Abstract
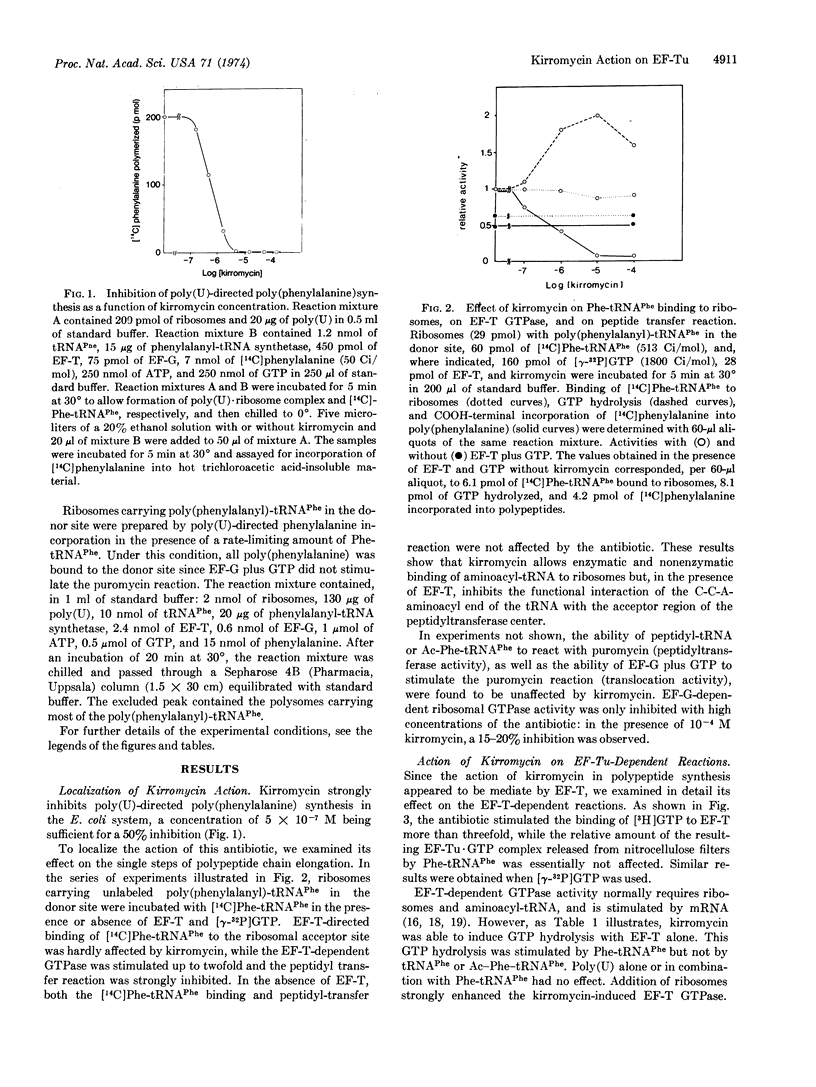
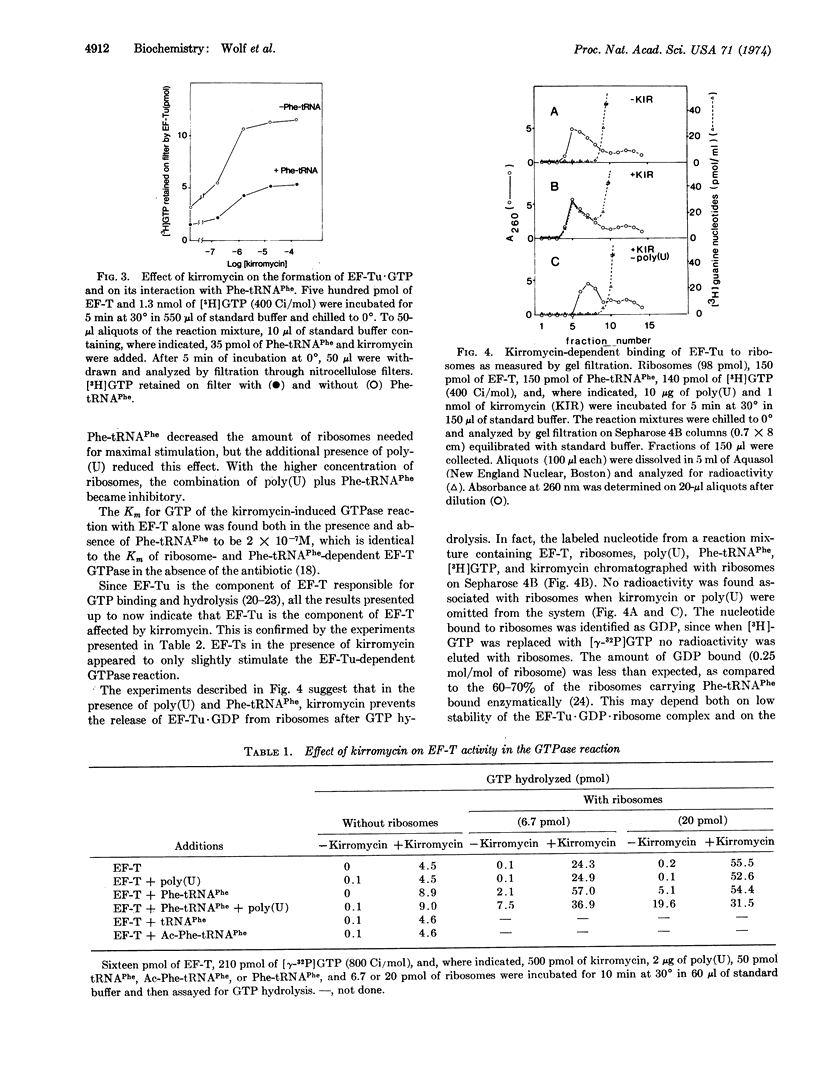
Kirromycin, a new inhibitor of protein synthesis, is shown to interfere with the peptide transfer reaction by acting on elongation factor Tu (EF-Tu). All the reactions associated with this elongation factor are affected. Formation of the EF-Tu·GTP complex is strongly stimulated. Peptide bond formation is prevented only when Phe-tRNAPhe is bound enzymatically to ribosomes, presumably because GTP hydrolysis associated with enzymatic binding of Phe-tRNAPhe is not followed by release of EF-Tu·GDP from the ribosome. This antibiotic also enables EF-Tu to catalyze the binding of Phe-tRNAPhe to the poly(U)·ribosome complex even in the absence of GTP. EF-Tu activity in the GTPase reaction is dramatically affected by kirromycin: GTP hydrolysis, which normally requires ribosomes and aminoacyl-tRNA, takes place with the elongation factor alone. This GTPase shows the same Km for GTP as the one dependent on Phe-tRNAPhe and ribosomes in the absence of the antibiotic. Ribosomes and Phe-tRNAPhe, but not tRNAPhe or Ac-Phe-tRNAPhe, stimulate the kirromycin-induced EF-Tu GTPase. These results indicate that the catalytic center of EF-Tu GTPase that is dependent upon aminoacyl-tRNA and ribosomes is primarily located on the elongation factor. In conclusion, kirromycin can substitute for GTP, aminoacyl-tRNA, or ribosomes in various reactions involving EF-Tu, apparently by affecting the allosteric controls between the sites on the EF-Tu molecule interacting with these components.
Keywords: EF-Tu GTPase, peptide bond formation, aminoacyl-tRNA binding, ribosomal complexes
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger J., Lehr H., Teitel S., Maehr H., Grunberg E. A new antibiotic X-5108 OF Streptomyces origin. I. Production, isolation and properties. J Antibiot (Tokyo) 1973 Jan;26(1):15–22. doi: 10.7164/antibiotics.26.15. [DOI] [PubMed] [Google Scholar]
- Chinali G., Parmeggiani A. Properties of elongation factor G: its interaction with the ribosomal peptidyl-site. Biochem Biophys Res Commun. 1973 Sep 5;54(1):33–39. doi: 10.1016/0006-291x(73)90884-x. [DOI] [PubMed] [Google Scholar]
- Chinali G., Parmeggiani A. Properties of the elongation factors from Escherichia coli. Exchange of elongation factor G during elongation of polypeptide chain. Eur J Biochem. 1973 Feb 1;32(3):463–472. doi: 10.1111/j.1432-1033.1973.tb02629.x. [DOI] [PubMed] [Google Scholar]
- Eckstein F., Kettler M., Parmeggiani A. Guanylylimidodiphosphate and its interaction with amino acid polymerization factors. Biochem Biophys Res Commun. 1971 Dec 3;45(5):1151–1158. doi: 10.1016/0006-291x(71)90139-2. [DOI] [PubMed] [Google Scholar]
- Gordon J. Hydrolysis of guanosine 5'-triphosphate associated wh binding of aminoacyl transfer ribonucleic acid to ribosomes. J Biol Chem. 1969 Oct 25;244(20):5680–5686. [PubMed] [Google Scholar]
- Hachmann J., Miller D. L., Weissbach H. Purification of factor Ts: studies on the formation and stability of nucleotide complexes containing transfer factor Tu. Arch Biochem Biophys. 1971 Dec;147(2):457–466. doi: 10.1016/0003-9861(71)90401-2. [DOI] [PubMed] [Google Scholar]
- Hamel E., Koka M., Nakamoto T. Requirement of an Escherichia coli 50 S ribosomal protein component for effective interaction of the ribosome with T and G factors and with guanosine triphosphate. J Biol Chem. 1972 Feb 10;247(3):805–814. [PubMed] [Google Scholar]
- Hill W. E., Anderegg J. W., Van Holde K. E. Effects of solvent environment and mode of preparation on the physical properties of ribosomes fron Escherichia coli. J Mol Biol. 1970 Oct 14;53(1):107–121. doi: 10.1016/0022-2836(70)90048-3. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lucas-Lenard J. Protein biosynthesis. Annu Rev Biochem. 1971;40:409–448. doi: 10.1146/annurev.bi.40.070171.002205. [DOI] [PubMed] [Google Scholar]
- Lucas-Lenard J., Tao P., Haenni A. L. Further studies on bacterial polypeptide elongation. Cold Spring Harb Symp Quant Biol. 1969;34:455–462. doi: 10.1101/sqb.1969.034.01.051. [DOI] [PubMed] [Google Scholar]
- Maehr H., Blount J. F., Evans R. H., Jr, Leach M., Westley J. W., Williams T. H., Stempel A., Büchi G. Antibiotic X-5108. II. Structure of goldinono-1,4-lactone-3,7-hemiketal, a degradation product of the antibiotic. Helv Chim Acta. 1972;55(8):3051–3054. doi: 10.1002/hlca.19720550837. [DOI] [PubMed] [Google Scholar]
- Maehr H., Leach M., Yarmchuk L., Stempel A. Antibiotic X-5108. V. Structures of antibiotic X-5108 and mocimycin. J Am Chem Soc. 1973 Dec 12;95(25):8449–8450. doi: 10.1021/ja00806a043. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Weissbach H. Studies on the purification and properties of factor Tu from E. coli. Arch Biochem Biophys. 1970 Nov;141(1):26–37. doi: 10.1016/0003-9861(70)90102-5. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A. Crystalline transfer factors from Escherichia coli. Biochem Biophys Res Commun. 1968 Mar 27;30(6):613–619. doi: 10.1016/0006-291x(68)90556-1. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A., Gottschalk E. M. Isolation and some properties of the amino acid polymerization factors from Escherichia coli. Cold Spring Harb Symp Quant Biol. 1969;34:377–384. doi: 10.1101/sqb.1969.034.01.044. [DOI] [PubMed] [Google Scholar]
- Ravel J. M., Shorey R. L., Froehner S., Shive W. A study of the enzymic transfer of aminoacyl-RNA to Escherichia coli ribosomes. Arch Biochem Biophys. 1968 May;125(2):514–526. doi: 10.1016/0003-9861(68)90609-7. [DOI] [PubMed] [Google Scholar]
- Ravel J. M., Shorey R. L., Garner C. W., Dawkins R. C., Shive W. The role of an aminoacyl-tRNA-GTP-protein complex in polypeptide synthesis. Cold Spring Harb Symp Quant Biol. 1969;34:321–330. doi: 10.1101/sqb.1969.034.01.039. [DOI] [PubMed] [Google Scholar]
- Sander G., Marsh R. C., Parmeggiani A. Isolation and characterization of two acidic proteins from the 50S subunit required for GTPase activities of both EF G and EF T. Biochem Biophys Res Commun. 1972 May 26;47(4):866–873. doi: 10.1016/0006-291x(72)90573-6. [DOI] [PubMed] [Google Scholar]
- Skoultchi A., Ono Y., Waterson J., Lengyel P. Peptide chain elongation. Cold Spring Harb Symp Quant Biol. 1969;34:437–454. doi: 10.1101/sqb.1969.034.01.050. [DOI] [PubMed] [Google Scholar]
- Weissbach H., Brot N., Miller D., Rosman M., Ertel R. Interaction of guanosine triphosphate with E. coli soluble transfer factors. Cold Spring Harb Symp Quant Biol. 1969;34:419–431. doi: 10.1101/sqb.1969.034.01.048. [DOI] [PubMed] [Google Scholar]
- Wolf H., Zähner H., Nierhaus K. Kirromycin, an inhibitor of the 30 S ribosomal subunits function. FEBS Lett. 1972 Apr 1;21(3):347–350. doi: 10.1016/0014-5793(72)80199-6. [DOI] [PubMed] [Google Scholar]
- Wolf H., Zähner H. Stoffwechselprodukte von Mikroorganismen. 99. Kirromycin. Arch Mikrobiol. 1972;83(2):147–154. [PubMed] [Google Scholar]



