Under the banner of such blue-sky thinking, it may even be possible one day to use gene therapy to repress expression of pathological, mutated, proteins and express healthy proteins in the proper cell-type specific manner to restore physiological cellular and network function for genetic epilepsies. For all approaches, achieving appropriate specificity will be a key factor for success.
The dreadded lightness of the future
Optogenetic and DREADD technology are two shining examples of tools of specificity – tools which allow scientists to manipulate neuronal circuits with unprecedented specificity, technologies which may one day provide clinicians with a means to control seizures without major negative side effects. While optogenetic and designer receptor technologies have already been implemented to begin to dissect neuronal circuits in epilepsy, the use of these tools in epilepsy research is still in its infancy, and will undoubtedly continue to make critical contributions to the field.
These advances in understanding of mechanisms in epileptogenesis and ictogenesis will indirectly improve patient care. Additionally, optogenetic and designer receptor technologies may someday become therapeutic options themselves, as the potential benefits are great and the hurdles in implementing these approaches in the clinic are not insurmountable. Together, optogenetics and designer receptor technology promise a brighter future for patients suffering from epilepsy.
Acknowledgments
This work was funded by a US National Institutes of Health grants NS35915 and NS74702 (to I.S.), a Citizens United for Research in Epilepsy (CURE) Taking Flight Award (to E.K-M.), and a US National Institutes of Health grant K99NS087110 (to E.K-M.).
Contributor Information
Esther Krook-Magnuson, Email: ekrookma@umn.edu.
Ivan Soltesz, Email: isoltesz@uci.edu.
References
- 1.England MJ, Liverman CT, Schultz AM, Strawbridge LM. Epilepsy across the spectrum: promoting health and understanding. A summary of the Institute of Medicine report. Epilepsy & behavior: E&B. 2012;25:266–276. doi: 10.1016/j.yebeh.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perucca P, Gilliam FG. Adverse effects of antiepileptic drugs. Lancet neurology. 2012;11:792–802. doi: 10.1016/S1474-4422(12)70153-9. [DOI] [PubMed] [Google Scholar]
- 3.Laxer KD, et al. The consequences of refractory epilepsy and its treatment. Epilepsy & behavior: E&B. 2014;37C:59–70. doi: 10.1016/j.yebeh.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Duchowny M, Bhatia S. Epilepsy: preserving memory in temporal lobectomy-are networks the key? Nature reviews Neurology. 2014;10:245–246. doi: 10.1038/nrneurol.2014.67. [DOI] [PubMed] [Google Scholar]
- 5.de Tisi J, et al. The long-term outcome of adult epilepsy surgery, patterns of seizure remission, and relapse: a cohort study. Lancet. 2011;378:1388–1395. doi: 10.1016/S0140-6736(11)60890-8. [DOI] [PubMed] [Google Scholar]
- 6.Engel J, Jr, et al. Practice parameter: temporal lobe and localized neocortical resections for epilepsy. Epilepsia. 2003;44:741–751. doi: 10.1046/j.1528-1157.2003.48202.x. [DOI] [PubMed] [Google Scholar]
- 7.Krook-Magnuson E, Ledri M, Soltesz I, Kokaia M. How might novel technologies such as optogenetics lead to better treatments in epilepsy? Advances in experimental medicine and biology. 2014;813:319–336. doi: 10.1007/978-94-017-8914-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Faust SM, Rabinowitz JE. The next step in gene delivery: molecular engineering of adeno-associated virus serotypes. Journal of molecular and cellular cardiology. 2011;50:793–802. doi: 10.1016/j.yjmcc.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Burger C, et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Molecular therapy: the journal of the American Society of Gene Therapy. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Visel A, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nature protocols. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnutgen F, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 13.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenno LE, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nature methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paz JT, et al. Closed-loop optogenetic control of thalamus as a tool for interrupting seizures after cortical injury. Nature neuroscience. 2013;16:64–70. doi: 10.1038/nn.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong C, Krook-Magnuson E, Oijala M, Soltesz I. Closed-loop optogenetic intervention in mice. Nature protocols. 2013;8:1475–1493. doi: 10.1038/nprot.2013.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nature communications. 2013;4:1376. doi: 10.1038/ncomms2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guettier JM, et al. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19197–19202. doi: 10.1073/pnas.0906593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakajima K, Wess J. Design and functional characterization of a novel, arrestin-biased designer G protein-coupled receptor. Molecular pharmacology. 2012;82:575–582. doi: 10.1124/mol.112.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzel D, Nicholson E, Schorge S, Walker MC, Kullmann DM. Chemical-genetic attenuation of focal neocortical seizures. Nature communications. 2014;5:3847. doi: 10.1038/ncomms4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sukhotinsky I, et al. Optogenetic delay of status epilepticus onset in an in vivo rodent epilepsy model. PloS one. 2013;8:e62013. doi: 10.1371/journal.pone.0062013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berglind F, et al. Optogenetic inhibition of chemically induced hypersynchronized bursting in mice. Neurobiology of disease. 2014;65:133–141. doi: 10.1016/j.nbd.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Chiang CC, Ladas TP, Gonzalez-Reyes LE, Durand DM. Seizure Suppression by High Frequency Optogenetic Stimulation Using In Vitro and In Vivo Animal Models of Epilepsy. Brain stimulation. 2014 doi: 10.1016/j.brs.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wykes RC, et al. Optogenetic and potassium channel gene therapy in a rodent model of focal neocortical epilepsy. Science translational medicine. 2012;4:161ra152. doi: 10.1126/scitranslmed.3004190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krook-Magnuson E, Szabo GG, Armstrong C, Oijala M, Soltesz I. Cerebellar Directed Optogenetic Intervention Inhibits Spontaneous Hippocampal Seizures in a Mouse Model of Temporal Lobe Epilepsy. eNeruo. 2014;1 doi: 10.1523/ENEURO.0005-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonnesen J, Sorensen AT, Deisseroth K, Lundberg C, Kokaia M. Optogenetic control of epileptiform activity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12162–12167. doi: 10.1073/pnas.0901915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selvaraj P, Sleigh JW, Freeman WJ, Kirsch HE, Szeri AJ. Open loop optogenetic control of simulated cortical epileptiform activity. Journal of computational neuroscience. 2014;36:515–525. doi: 10.1007/s10827-013-0484-2. [DOI] [PubMed] [Google Scholar]
- 30.Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krook-Magnuson E, Varga C, Lee SH, Soltesz I. New dimensions of interneuronal specialization unmasked by principal cell heterogeneity. Trends in neurosciences. 2012;35:175–184. doi: 10.1016/j.tins.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SH, et al. Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron. 2014;82:1129–1144. doi: 10.1016/j.neuron.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown SP, Hestrin S. Cell-type identity: a key to unlocking the function of neocortical circuits. Current opinion in neurobiology. 2009;19:415–421. doi: 10.1016/j.conb.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bragin A, Engel J, Jr, Wilson CL, Vizentin E, Mathern GW. Electrophysiologic analysis of a chronic seizure model after unilateral hippocampal KA injection. Epilepsia. 1999;40:1210–1221. doi: 10.1111/j.1528-1157.1999.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 35.Haussler U, Bielefeld L, Froriep UP, Wolfart J, Haas CA. Septotemporal position in the hippocampal formation determines epileptic and neurogenic activity in temporal lobe epilepsy. Cereb Cortex. 2012;22:26–36. doi: 10.1093/cercor/bhr054. [DOI] [PubMed] [Google Scholar]
- 36.Bezaire MJ, Soltesz I. Quantitative assessment of CA1 local circuits: Knowledge base for interneuron-pyramidal cell connectivity. Hippocampus. 2013 doi: 10.1002/hipo.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiruska P, et al. Synchronization and desynchronization in epilepsy: controversies and hypotheses. The Journal of physiology. 2013;591:787–797. doi: 10.1113/jphysiol.2012.239590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coulter DA. Algal proteins illuminate epilepsy. Epilepsy currents/American Epilepsy Society. 2013;13:221–223. doi: 10.5698/1535-7597-13.5.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osawa S, et al. Optogenetically induced seizure and the longitudinal hippocampal network dynamics. PloS one. 2013;8:e60928. doi: 10.1371/journal.pone.0060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madisen L, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nature neuroscience. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nature neuroscience. 2014 doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nature neuroscience. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nature neuroscience. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–424. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simonato M, et al. Progress in gene therapy for neurological disorders. Nature reviews Neurology. 2013;9:277–291. doi: 10.1038/nrneurol.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartus RT, Weinberg MS, Samulski RJ. Parkinson’s disease gene therapy: success by design meets failure by efficacy. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22:487–497. doi: 10.1038/mt.2013.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartus RT, et al. Safety/feasibility of targeting the substantia nigra with AAV2-neurturin in Parkinson patients. Neurology. 2013;80:1698–1701. doi: 10.1212/WNL.0b013e3182904faa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy AM, Rabkin SD. Current status of gene therapy for brain tumors. Translational research: the journal of laboratory and clinical medicine. 2013;161:339–354. doi: 10.1016/j.trsl.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worgall S, et al. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Human gene therapy. 2008;19:463–474. doi: 10.1089/hum.2008.022. [DOI] [PubMed] [Google Scholar]
- 51.Kullmann DM, Schorge S, Walker MC, Wykes RC. Gene therapy in epilepsy-is it time for clinical trials? Nature reviews Neurology. 2014;10:300–304. doi: 10.1038/nrneurol.2014.43. [DOI] [PubMed] [Google Scholar]
- 52.Duque S, et al. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Molecular therapy: the journal of the American Society of Gene Therapy. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray SJ, et al. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Molecular therapy: the journal of the American Society of Gene Therapy. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drexel M, Kirchmair E, Wieselthaler-Holzl A, Preidt AP, Sperk G. Somatostatin and neuropeptide Y neurons undergo different plasticity in parahippocampal regions in kainic acid-induced epilepsy. J Neuropathol Exp Neurol. 2012;71:312–329. doi: 10.1097/NEN.0b013e31824d9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han X, et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Frontiers in systems neuroscience. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pei Y, Rogan SC, Yan F, Roth BL. Engineered GPCRs as tools to modulate signal transduction. Physiology. 2008;23:313–321. doi: 10.1152/physiol.00025.2008. [DOI] [PubMed] [Google Scholar]
- 57.Kim TI, et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stark E, Koos T, Buzsaki G. Diode probes for spatiotemporal optical control of multiple neurons in freely moving animals. Journal of neurophysiology. 2012;108:349–363. doi: 10.1152/jn.00153.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cook MJ, et al. Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: a first-in-man study. Lancet neurology. 2013;12:563–571. doi: 10.1016/S1474-4422(13)70075-9. [DOI] [PubMed] [Google Scholar]
- 60.Heck CN, et al. Two-year seizure reduction in adults with medically intractable partial onset epilepsy treated with responsive neurostimulation: final results of the RNS System Pivotal trial. Epilepsia. 2014;55:432–441. doi: 10.1111/epi.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wess J, Nakajima K, Jain S. Novel designer receptors to probe GPCR signaling and physiology. Trends in pharmacological sciences. 2013;34:385–392. doi: 10.1016/j.tips.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jann MW, Lam YW, Chang WH. Rapid formation of clozapine in guinea-pigs and man following clozapine-N-oxide administration. Archives internationales de pharmacodynamie et de therapie. 1994;328:243–250. [PubMed] [Google Scholar]
- 63.Collaborative E, et al. The epilepsy phenome/genome project. Clinical trials. 2013;10:568–586. doi: 10.1177/1740774513484392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, et al. Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Annals of neurology. 2013;74:128–139. doi: 10.1002/ana.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henderson KW, et al. Long-term seizure suppression and optogenetic analyses of synaptic connectivity in epileptic mice with hippocampal grafts of GABAergic interneurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:13492–13504. doi: 10.1523/JNEUROSCI.0005-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vezzani A. The promise of gene therapy for the treatment of epilepsy. Expert review of neurotherapeutics. 2007;7:1685–1692. doi: 10.1586/14737175.7.12.1685. [DOI] [PubMed] [Google Scholar]
- 67.Sorensen AT, et al. Hippocampal NPY gene transfer attenuates seizures without affecting epilepsy-induced impairment of LTP. Experimental neurology. 2009;215:328–333. doi: 10.1016/j.expneurol.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 69.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 70.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gradinaru V, et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Motta-Mena LB, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature chemical biology. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. Journal of neuroscience methods. 1997;71:3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]