Abstract
The spectrum of nonalcoholic fatty liver disease (NAFLD) includes steatosis, nonalcoholic steatohepatitis (NASH), and cirrhosis. Recognition and timely diagnosis of these different stages, particularly NASH, is important for both potential reversibility and limitation of complications. Liver biopsy remains the clinical standard for definitive diagnosis. Diagnostic tools minimizing the need for invasive procedures or that add information to histologic data are important in novel management strategies for the growing epidemic of NAFLD. We describe an “omics” approach to detecting a reproducible signature of lipid metabolites, aqueous intracellular metabolites, SNPs, and mRNA transcripts in a double-blinded study of patients with different stages of NAFLD that involves profiling liver biopsies, plasma, and urine samples. Using linear discriminant analysis, a panel of 20 plasma metabolites that includes glycerophospholipids, sphingolipids, sterols, and various aqueous small molecular weight components involved in cellular metabolic pathways, can be used to differentiate between NASH and steatosis. This identification of differential biomolecular signatures has the potential to improve clinical diagnosis and facilitate therapeutic intervention of NAFLD.
Keywords: diagnostic tools, mass spectrometry, phospholipids, sphingolipids, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis
Nonalcoholic fatty liver disease (NAFLD) is rapidly becoming one of the most common forms of liver disease in the United States and worldwide (1). NAFLD is associated with the metabolic syndrome and represents a spectrum of liver disease that includes steatosis and nonalcoholic steatohepatitis (NASH) that can progress to end-stage liver disease and cirrhosis, which can be further complicated by hepatocellular carcinoma (HCC). Approximately 30–50% of NAFLD patients have NASH at the time of diagnosis (1), and over 80% when patients additionally present with certain lipodystrophies (2). Between 10 and 29% of subjects with NASH develop cirrhosis within 10 years, and a smaller subset of NASH patients develops HCC (3). Biopsy is currently the only conclusive means of diagnosis of any stage of the disease, and the presence of inflammation at the time of biopsy for a NAFLD patient is associated with a subsequent risk of clinically significant fibrosis (4, 5). Tools to better define progression of disease in NAFLD, as well as to stratify risk of disease progression noninvasively or in conjunction with a liver biopsy, will have great clinical significance.
The relationship of inflammatory stress, lipotoxicity, and lipid signaling in NASH etiology and progression is under study. While cellular ballooning, degeneration, and inflammation are histologic hallmarks of the disease, mechanistic connections to inflammatory lipids in particular are only now being examined (6). Several lipid mediators typically associated with lipotoxicity such as diacylglycerols (DAGs), oxysterols, ceramides, and FFAs are suggested as potential key links in the mechanism of disease progression toward NASH. Lipotoxicity involving these lipids is commonly found in metabolic syndrome, insulin resistance, and type 2 diabetes (7), which are frequent comorbidities associated with NAFLD (8, 9). Further, while the role of lipids in fibrosis is often discussed in the context of repair (10), their ability to regulate oxidative stress and key transcription factors appears more influential and may have significant effects on the progression of NAFLD (11, 12). Lipids are also important mediators of inflammatory signals, and previous studies have yielded information about plasma and liver lipids in the setting of NASH. For instance, FA analysis revealed that the profiles of nonalcoholic fatty liver and NASH are relatively similar, but that NASH and simple steatosis differ in their content of lipoxygenase-derived eicosanoids (13, 14). Lipidomic analyses of human serum indicate that ether-linked phospholipids, certain neutral lipids, and FAs show suggestive differences between NASH and steatosis for some classes of patients by degree of obesity (15). Ceramides have been proposed to participate in the development and progression of NAFLD (16).
Genetic and epigenetic mutations have also been investigated in association with NASH and NAFLD (3, 10, 17). Variants of PNPLA3 correlate with differences in NAFLD prevalence across different ethnicities (Hispanics > Caucasians > African Americans). PNPLA3 encodes triacylglycerol (TAG) lipase and expression of a particular mutant (I148M) is associated with the severity of liver disease, including NASH (18). The capacity of genetic factors to modulate T-cell responses and epigenetic suppression of PPARγ expression to affect NAFLD has also been described (10). NAFLD is fundamentally a disease characterized by marked derangements in lipid storage and metabolism. A comprehensive examination of lipid species through the advanced stages of NAFLD can provide insights into mechanisms of disease progression and can identify noninvasive biomarkers of different stages. To date, no molecular species-level study examining all major lipid classes and metabolites has been conducted on the spectrum of NAFLD patients.
To address this need, we profiled lipids from liver biopsies, plasma, and urine samples in a double-blinded study from 88 individuals classified on the basis of liver histology as either normal (n = 31), steatotic (n = 17), NASH (n = 20), or cirrhotic (n = 20). Additionally, metabolites of the citric acid cycle, glycolytic pathway, nucleotides, and CoA-derivatives were profiled. Finally, gene expression from liver samples was analyzed by RNA sequence (RNA-Seq) and the results correlated with both lipid and intermediary metabolite levels on a sample-by-sample basis.
Data from these analyses was sufficient to allow clinically significant discrimination between histologically defined categories of NAFLD. Cirrhosis was identified by many individual analytes and transcripts, while discrimination between other disease states required broader panels of analyte species for efficient classification. Among the classes, sphingolipids and glycerophospholipids (GPLs) were most predictive of a given disease stage. A diverse panel of just 20 plasma lipid and aqueous metabolites efficiently separated all disease states by linear discriminant analysis (LDA).
MATERIALS AND METHODS
Study design
Ninety-one patients, selected on an “all-comers” approach, were included in this double-blind lipidomic study from patients who presented to the Vanderbilt Liver Disease Clinic, General Surgery Clinic, and Center for Surgical Weight Loss or were multiorgan donors. All patients subsequently underwent surgical procedures during which clinical indications dictated the need for liver biopsy. The list of surgical procedures during which liver biopsies were obtained include: 43 gastric bypasses, 18 liver transplants, 3 multiorgan donors, 16 ventral hernia repairs, 2 liver resections, 1 cholecystectomy, 1 gastric band removal, 2 gastric pacemakers, and 2 hiatal hernias. Clinical parameters were generally available based upon medical assessments as ordered by primary physicians. Consistent with the intent of the study, those patients diagnosed as having hepatitis or other viral-based liver diseases, HCC, or liver disease, likely due to excessive alcohol consumption as defined by established clinical criteria, were excluded from the study in order to focus on NAFLD patients.
Ninety-one subjects underwent a liver biopsy for clinical indications, and a histological diagnosis was made in a blinded fashion by GI pathologists at Vanderbilt University according to standard of care practice and using established criteria (19). Of these, 88 were classified as belonging to one of these four conditions: normal (n = 31), steatosis (n = 17), steatohepatitis (n = 20), or cirrhosis (n = 20), Three patients were excluded from subsequent analysis as they did not definitively fall into one of these four categories. Aqueous metabolites and lipidomic analysis were performed on appropriate extracts of liver, plasma, and urine, and transcriptomic polymorphisms and SNPs were profiled from liver (wedge) biopsies. All samples were immediately snap-frozen and stored at −80°C following collection according to a preestablished standard operating procedure (SOP).
All sample data were deidentified from patient records. A strict double-blind firewall was maintained during the data collection phase, meaning the investigators involved in sample analyses were unaware of the clinical diagnosis associated with the liver, plasma, and urine samples or the demographics of any of the sample subjects. Similarly, the clinicians were not shown any of the analytical measurements during the data collection phase. Once the final samples were collected and analysis completed, the data set was locked. Only at time of manuscript preparation was the diagnosis of individual clinical samples revealed. Data analyses and procedures were explicitly written in a SOP document and approved by the Vanderbilt Protection of Human Subjects Committee.
A test was performed to ensure no gender driven differences were present by applying a false discovery rate (FDR) limitation (FDR <5%) within any histological categories. Analytes (lipids, aqueous metabolites, SNPs, mRNA transcripts) were subjected to ANOVA followed by FDR (<5%) and only two species in urine met these criteria. Based on that analysis, only data from liver and plasma samples were included in the subsequent analysis.
Ethics statement
Human samples were collected according to a protocol approved by Vanderbilt University Medical Center’s Internal Review Board (#120829) and under informed written patients’ consent prior to inclusion in this study. Sample sizes were selected to minimize the invasive procedures. Plasma samples were obtained from patients’ blood collected during standard of care surgical procedures. Urine samples were collected from patients’ Foley catheters placed for standard of care procedure. Liver samples were obtained from the excess tissue collected as part of the standard of care liver biopsies performed at the time of surgery that would otherwise be discarded. Subsequently, studies at University of California, San Diego were conducted under further auspices of University of California, San Diego Internal Review Board #121220.
Materials
Lipid standards used in lipid quantitation were acquired from Avanti Polar Lipids, Inc. (Alabaster, AL) and Cayman Chemicals (Ann Arbor, MI). D4-citrate (CDN, Pointe-Claire, Quebec Canada), 5-(3-aminoallyl) uridine-5′-triphosphate (Sigma-Aldrich), 2-deoxy-D-glucose-6-phosphate (Santa Cruz, Dallas, TX), and n-propionyl CoA (Sigma-Aldrich) were used in the standard mixture for metabolite mass spectrometric analysis. HPLC grade solvents were purchased from VWR (West Chester, PA) and used without further purification.
Extraction methods
GPLs.
GPLs from liver samples were extracted and analyzed by MS essentially as described in (20, 21). Extraction and analysis of plasma samples was according to previously published procedures (22).
Cardiolipin, coenzyme Q, and dolichol.
Lipid extractions were performed based on the Bligh and Dyer method with minor modifications (23–25).
FAs and eicosanoids.
FFAs were extracted essentially as previously described after supplementation with deuterated internal standards (Cayman Chemicals) (26, 27). Eicosanoids were isolated via solid phase extraction, utilizing 25 deuterated internal standards (28, 29).
Sterols and oxysterols.
Sterols and oxysterols were extracted using previously described methods (30).
Neutral lipids.
Cholesteryl esters (CEs), TAGs, and DAGs were extracted from weighed liver tissue (0.5–1 mg) suspended in 0.5 ml PBS that had been homogenized by sonication. Extractions of plasma (0.05 ml diluted to 0.1 ml with PBS), urine (1 ml), and tissue sonicates were carried out using 1 ml hexane:methyl t-butyl ether (1:1, v/v), essentially as previously described (31).
Sphingolipids.
Sphingolipids from liver, plasma, and urine were extracted following previously published procedures (32, 33), with the exception that methylene chloride was substituted for chloroform for the single-phase extraction of sphingoid bases.
Aqueous metabolites.
A mixture of 5 mM D4-citrate:4 mM 5-(3-aminoallyl) uridine-5′-triphosphate:2.5 mM 2-deoxy-D-glucose-6-phosphate:2 mM n-propionyl CoA (1:1:1:1) was used as an internal standard mix and added to each sample during the extraction procedure. Before analysis, samples were dissolved in different volumes of resuspension solvent consisting of aqueous 2 mM NH4OCOCH3 and 3 mM hexylamine (pH 9.2, adjusted with acetic acid).
Liver sample extraction.
Approximately 10 mg of frozen liver was homogenized in 500 μl of cold (−20°C) 70% CH3OH using a tight-fit glass homogenizer (Kimble/Kontes Glass Co., Vineland, NJ) for about 1 min on ice in the presence of 4 μl of internal standard mix. The suspension was vortexed for 10 s and left in an ice bath for 30 min. After mixing by vortex at 4°C for 1 min and centrifugation (4°C, 18,000 g, 10 min), the supernatant was collected and the solvent was evaporated. The residue was dissolved in 200 μl of resuspension solvent, vortexed to mix (1 min at 4°C), and centrifuged (4°C, 18,000 g, 10 min) to remove any insoluble material.
Plasma sample extraction.
One hundred microliters of thawed plasma containing 4 μl of internal standard mix were extracted with 500 μl cold (−20°C) 70% CH3OH by incubation in an ice bath for 30 min. After mixing by vortex at 4°C for 1 min and centrifugation (4°C, 18,000 g, 10 min), the solvent was evaporated from the supernatant. The residue was dissolved in 200 μl of resuspension solvent, vortexed to mix (1 min at 4°C), and centrifuged (4°C, 18,000 g, 10 min) to remove any insoluble material.
Urine sample extraction.
Five hundred microliters of thawed urine containing 4 μl of internal standard mix were extracted with 500 μl cold (−20°C) CH3OH by incubation in an ice bath for 30 min. After mixing by vortex at 4°C for 1 min and centrifugation (4°C, 18,000 g, 10 min), the solvent was evaporated from the supernatant and the residue was dissolved in 400 μl of resuspension solvent, vortexed to mix (1 min at 4°C), and centrifuged (4°C, 18,000 g, 10 min) to remove any insoluble material.
LC-MS methods
GPLs.
Extracted GPLs from liver and plasma were analyzed by MS, as described elsewhere (20, 21).
Cardiolipin, coenzyme Q, and dolichol.
Cardiolipin analysis was achieved with normal phase LC coupled with high-resolution MS performed on a TripleTOF 5600 system (AB SCIEX, Foster City, CA) (24). Dolichol and coenzyme Q were analyzed by reverse phase LC coupled with multiple reaction monitoring (MRM) MS utilizing a 4000 Q-Trap hybrid triple quadrupole linear ion-trap mass spectrometer (AB SCIEX) (22, 23). For dolichol analysis, MRM was performed in negative ion mode, with the precursor ions being the (M+acetate)− adduct ions and the product ions being the acetate ions (m/z 59). For coenzyme Q analysis, MRM was carried out in positive ion mode, with ammonium adducts (M+NH4)+ as precursor ions and the proton adducts of the quinone ring of coenzyme Q (m/z 197) as product ions. For quantitation, an internal standard mixture composed of a cardiolipin mix (Avanti Polar Lipids, Inc.), nor-dolichol (13–22) (Avanti Polar Lipids, Inc.), and yeast coenzyme Q6 (Sigma) was added during the first step of lipid extraction (22).
FAs and eicosanoids.
FFAs were analyzed by stable isotope dilution GC-MS after derivatization, essentially as described previously (26, 27). This method quantifies 33 FAs including all major and minor saturated FAs, monounsaturated FAs, and PUFAs containing 12 to 26 carbons.
Eicosanoids were analyzed by a stable isotope dilution LC/MS method utilizing 26 deuterated internal standards (28, 29). The metabolites were quantified after separation by reverse phase chromatography on a 2.1 × 100 mm BEH Shield column, 1.7 μM (Waters, Milford, MA) employing an Acquity UPLC system (Waters). Detection and quantification were performed on an AB SCIEX 6500 QTrap mass spectrometer equipped with an IonDrive Turbo V source (AB SCIEX, Framingham, MA), operated in negative ionization mode via MRM, using standard curves generated from 145 authentic quantification standards (34). The method analyzes an additional 13 metabolites based on authentic primary standards, but which cannot be quantified due to the lack of appropriate internal standards. Data analysis was performed using MultiQuant 2.1 software (AB SCIEX).
Sterols and oxysterols.
Sterols and oxysterols were measured using methods previously described (30). Plasma total cholesterol was measured using a Vitros 250 chemistry system (Ortho-Clinical Diagnostics, Rochester, NY). Plasma free cholesterol and liver free and total cholesterol were measured using methods adapted from (30).
Neutral lipids.
The organic solvent extraction layer containing CEs, TAGs, and DAGs was taken to dryness, then derivatized with 2,5-difluorophenylisocyanate to convert DAGs to urethane derivatives (35). The derivatized extract was separated by normal phase LC as previously described (35). The CEs eluting first from the LC column were detected by 20 specific MRM transitions corresponding to each [M+NH4]+ ion being collisionally activated to m/z 369.3. During elution of TAGs, the mass spectrometer was set to carry out full mass scanning from m/z 400–1,000. The DAGs were detected by neutral loss scanning of 190 Da.
Sphingolipids.
Sphingolipids were analyzed by LC-MS/MS essentially as described in (32, 33) with minor modifications to include the 1-deoxy-sphingolipids as generally described in (36).
TCA /glycolysis metabolites.
For the metabolites analysis, an MDS SCIEX 4000 Q TRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA) was utilized, coupled to a Waters Acquity I-class UPLC system, using a modified method from (37). Samples were separated on an Acquity HSS T3 column (Waters) (2.1 × 100 mm, 1.8 μm particle size) using a 10 μl sample injection. Metabolites were separated using a binary gradient program consisting of 5 mM hexylamine (pH 6.3 with acetic acid) (mobile phase A) and methanol:10 mM NH4OCOCH3(aq) (9:1) (pH 8.5 with NH4OH) (mobile phase B) at a flow rate of 0.4 ml/min.
The MS spectra were acquired in negative ionization mode using a turbo spray source with target scan time of 50 msec in MRM scan type. Parameters of the MRM scan used for these metabolites are detailed in Table 1.
TABLE 1.
Parameters of MRM scans for TCA/glycolysis metabolites MS analysis
| Compound | Q1 Mass (Da) | Q3 Mass (Da) |
| G6P | 259 | 97 |
| F-1,6-P2 | 339 | 79 |
| DHAP | 169 | 79 |
| PEP | 167 | 79 |
| PGA | 185 | 79 |
| 2DG6P (IS) | 243 | 225 |
| α-Keto | 145 | 101 |
| Malate | 133 | 115 |
| Succinate | 117 | 73 |
| Aconitate | 173 | 129 |
| Oxaloacetate | 131 | 87 |
| Citrate/isocitrate | 191 | 111 |
| Glutamate | 146 | 128 |
| 6-Phosphogluconate | 275 | 257 |
| D4-citrate (IS) | 195 | 113 |
G6P, glucose-6-phosphate; F-1,6-P2, fructose-1,6-bisphosphate; DHAP, dihydroxy acetone phosphate; PEP, phosphoenol pyruvate; PGA, phosphoglyceric acid; 2DG6P, 2-deoxy-D-glucose-6-phosphate; IS, internal standard.
Nucleotides and CoA.
For the nucleotide analysis, an MDS SCIEX 4000 Q TRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems) was utilized, coupled to a Waters Acquity I-class UPLC system and a method modified from (38). Samples were separated on an Acquity BEH C18 column (Waters) (2.1 × 50 mm, 1.7 μm particle size) using a 10 μl sample injection. Metabolites were separated using a binary gradient program consisting of 2 mM NH4OCOCH3 and 3 mM hexylamine (to pH 9.2 with acetic acid) (mobile phase A) and 100% acetonitrile (mobile phase B) at a flow rate of 0.6 ml/min.
The MS spectra were acquired in negative ionization mode using a turbo spray source with target scan time of 50 msec in MRM scan type using the MRM transitions detailed in Table 2.
TABLE 2.
MRM transitions for nucleotide MS analysis
| Compound | Q1 Mass (Da) | Q3 Mass (Da) |
| ATP | 506 | 159 |
| ADP | 426 | 159 |
| AMP | 346 | 79 |
| 5-(3AA) UTP (IS) | 538 | 159 |
| Acetyl CoA | 808 | 728 |
| Succinyl CoA | 866 | 408 |
| Prop CoA (IS) | 822 | 408 |
| NADH | 664 | 408 |
| NADP+ | 742 | 620 |
| NAD+ | 662 | 540 |
IS, internal standard.
RNA-Seq and SNP arrays
DNA and RNA isolation/library preparation.
Liver homogenates were prepared from approximately 10 mg frozen liver by the cryopulverization method using a BioSpec 59012MS 316 stainless steel 12-well multisample biopulverizer under liquid nitrogen and dry-ice cooled conditions. Pulverized tissue was immediately resuspended in RLT-Plus buffer (Qiagen) supplemented with β-mercaptoethanol. Genomic DNA and total RNA were extracted from liver homogenate using the AllPrep DNA/RNA Mini kit (Qiagen) according to the manufacturer’s instructions. Approximately 200 ng genomic DNA was used for SNP detection using the HumanCoreExome-12v1.1 BeadChIP platform (542585 SNPs) (Illumina). The SNP array contains common variant SNPs and extended coverage of exonic regions accounting for about 19,000 genes. PolyA-RNA was selected from 5 μg total RNA using the MicroPoly(A)Purist kit (Ambion) according to the manufacturer’s instructions. Isolated RNA was hydrolyzed in a total volume of 20 μl with 2 μl RNA fragmentation buffer (Ambion) for 10 min at 70°C. The reaction was terminated with stop buffer, and buffer was exchanged to Tris (pH 8.5) using P30 size-exclusion columns (Bio-Rad). The fragmented RNA was then used for preparation of Illumina-compatible indexed sequencing libraries as described previously (39). Purified libraries were quantified by Qubit dsDNA HS assay kit (Invitrogen) and sequenced for 51 (insert) + 7 (index) cycles on a HiSeq 2000 sequencer (Illumina) with sRNA sequencing primer 5′-CGACAGGTTCAGAGTTCTACAGTCCGACGATC-3′ and TruSeq index sequencing primer (Illumina).
Mapping of the RNA-Seq reads to human genome and transcriptome.
Omicsoft sequence aligner (40) was used to map the reads (single-end sequencing) to human genome (version B37.3) and transcriptome (Ensembl R66). Average read count for the samples was 30 million and 50–70% reads were uniquely mapped for most samples. Some samples were sequenced twice to achieve a higher sequencing depth.
Statistical analysis
Lipids and aqueous metabolites.
Lipid and metabolite values were analyzed by ANOVA across disease states with a Benjamini-Hochberg FDR <5%. Subsets of analytes were used in LDA to cluster the results and examine LDA coefficient loading vectors. Analytes with less than 10 patient values greater than the limit of detection were eliminated prior to LDA. A table showing average lipid concentrations by class for the various groups is provided in the supplementary information (supplementary Table 1).
Differential expression analysis for transcript sequence count data.
Differential expression analysis for transcript sequence count data was used for data normalization and to identify significantly regulated transcripts out of 43,279 transcripts matching entries in the Ensembl database, employing a variance modeling-based approach with a negative binomial distribution (41). A transcript was identified as differentially regulated across conditions (normal, steatosis, steatohepatitis, cirrhosis) if P < 0.05. ANOVA was also carried out on the normalized read counts.
Pathway based analysis.
We identified statistically enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in gene expression data sets using a hypergeometric distribution-based approach through the software Database for Annotation, Visualization, and Integrated Discovery (DAVID) (42, 43). DAVID also allows functional/disease/gene ontology (GO)-based enrichment. Similar approaches are implemented elsewhere, such as VAMPIRE-GOBY (44). The enrichment factor (EF) is defined as follows:
| (Eq. 1) |
where b is the total number of genes in the pathway of interest, s is the number of genes of interest in this pathway, N is the total number of genes in the microarray experiment, and k is the total number of genes of interest (significantly regulated genes). P value, the probability of finding at least s genes of interest in this pathway by assuming hypergeometric distribution, was computed as follows:
| (Eq. 2) |
GO term enrichment was performed similarly. The results of KEGG and GO term enrichment for pathways with ANOVA P value <0.05 between steatosis and NASH were considered significant.
Pearson correlations were used to assess association between analytes and expression of particular genes in select pathways identified as above and in the curated gene lists of WikiPathways (45).
SNPs.
PLINK software (46) was used for analysis of SNP data, including quality control and association test; allele-AB data was used to generate PLINK ped files. For quality control, individuals with more than 10% missing genotypes and SNPs with less than 90% genotyping rate across all individuals were removed. Furthermore, SNPs that failed to meet the Hardy-Weinberg equilibrium threshold (exact test P < 0.001) were removed. A chi-squared test was then applied for case/control association tests on pairs of histological conditions (e.g., normal vs. steatosis; steatosis vs. NASH; NASH vs. cirrhosis) for PNPLA3 and other mutations.
Two-level LDA for steatosis and NASH samples.
To design an eventual LDA with n variables, one LDA was performed on a somewhat larger number (say, 2n) of variables. The discriminant coefficients were examined and rank-ordered according to their contribution (in this case, taken as LD coefficient × center of all samples). Next, the top n variables were used to perform another LDA of the data. This two-level LDA is referred to as LDA2. Once a LDA model is built using training data, when a new test sample arrives, one can compute the discriminant value as, D = xT · c, where x is a vector of the values of the variables (e.g., lipids) for the test sample and c is the vector of LD coefficients. Using D and the population means for the two classes, the posterior probabilities and the predicted class are computed using the “predict” or “predict.lda” function from the MASS package in R.
RESULTS
Clinical demographic profile
Ninety-one patients who underwent liver biopsy were included in this study. Among these, 88 had a definitive histological diagnosis of either normal (n = 31), steatosis (n = 17), steatohepatitis (n = 20), or cirrhosis (n = 20). Three patients who did not fall definitively within these pathologic categorizations were excluded from further analysis. Table 3 indicates the clinical characteristics by disease state within the study population and includes several commonly used methods of assessing liver function, such as alanine aminotransferase levels and total bilirubin. None of the clinical laboratory data or other demographic information (i.e., gender, ethnicity, age) showed significant differences between steatosis and NASH. This finding further reinforces the difficulty of using traditional liver function indicators to distinguish NASH from steatosis, as others have demonstrated as well. The majority of individuals with cirrhosis were male (75%) in contrast to the overall study population. Therefore, a test was performed to ensure that the profile of analytes varying in cirrhosis was not merely driven by gender differences. No analytes were found to vary significantly between genders after FDR limitation <5% within any histological categories.
TABLE 3.
Clinical laboratory data for assessing liver function
| Normal (n = 31) | Steatosis (n = 17) | NASH (n = 20) | Cirrhosis (n = 20) | P (Over Four States) | Post hoc (Steatosis vs. NASH) | Post hoc (NASH vs. Cirrhosis) | |
| Gender (% female) | 80.6 | 70.6 | 55.0 | 25.0 | 8.0E-04 | NS | NS |
| Ethnicity (% white) | 67.7 | 100.0 | 80.0 | 90.0 | NS | NS | NS |
| BMI (kg/m2) | 40.1 ± 1.7 | 46.4 ± 2.7 | 47.5 ± 2.9 | 31.8 ± 1.5 | 2.3E-05 | NS | a |
| Age (years) | 48.1 ± 2.6 | 48.4 ± 2.8 | 47.6 ± 2.7 | 58.1 ± 2.0 | 1.8E-02 | NS | b |
| Glucose (mg/dl) | 110.3 ± 4.7 | 118.2 ± 10.9 | 128.9 ± 8.7 | 122.5 ± 10.6 | NS | NS | NS |
| AST (U/l) | 30.3 ± 4.6 | 26.4 ± 3.8 | 26.8 ± 2.2 | 62.7 ± 11.1 | 4.5E-03 | NS | c |
| ALT (U/l) | 28.5 ± 4.5 | 24.5 ± 2.9 | 31.0 ± 4.0 | 36.4 ± 4.6 | NS | NS | NS |
| AlkP (U/l) | 83.7 ± 5.8 | 74.1 ± 4.4 | 76.8 ± 5.1 | 116.6 ± 14.9 | 2.9E-02 | NS | c |
| Total bilirubin (mg/dl) | 0.45 ± 0.05 | 0.53 ± 0.04 | 0.43 ± 0.06 | 6.3 ± 2.0 | 3.8E-03 | NS | c |
Data are expressed as mean ± SEM. Indicators, such as aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (AlkP), and total bilirubin failed to show a strong difference between steatosis and NASH. Statistical testing across all four histological types was by chi-squared test for gender and ethnicity and ANOVA for all others. Post hoc testing of steatosis versus NASH and NASH versus cirrhosis for clinical data was conducted by Welch test (to control for unequal variances) for continuous variables and by chi-squared test for gender and ethnicity. Bonferroni corrected significance of the post hoc tests is indicated.
P < 0.001.
P < 0.01.
P < 0.05.
Identification of potential biomarkers
Molecular species in each sample type (i.e., liver, plasma, urine) were subjected to ANOVA followed by FDR (<5%). A total of 186 species showed significant variation across the four histological categories in the liver, and 132 in plasma. The critical P values from the FDR procedure were 0.016, 0.014, and 6.3E-5 in the liver, plasma, and urine, respectively. These lists of species are indicated in supplementary Data File 1. Only two species survived this criterion in urine [leukotriene E4 (LTE4) and 12,13-dihydroxy octadecenoic acid (12,13diHOME)], where variation between samples within disease states was largest. As such, subsequent analysis focused primarily on data obtained from the liver and plasma.
A broad cross-section of lipids was represented in the candidate biomarker lists. In the liver, most of the major CEs and TAGs showed significant variation across disease states, as well as several sphingolipids and GPLs. In plasma, differences across histological categories occurred for many major phosphatidylcholines (PCs), ether phosphatidylethanolamines (PEs), phosphatidylinositols, and sphingolipids, including several 1-deoxy- and dihydro- species of sphingomyelin and ceramide. Many of the eicosanoid species discriminated primarily between cirrhosis and the other categories (supplementary Fig. 1, supplementary Data File 1).
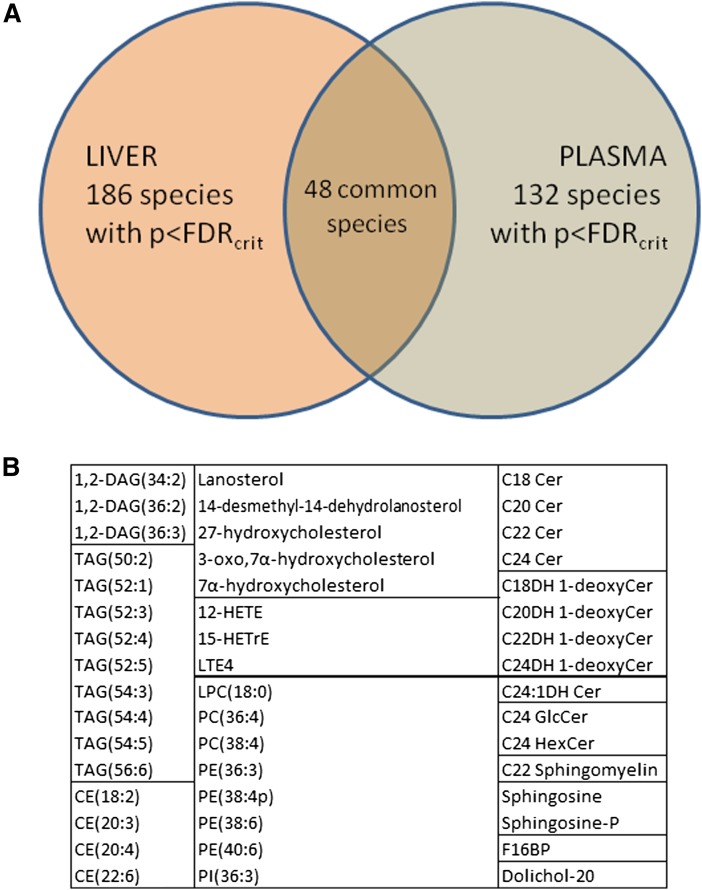
There were a total of 48 species showing overlap between significant liver and plasma analytes (Fig. 1A). These included several DAGs, TAGs, CEs, and GPLs, which generally contain PUFAs (Fig. 1B). Such a pattern is in accordance with a previous study finding differences between the livers of normal, steatotic, and cirrhotic patients in PUFA-containing GPLs (47). Most of the sphingolipid species that differed in both the liver and plasma across histological categories were longer chain ceramides, dihydroceramides, or 1-deoxy-dihydroceramides (Fig. 1B). The fact that there are many species with significant differences between categories in both the liver and plasma further encourages the notion that a less invasive method for diagnosing disease state is possible using mass spectrometric methods.
Fig. 1.
Common analytes between liver and plasma. A: Venn diagram of species with FDR-adjusted significant P values overlapping in the liver and plasma. B: Listing of common analytes indicates major species in several lipid classes. DAGs, TAGs, CEs, and GPLs listed are almost all polyunsaturated while sphingolipids are primarily longer chain species.
Profiles of differences between primary disease states
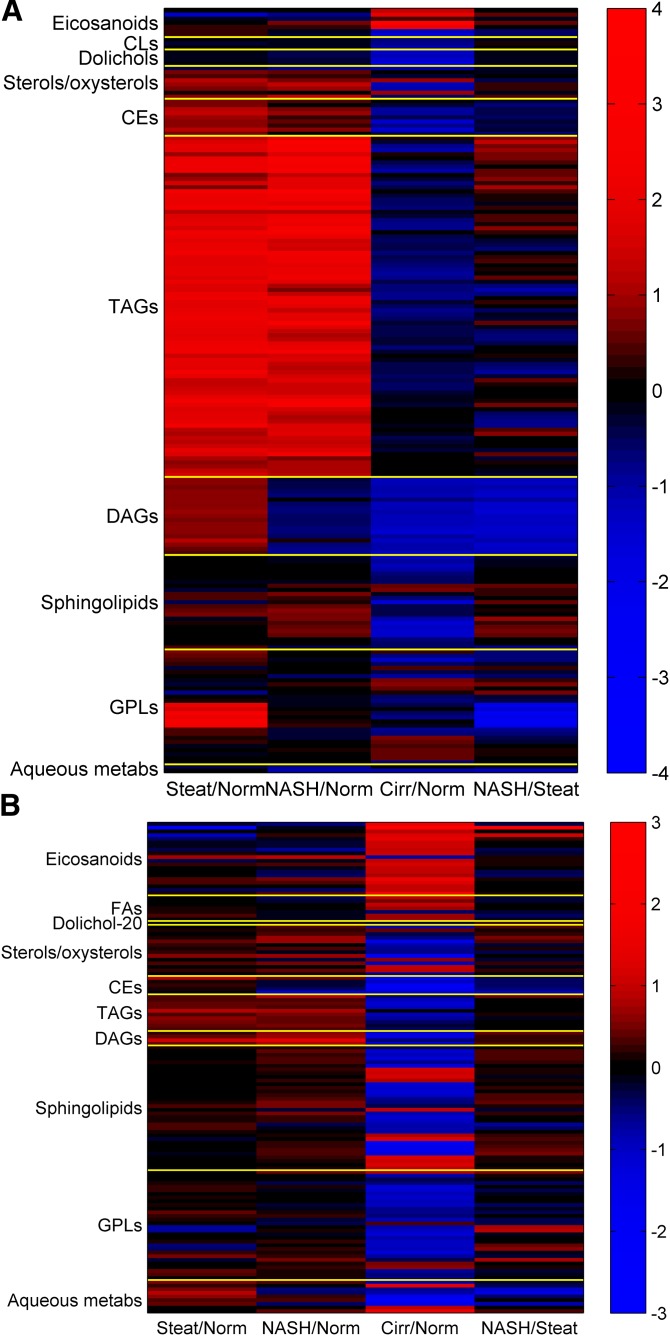
Heatmaps of the log2-fold changes between the primary histological categories (i.e., normal, steatotic, NASH) are displayed in Fig. 2A, B for the liver and plasma, respectively. In the liver, TAG species were nearly uniformly higher in NASH and steatosis compared with normal, as expected. In transcriptomic data, levels of DGAT1, which encodes the enzyme for converting DAG into TAG (ENST00000528718 and ENST00000524965), is somewhat higher in NASH and steatosis samples relative to normal samples (supplementary Fig. 2A, B). Differences between NASH and steatosis in TAG acyl chain composition followed a pattern of higher amounts of short and saturated fatty acyl chain-containing species in NASH and lower amounts of PUFA-containing TAGs. Transcriptomic data partially support this statement, as levels of elongase ELOVL2 (ENST00000354666) are reduced in NASH versus steatosis (supplementary Fig. 2C). On the contrary, levels of the elongase ELOVL5 [ENST00000370918 and ENST00000542638 (supplementary Fig. 2D, E)] and the desaturase FADS2 [ENST00000257261 and ENST00000278840 (supplementary Fig. 2F, G)] are either unchanged or slightly increased (supplementary Fig. 2). DAGs showed far less abundance in NASH compared with steatosis for virtually all species in the class, and slightly lower amounts in NASH compared with normal. The DAG trends for NASH were similar to cirrhosis. Several CE species were also lower in NASH versus steatosis. The GPL species with the strongest discriminatory power between the NASH and steatosis in the liver were PUFA lysophosphatidylethanolamines (LPEs), which were strongly elevated in steatosis versus normal, but not in NASH, suggesting increased lipid turnover (more so than hydrolysis), as previously implied for steatotic versus nonsteatotic liver based on metabolomics analysis (48). However, the mRNA levels for several PLA2s catalyzing the PE hydrolysis are contrary to this trend. For example, steatosis/normal (S/N) fold-change (of medians) for PLA2G2A (ENST00000491964, supplementary Fig. 2H) is 0.4 (supplementary Data File 2, sheet “GPLs HbyS”). Similar observation is made for PLA2G4C (ENST00000354276, supplementary Fig. 2I) with S/N fold-change of 0.81 [NASH/steatosis (H/S) fold-change of 1.44 and P value of 0.03] and PLA2G16 (ENST0000323646, supplementary Fig. 2J) with S/N fold-change of 0.76. An exception is PLA2G15 (ENST00000565460, supplementary Fig. 2K) for which S/N is greater than 1 but less than 1.25 (not listed); low abundance requires caution (supplementary Fig. 2). The transcripts for genes for enzymes that convert LPE into PE (e.g., AGPAT5, MBOAT2) or LPC into PC (e.g., LPCAT4) are either in low abundance or are not significantly different across the four conditions. In the easily distinguishable cirrhotic phenotype, almost all lipids except for eicosanoids and certain GPLs (e.g., phosphatidylinositols, ether-linked PEs) were lower than in normal samples. The aqueous metabolites, malate and fructose-1,6-bisphosphate (F16BP), demonstrated lower levels in NASH in particular, and several other citric acid cycle and glycolytic metabolites, such as succinate (P = 0.025), citrate (P = 0.020), glutamate (P = 0.026), and 6-phosphogluconate (P = 0.048) shown in supplementary Fig. 3, were modestly lower as well. In contrast, the levels of genes of the TCA cycle, such as fumarate hydratase (FH; enzyme for malate production) and pyruvate kinase (PKL; enzyme for F16BP synthesis) are nearly unchanged (not shown). The same is true for the transcripts of succinate dehydrogenase enzymes (SDH) subunits A, B, C, and D (SDHA, SDHB, SDHC, SDHD), the enzymes involved in fumarate production (not shown).
Fig. 2.
Fold changes (log2) for species with FDR-adjusted significant P values in liver and plasma. A: Fold changes (log2) for species with FDR-adjusted significant P values in liver. A complete listing of the fold change values depicted above by individual species is found in supplementary Data File 1. B: Fold changes (log2) for species with FDR-adjusted significant P values in plasma. A complete listing of the fold change values depicted above by individual species is found in supplementary Data File 1.
In plasma (Fig. 2B), the heatmap of fold-differences in lipid or aqueous metabolites between histological categories is dominated by the contrast between cirrhotic samples and all other states. Several eicosanoids and FAs were elevated in cirrhosis, but not NASH, and many plasma neutral lipids, GPLs, and some sterols were lower in cirrhosis, but not in NASH. Sphingolipids and aqueous metabolites showed cirrhosis-specific differences for most analytes in those classes. In particular, most sphingomyelins, long-chain bases, and glycolytic/TCA cycle metabolites were lower in cirrhosis, while a handful of glucosylceramide species and nucleotides were higher in cirrhosis (supplementary Data File 1). A total of 48 species showed significant differences in amounts across disease states in both plasma and liver, including some of the major DAG, TAG, and CE species, PUFA GPLs, sphingosine and sphingosine 1-phosphate, and several oxysterols. Subsequent analysis focused on plasma as a more relevant source of material for measuring biomarkers in the clinic.
Plasma lipids distinguish NASH from steatosis
We have shown differences in metabolites corresponding to different disease stages in liver biopsies, but of greater importance for the purpose of this study is the evaluation of these markers in an easily accessible body fluid, such as plasma. Several lipid classes in plasma showed differences distinguishing NASH from steatosis. These were largely encompassed by CEs, some sphingolipids and GPLs, and the aqueous metabolite F16BP, but without the trends for malate or succinate (not shown) that were observed in the liver. In particular among GPLs, PUFA PEs and notably those containing ether-linked moieties were elevated in NASH compared with steatosis (supplementary Data File 1). Although NASH/steatosis contrasts in plasma LPEs were not statistically significant, there was a trend toward lower levels with progression from steatosis to NASH and cirrhosis. Together with the lower liver LPE levels found in NASH versus steatosis, this finding suggests decreased hydrolysis of ether-linked PEs in individuals with NASH.
Plasma sphingolipids exhibited elevated ceramides, dihydroceramides, and 1-deoxy- dihydroceramides and -ceramides associated with NASH with differences across histological states for numerous molecular subspecies (supplementary Data File 1). The liver also displayed higher amounts of many of these species in NASH (interestingly with elevation of sphinganine but not ceramide) (supplementary Data File 1), and the greatest distinction between NASH versus steatosis was in very-long-chain dihydroceramides and 1-deoxydihydroceramides. Ceramides have been previously associated with NASH and hypothesized to contribute to disease development (16), and this study expanded the scope of the perturbation of sphingolipid metabolism to include dihydro- and 1-deoxy- species.
Broad panel of species distinguishes disease states via LDA
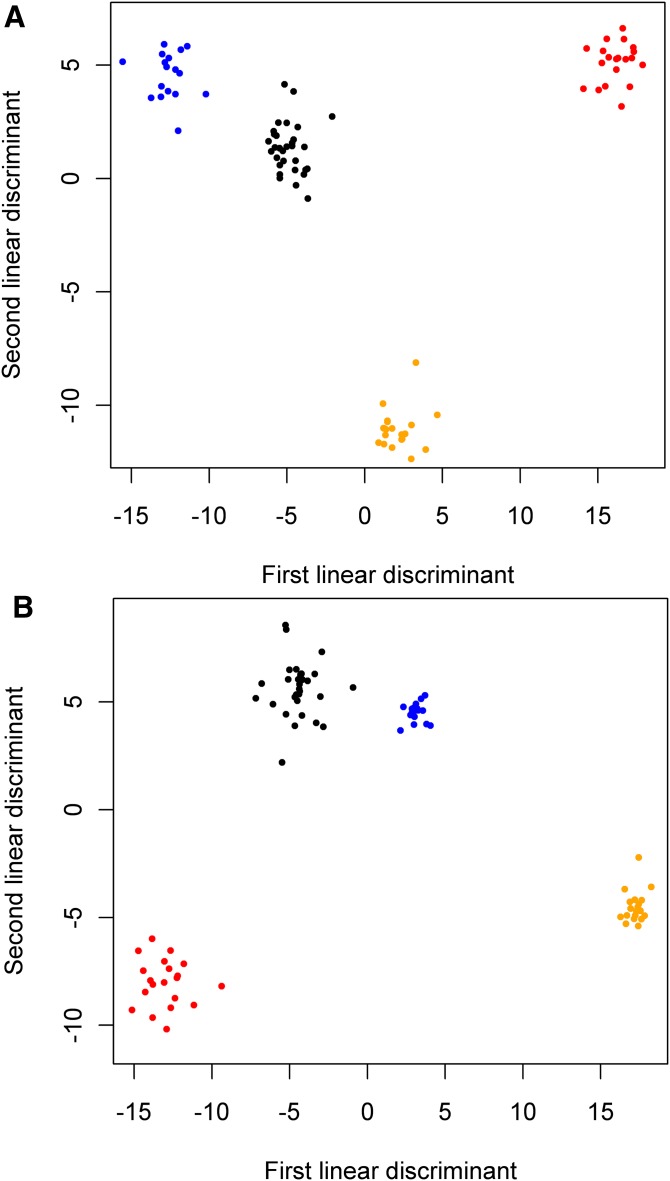
Given that the diversity of lipids with strong differences across disease states is rather broad (Fig. 2), LDA was performed across all lipid classes using analytes with significant differences chosen by their ranking of ANOVA scores (species in bold in supplementary Data File 1). When the top 80 such plasma lipids and aqueous metabolites were used in this way, effective clustering of four disease states in LDA was achieved (Fig. 3A). Decreasing the number of analytes used in this manner to 50 resulted in overlap of clusters (not shown). With respect to liver analytes, discrimination between disease states reached a similar degree of separation between clusters using the top 80 liver analytes by ANOVA (Fig. 3B), but with even tighter clustering of data for both the NASH and steatotic states than in plasma (Fig. 3A).
Fig. 3.
LDA on plasma (A) and liver species (B) with top 80 ANOVA scores classified samples by disease status. Species used in LDA are listed in bold in supplementary Data File 1. In liver (B), the clusters for steatosis and steatohepatitis are noticeably tighter than for normal and cirrhosis. Blue, steatosis; black, normal; orange, steatohepatitis; red, cirrhosis.
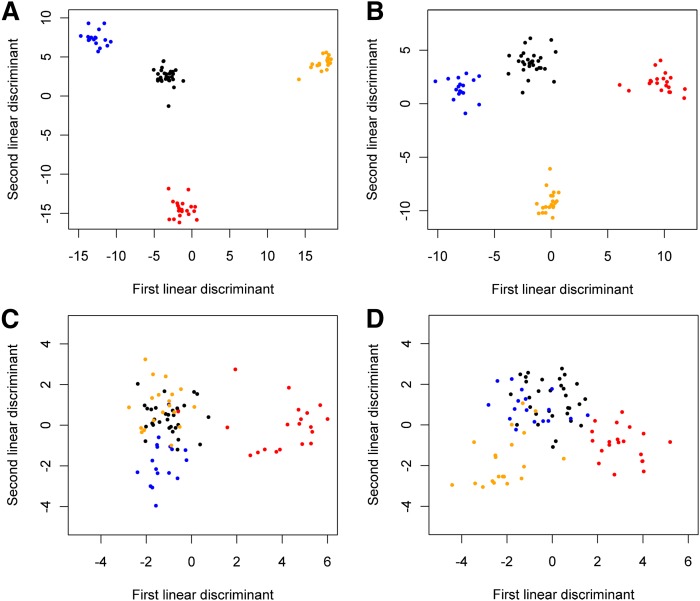
The two lipid classes in the plasma dataset that individually provided the best clustering by LDA were sphingolipids (Fig. 4A) and GPLs (Fig. 4B). In each case, LDA was applied to all species in the class, not just to those with significant P values and fold changes. Results from other plasma lipid classes, such as eicosanoids and neutral lipids, showed less separation between groups (Fig. 4C, D, respectively). Eicosanoids showed relatively strong clustering of cirrhosis samples from the other histological categories, but little contrast between normal, steatosis, and NASH (Fig. 4C). While neutral lipids encompass a larger number of plasma lipid species than either sphingolipids or GPLs, it should be noted that just over 25% (42 of 166) of the plasma DAGs and TAGs were above the limit of detection for more than 10 of the patient samples, and only these species were utilized to allow LDA to proceed without errors due to colinearity. Nonetheless, while some contrast between cirrhosis and steatohepatitis can be seen for neutral lipids (Fig. 4D), this difference was not as strongly resolved.
Fig. 4.
Lipid classes in plasma distinguished between histological states by LDA. A: Sphingolipids best discriminated between histological states via LDA. B: GPLs with similarly strong discrimination between histological states. C: LDA on eicosanoids shows cirrhosis separating from other categories but normal, steatosis, and steatohepatitis clusters are merged. D: LDA on neutral lipids does not show a strong separation between disease stages. Blue, steatosis; orange, steatohepatitis; red, cirrhosis; black, normal.
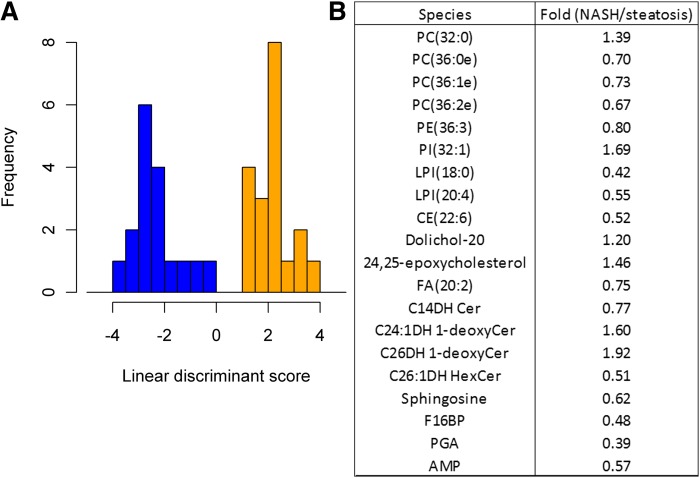
While the detection of unsuspected early cirrhosis has prognostic implications for patients, diagnosis of cirrhosis can often be made from other types of clinical data. A more critical and perhaps subtle relevant clinical distinction was sought between the two potentially reversible disease stages, NASH and steatosis. To identify metabolites responsible for the delineation between NASH and steatosis, LDA was performed on samples from just these disease states. A subset of the top 20 plasma lipids and aqueous metabolites by P value that showed contrast between these categorizations was used to efficiently classify NASH versus steatosis (Fig. 5A). This grouping resulted in a diverse set of species from a variety of classes and included 1-deoxy-ceramide species, ether-linked GPLs, lysophosphatidylinositols, and aqueous metabolites (Fig. 5B). Leave-out-one cross-validation of the classification error of the LDA for NASH versus steatosis was 19%. Distribution of the levels of these 20 key species across histological states can be viewed in supplementary Fig. 1.
Fig. 5.
Analysis of the results from LDA. A: Histogram of LDA results from the 20 plasma metabolites having the most statistically significant differences between NASH and steatotic samples as demonstrated by P values. When these species are used for classification, the two disease states are clearly distinguished. Blue, steatosis; orange, steatohepatitis. B: The list of 20 species used in (A) shows that a diverse spectrum of lipids and aqueous metabolites make up the signature distinguishing steatosis and NASH.
Transcriptomic analysis correlates gene expression with clinical state
Gene expression of endoplasmic reticulum stress-related, unfolded protein response, lipogenesis, autophagy, apoptosis, and other genes responsible for absorption, distribution, metabolism, and elimination of drugs have been analyzed for disease progression (49, 50). Certain clusters of genes were associated with disease progression and showed coordinated regulation in response to hepatic endoplasmic reticulum stress (50). On the other hand, there is a regulated uptake transporter gene expression related to drug uptake, indicative of a hepatoprotective mechanism, preventing metabolite accumulation in compromised hepatocytes (49).
In this study, we analyzed the relation between metabolite changes and gene expression during disease progression, and more specifically, whether they can distinguish between NASH and steatotic livers. RNA-Seq data were acquired from liver samples. When the top transcripts showing differences across all histological states are considered (P < 0.01 by ANOVA, 4,818 transcripts out of total of 43,279), a number of genes in GO and KEGG pathways are highlighted as being coordinately upregulated. Most of these differences arose due to strong up- or downregulation in the cirrhotic condition only. This finding alone does not assist in distinguishing NASH and steatosis from a clinical perspective. Only 1,210 transcripts showed ANOVA P < 0.05 for NASH versus steatosis whereas 174 transcripts showed ANOVA P < 0.01. These results were organized into certain defined functional pathways. Using the 1,210 transcripts, the pathways in Table 4 were determined to have a KEGG and/or GO pathway enrichment with P < 0.05. Among these pathways were clusters of upregulated genes for collagen fibril organization (4.2-fold), focal adhesion (1.92-fold), and extracellular matrix (ECM)-receptor interaction (2.1-fold). Upregulation of ECM degradation has been previously related to both NAFLD and liver injury states (10). Along with the other entries for adhesion network pathway and complement and coagulation cascades, this result further reinforces the concept that vascular repair networks are activated as liver disease progresses. Transcripts from key collagen genes that participate in ECM deposition, fibril organization, and inflammatory response generally showed upregulation that followed the disease state progression (Fig. 6; supplementary Figs. 4, 5). As suggested by previous studies based on smaller sample size (51, 52), genes related to lipid metabolism and ECM remodeling are differentially expressed in samples from NASH patients compared with controls, and thus correlation between these and metabolites may be established. Certain matrix metalloproteinase (MMP) genes involved in processing ECM, including collagen, showed upregulation in NASH versus steatosis as well (supplementary Figs. 5, 6). Increases in the expression of several genes involved in fibrosis were observed in samples from patients who exhibited NASH (supplementary Fig. 5). An examination of genes activated by NLRP3, TIMP1 (not shown), interleukin IL-1β (panel A), transforming growth factor β1 (TGFβ1) (panel B), chemokine (C-C motif) ligand 2 (CCL2) (panel C), chemokine (C-X-C motif) ligand 1 (CXCL1) (panel D), and MMP9 (panel E), MMP14 (panel F) (supplementary Fig. 5A–F), showed that genes involved in fibrosis exhibited enhanced expression in the progression from NAFLD to NASH to cirrhosis.
TABLE 4.
KEGG and GO pathways with fold enrichment of transcripts
| Count | P | Fold | |
| KEGG term | |||
| hsa04510:focal adhesion | 24 | 2.99E-03 | 1.9 |
| hsa04115:p53 signaling pathway | 10 | 2.37E-02 | 2.4 |
| hsa04512:ECM-receptor interaction | 11 | 3.42E-02 | 2.1 |
| GO Term | |||
| GO:0051301, cell division | 30 | 3.34E-03 | 1.8 |
| GO:0030199, collagen fibril organization | 7 | 5.36E-03 | 4.2 |
| GO:0043067, regulation of programmed cell death | 64 | 8.57E-03 | 1.4 |
| GO:0009611, response to wounding | 45 | 8.83E-03 | 1.5 |
| GO:0001649, osteoblast differentiation | 8 | 9.31E-03 | 3.3 |
| GO:0010941, regulation of cell death | 64 | 9.35E-03 | 1.4 |
| GO:0006766, vitamin metabolic process | 11 | 1.03E-02 | 2.6 |
| GO:0042981, regulation of apoptosis | 63 | 1.05E-02 | 1.4 |
| GO:0017144, drug metabolic process | 5 | 1.13E-02 | 5.4 |
| GO:0007009, plasma membrane organization | 5 | 1.13E-02 | 5.4 |
| GO:0034613, cellular protein localization | 36 | 1.28E-02 | 1.5 |
| GO:0042060, wound healing | 20 | 1.38E-02 | 1.8 |
| GO:0070727, cellular macromolecule localization | 36 | 1.42E-02 | 1.5 |
| GO:0033692, cellular polysaccharide biosynthetic process | 6 | 1.48E-02 | 4 |
| GO:0006886, intracellular protein transport | 33 | 1.59E-02 | 1.5 |
| GO:0007155, cell adhesion | 55 | 1.60E-02 | 1.4 |
| GO:0022610, biological adhesion | 55 | 1.65E-02 | 1.4 |
| GO:0006917, induction of apoptosis | 29 | 1.76E-02 | 1.6 |
| GO:0012502, induction of programmed cell death | 29 | 1.84E-02 | 1.6 |
| GO:0051336, regulation of hydrolase activity | 30 | 1.94E-02 | 1.6 |
Data showing ANOVA P < 0.05 for contrast distinguishing between steatosis and NASH.
Fig. 6.
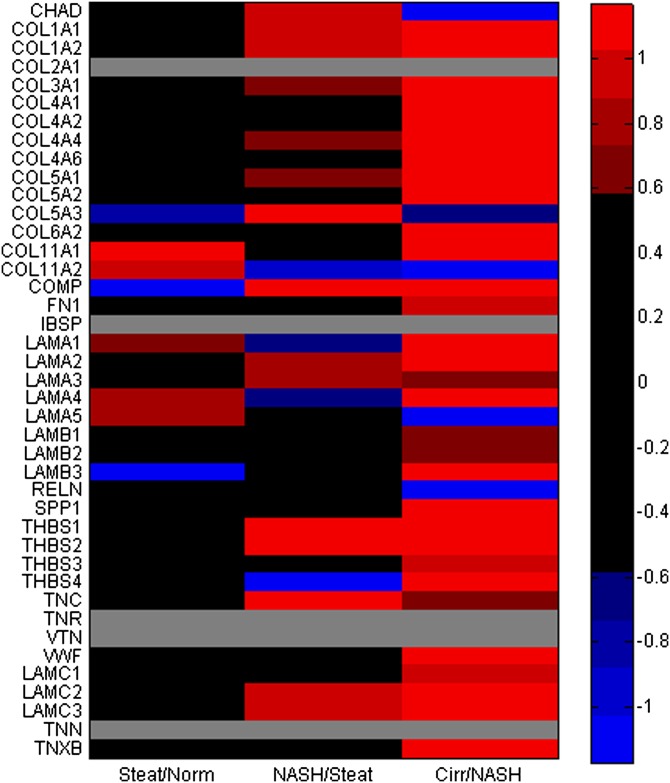
Comparison of log2 (fold-expression) levels of transcripts for genes related to ECM interaction between progressive disease states, Steatosis/normal (Norm), NASH/steatosis (Steat), and Cirrhosis (Cirr)/NASH. Differential expression of transcripts for several collagen genes is present in the transition between steatosis and NASH and intensifies in cirrhosis. Genes listed are curated from ECM receptor interaction module feeding the focal adhesion network in WikiPathways. When more than one transcript for a gene was detected, the one with the highest average abundance was used. Color key: values shaded red correspond to >1.5-fold differences, blue for <0.67-fold, and gray for transcripts not detected in the array. All genes had P < 0.05 by ANOVA analysis except LAMB1, LAMB3 (P = 0.058), COL4A6, and COMP.
Few of the networks highlighted in Table 4 show a strong relationship to lipid metabolic processes. In order to further elucidate connections with lipid metabolites, genes participating in the highlighted networks were cross-correlated against the liver analyte data to assess associations of potential importance, with cirrhotic patients exempted to avoid overwhelming signals due to data from that category alone. Collagen-encoding genes were selected for further investigation, given their upregulation is crucial to progressive liver disease characterized by fibrosis and is typically only activated after stellate cells shed lipid droplets (10). Of note, several of these transcripts were found to correlate with |r| > 0.4 with lipid species (supplementary Table 2). Some of the same classes of species noted in Fig. 1B as having differences across histological categories demonstrated correlation with collagen gene expression, including long-chain FAs, ceramides, ether-linked GPLs, CEs, TAGs, and dolichol-20. This association represents an avenue by which the transcriptional network could be interacting in an unknown manner with PUFA-containing lipid species to produce the observed patterns across different disease states. There are also correlations between MMP genes and many of the same lipids (supplementary Data File 3).
To assess the effect of genotype on the observed correlations, SNPs were assessed for all patients. Illumina’s HumanCoreExome-12v1.1 BeadChIP platform (542,585 SNPs) was used. A few significant SNPs with contrast between steatosis and NASH were noted, but only seven SNPs showed P < 0.001 by analysis with PLINK (45) between steatosis and NASH (supplementary Table 3); in contrast, several SNPs have P < 0.0001 in cirrhosis versus normal comparison (supplementary Table 4). An assessment of the common mutations [especially rs738409 (I148M)] of the gene PNPLA3 showed that there was a significant odds ratio for the difference between cirrhosis and the other histological categories (supplementary Data File 4, sheet “plink HC SNP PNPLA3”), suggesting that PNPLA3 status was not a factor in the observed differences between steatosis and NASH. This outcome is expected given that variations in this gene are strongly related to liver disorders in individuals of Hispanic origin (53), which have very low representation in the present study.
DISCUSSION
The distinction of NASH in NAFLD patients currently requires biopsy for conclusive diagnosis. The development of NASH portends further progression of liver disease to cirrhosis and can incur greater complications for patients than simple steatosis. In fact, cirrhosis resulting from NASH is becoming one of the most common indications for liver transplantation in the United States. The focus of this study was to determine whether a panel of lipid and aqueous metabolites measured in an accessible body fluid, such as plasma, could be correlated with liver histology to distinguish these two disease states within the spectrum of NAFLD. To this end, we identified a signature of 20 plasma lipid species that are capable of distinguishing steatosis from NASH (Fig. 5). In addition to providing focused areas for further study, the internal leave-out-one cross-validation of steatosis versus NASH determination resulted in error of only 19%, and this stringency may aid in noninvasive prediction of which patients are at risk for the development of NASH or even end-stage liver disease.
A similar set of 20 plasma metabolites was obtained by using the two-level LDA analysis starting with 30 plasma lipids at level 1 of LDA [as described in the Statistical Analysis of the Methods section (supplementary Data File 5)]. When these 20 analytes are measured in a patient’s plasma, the sample can clearly be classified into NASH or steatosis (supplementary Fig. 7), making this metabolite signature a powerful biomarker of disease progression in NAFLD (supplementary Data File 5; sheet “posterior”).
While the cohort described in this work represents the most comprehensive analysis of lipid species and analytes to date, the number of individuals is relatively small and thus the results are subject to limitations, including balance within groups for gender and race. The study design encompassed “all-comers” to appropriate clinical services, and did not specifically aim to include or exclude patients with known genetic predispositions to NASH (e.g., PNPLA3 mutations). Despite these limitations, the current findings identify a diagnostic signature that can be the basis for similar studies in larger patient cohorts, and identifies new molecular species that have not previously been associated with diagnosis of NAFLD.
The absence of many major analytes known to participate in lipid droplet formation in NAFLD progression (Fig. 5B), such as the TAGs and DAGs which are prominent in steatosis (Fig. 2), is an indication that the mediators involved in the transition from steatosis to NASH may be quite diverse. The specific metabolite species highlighted as having power to discriminate between NASH and steatosis fall into several categories with known connections to metabolism. These include dihydrosphingolipids, ether PCs, and aqueous metabolites in the citric acid cycle and glycolysis pathways. Specifically, dihydrosphingolipids appear to have some of the strongest fold differences for distinguishing these disease states (Fig. 5B), and plasma sphingolipids alone provided the most discriminatory power of all lipid classes in separating the four histological categories (Fig. 4A). Changes in sphinganine and dihydroceramides are revealing because sphinganine is acylated to dihydroceramides that in turn are desaturated to ceramides; therefore, these dihydrosphingolipids typically reflect the status of de novo sphingolipid biosynthesis rather than turnover (54). Elevations in 1-deoxy-sphingolipids are also reflective of altered biosynthesis because they are produced when there is a shift in the availability of alanine versus serine, as both are utilized by serine palmitoyltransferase (54), and have been associated with diabetes (55). The mRNA level of SPTLC1 is unchanged and SPTLC2 was not detected in transcriptomic data. Thus, the above metabolic shift is likely to be due to change in the availability of upstream metabolites. Furthermore, 1-deoxy-sphingolipids are very poorly miscible in membrane bilayers and, thus, have been predicted to impact membrane integrity and biologic function (56). Plasma ceramides are thought to originate predominantly from the liver (57) due to their secretion in association with very low density lipoproteins (58), which might account for the close association of these compounds as biomarkers with altered hepatic function in NASH.
To reinforce the idea that these and other closely related species correlate with overall disease progression, six of the analytes used to generate the data in Fig. 5 (steatosis vs. NASH segregation) also appear in the list of species found in both liver and plasma that differ over the spectrum of histological categories (Fig. 1B, supplementary Data File 1). For example, the aqueous liver metabolites participating in metabolic energy production (supplementary Fig. 3) demonstrate essentially their lowest levels in NASH when compared with other disease states. After the development of more severe disease, there are lower levels of these metabolic intermediates needed to sustain growth compared with steatosis. There is existing evidence that significant succinylation of proteins during inflammation can occur (59), which could explain the observed lower levels of succinate in NASH specimens. Increased succinate levels lead to activation of hypoxia-inducible factors, and are associated with steatosis (60, 61). Transcriptomic data partially support this finding, as the mRNA levels of the gene SUCLG2 (ENST00000460567) are slightly increased in steatosis versus normal samples; but they decrease slightly in NASH and cirrhosis samples (supplementary Fig. 2L). The two proposed mechanisms involve TCA cycle activation and TAG synthesis (62). Excess FAs cannot be accommodated by β-oxidation, the TCA cycle, or the respiratory chain and are directed toward TAG synthesis, which leads to cell enlargement. Hepatocyte swelling, in turn, reduces oxygen supply and impairs β-oxidation and the TCA cycle. These results suggest that once disease has progressed to NASH, TAG synthesis continues, but that the TCA cycle activity is slowed (Fig. 2A, supplementary Fig. 3, supplementary Data File 1).
HCC can characterize the later stages of NAFLD (63). Signals involved in tumor initiation and progression may be uncovered in earlier stages of NAFLD, such as NASH. In fact, clinical reports are increasingly identifying HCC in patients with NASH prior to the development of cirrhosis. Given that perturbed phosphocholine metabolism, including changes to the fatty acyl composition of the PC pool, is often associated with paradigms of proliferative cell growth and cell death found in cancer (64, 65), it is interesting to note that changes to both ether-linked PCs and 32:0 PC appear in the transition between steatosis and NASH as highlighted by the LDA-based analysis. The increased levels of 32:0 PC in NASH are likely a result of increased de novo lipid synthesis (66) and cellular proliferation. Ether-linked GPLs affect membrane fluidity and are associated with several cellular dysfunctions and malignancies. The presence of these lipid species is typically correlated to metastatic properties of human cancers (67) and may be involved in the intrahepatic metastasis of HCC.
Fibrosis is a defining feature of end-stage liver disease and an important point of distinction evidenced from transcriptional changes. Pathways such as focal adhesion and the network of collagen genes were upregulated in NASH as compared with steatosis. These pathways may be important to ECM degradation and development of fibrosis as the inflammation-related effects of advanced NAFLD progress (10). Further, the significant correlation of collagen gene expression with longer chain and PUFA-containing lipids of many different classes suggests that some lipids may be involved in signaling or regulation of these pathways, and that these lipids should be further examined for their ability to serve as functional indicators of collagen regulation. The enhanced expression of several genes involved in fibrosis was also observed in patients who exhibit NASH. A recent study (68) provided histological evidence that increases in NLRP3, a protein responsible for inflammasome activation, are characteristic of fibrosis. Genes activated by NLRP3 that are involved in the fibrotic pathway (e.g., interleukin IL-1β, TGFβ1, CCL2, CXCL1, MMP9, and MMP14) showed enhanced expression in the progression from NAFLD to NASH to cirrhosis (supplementary Fig. 5). Consistent with our observations, a pathway in which activation of the inflammasome followed by activation of IL1β together with lipid-driven activation of macrophages leading to inflammation was found to contribute to fibrosis (69). Further analysis of the gene expression data from the liver is likely to shed more light on other mechanisms involved in fibrosis.
The results presented here demonstrate that concise “signatures” of lipids and aqueous metabolites can serve as biomarkers of disease progression in NAFLD and may be useful markers of response to therapy in treating this complex disease. The analytical methods used for the measurements have been well-developed, and with additional validation could be adapted to a routine clinical test. A larger, more diverse, and comprehensive cohort of patients with different presenting conditions (including wider ethnic variation, control for gender and age) will be important to further validate these important initial findings and extend this biomarker signature to use in clinical diagnosis of a broad spectrum of subjects with NAFLD.
Supplementary Material
Acknowledgments
The authors thank Jennifer Lee, Michelle Vu, Andrew Warren, and Courtney Widjaja for excellent technical assistance to the sphingolipids core. We also thank Drs. Sarah Dunsmore and Jean Chin at the National Institute of General Medical Sciences for advice and insights on the study, particularly guidance on the SOP and human subject aspects.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- DAG
- diacylglycerol
- ECM
- extracellular matrix
- FDR
- false discovery rate
- F16BP
- fructose-1,6-bisphosphate
- GO
- gene ontology
- GPL
- glycerophospholipid
- HCC
- hepatocellular carcinoma
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LDA
- linear discriminant analysis
- LPE
- lysophosphatidylethanolamine
- MMP
- matrix metalloproteinase
- MRM
- multiple reaction monitoring
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- RNA-Seq
- RNA sequence
- S/N
- steatosis/normal
- SOP
- standard operating procedure
- TAG
- triacylglycerol
- TCA
- the citric acid or tricarboxylic acid cycle
This work was primarily supported by National Institutes of Health Grant GM U54069338 to the LIPID MAPS Consortium. The authors declare no competing interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures, four tables, and five data files.
REFERENCES
- 1.Williams C. D., Stengel J., Asike M. I., Torres D. M., Shaw J., Contreras M., Landt C. L., Harrison S. A. 2011. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 140: 124–131. [DOI] [PubMed] [Google Scholar]
- 2.Safar Zadeh E., Lungu A. O., Cochran E. K., Brown R. J., Ghany M. G., Heller T., Kleiner D. E., Gorden P. 2013. The liver diseases of lipodystrophy: the long-term effect of leptin treatment. J. Hepatol. 59: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen J. C., Horton J. D., Hobbs H. H. 2011. Human fatty liver disease: old questions and new insights. Science. 332: 1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gan L., Chitturi S., Farrell G. C. 2011. Mechanisms and implications of age-related changes in the liver: nonalcoholic fatty liver disease in the elderly. Curr. Gerontol. Geriatr. Res. 2011: 831536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell S., Argo C. 2010. The natural history of non-alcoholic fatty liver disease. Dig. Dis. 28: 162–168. [DOI] [PubMed] [Google Scholar]
- 6.Syn W. K., Choi S. S., Diehl A. M. 2009. Apoptosis and cytokines in non-alcoholic steatohepatitis. Clin. Liver Dis. 13: 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhi H., Gores G. J. 2008. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin. Liver Dis. 28: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams L. A., Waters O. R., Knuiman M. W., Elliott R. R., Olynyk J. K. 2009. NAFLD as a risk factor for the development of diabetes and the metabolic syndrome: an eleven-year follow-up study. Am. J. Gastroenterol. 104: 861–867. [DOI] [PubMed] [Google Scholar]
- 9.Marchesini G., Bugianesi E., Forlani G., Cerrelli F., Lenzi M., Manini R., Natale S., Vanni E., Vilanova N., Melchionda N., et al. 2003. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 37: 917–923. [DOI] [PubMed] [Google Scholar]
- 10.Pellicoro A., Ramachandran P., Iredale J., Fallowfield J. 2014. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 14: 181–194. [DOI] [PubMed] [Google Scholar]
- 11.Teratani T., Tomita K., Suzuki T., Oshikawa T., Yokoyama H., Shimamura K., Tominaga S., Hiroi S., Irie R., Okada Y., et al. 2012. A high-cholesterol diet exacerbates liver fibrosis in mice via accumulation of free cholesterol in hepatic stellate cells. Gastroenterology. 142: 152–164. [DOI] [PubMed] [Google Scholar]
- 12.Moustafa T., Fickert P., Magnes C., Guelly C., Thueringer A., Frank S., Kratky D., Sattler W., Reicher H., Sinner F., et al. 2012. Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology. 142: 140–151. [DOI] [PubMed] [Google Scholar]
- 13.Puri P., Wiest M. M., Cheung O., Mirshahi F., Sargent C., Min H. K., Contos M. J., Sterling R. K., Fuchs M., Zhou H., et al. 2009. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 50: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldstein A. E., Lopez R., Tamimi T. A., Yeian L., Chung Y. M., Berk M., Zhang R., McIntyre T. M., Hazen S. L. 2010. Mass spectrometric profiling of oxidized lipid products in human nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J. Lipid Res. 51: 3046–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr J., Vazquez-Chantada M., Alonso C., Perez-Cormenzana M., Mayo R., Galan A., Caballeria J., Martin-Duce A., Tran A., Wagner C., et al. 2010. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J. Proteome Res. 9: 4501–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagadala M., Kasumov T., McCullogh A. J., Zein N. N., Kirwan J. P. 2012. Role of ceramides in nonalcoholic fatty liver disease. Trends Endocrinol. Metab. 23: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musso G., Gambino R., De Michieli F., Durazzo M., Pagano G., Cassader M. 2008. Adiponectin gene polymorphisms modulate acute adiponectin response to dietary fat: Possible pathogenetic role in NASH. Hepatology. 47: 1167–1177. [DOI] [PubMed] [Google Scholar]
- 18.Smagris E., BasuRay S., Li J., Huang Y., Lai K. M., Gromada J., Cohen J. C., Hobbs H. H. 2015. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 61: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleiner D. E., Brunt E. M., Van Natta M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., et al. ; Nonalcoholic Steatohepatitis Clinical Research Network. 2005. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 41: 1313–1321. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., Brown H. A. 2007. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432: 21–57. [DOI] [PubMed] [Google Scholar]
- 21.Myers D. S., Ivanova P. T., Milne S. B., Brown H. A. 2011. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim. Biophys. Acta. 1811: 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quehenberger O., Armando A. M., Brown H. A., Milne S. B., Myers D. S., Merrill A. H., Jr, Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan Z., Li S., Smith D., Shaw W., Raetz C. 2007. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry. 46: 14500–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan B. K., Bogdanov M., Zhao J., Dowhan W., Raetz C. R., Guan Z. 2012. Discovery of cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. USA. 109: 16504–16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen R., Lam B., Guan Z. 2013. Aberrant dolichol chain lengths as biomarkers for retinitis pigmentosa caused by impaired dolichol biosynthesis. J. Lipid Res. 54: 3516–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quehenberger O., Armando A., Dumlao D., Stephens D. L., Dennis E. A. 2008. Lipidomics analysis of essential fatty acids in macrophages. Prostaglandins Leukot. Essent. Fatty Acids. 79: 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quehenberger O., Armando A. M., Dennis E. A. 2011. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta. 1811: 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deems R., Buczynski M. W., Bowers-Gentry R., Harkewicz R., Dennis E. A. 2007. Detection and quantitation of eicosanoids via high performance liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 432: 59–82. [DOI] [PubMed] [Google Scholar]
- 29.Dumlao D. S., Buczynski M. W., Norris P. C., Harkewicz R., Dennis E. A. 2011. High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines. Biochim. Biophys. Acta. 1811: 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald J. G., Smith D. D., Stiles A. R., Russell D. W. 2012. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53: 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchins P. M., Barkley R. M., Murphy R. C. 2008. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 49: 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullards M. C., Liu Y., Chen Y., Merrill A. H., Jr 2011. Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS). Biochim. Biophys. Acta. 1811: 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Armando A. M., Quehenberger O., Yan C., Dennis E. A. 2014. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J. Chromatogr. A. 1359: 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiker T. J., Barkley R. M., Murphy R. C. 2011. Analysis of diacylglycerol molecular species in cellular lipid extracts by normal-phase LC-electrospray mass spectrometry. Int. J. Mass Spectrom. 305: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., et al. 2009. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284: 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coulier L., Bas R., Jespersen S., Verheij E., van der Werf M. J., Hankemeier T. 2006. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography - electrospray ionization mass spectrometry. Anal. Chem. 78: 6573–6582. [DOI] [PubMed] [Google Scholar]
- 38.Fromentin E., Gavegnano C., Obikhod A., Schinazi R. 2010. Simultaneous quantification of intracellular natural and antiretroviral nucleosides and nucleotides by liquid chromatography- tandem mass spectrometry. Anal. Chem. 82: 1982–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinz S., Romanoski C. E., Benner C., Allison K. A., Kaikkonen M. U., Orozco L. D., Glass C. K. 2013. Effect of natural genetic variation on enhancer selection and function. Nature. 503: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J., Ge H., Newman M., Liu K. 2012. OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics. 28: 1933–1934. [DOI] [PubMed] [Google Scholar]
- 41.Anders S., Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W., Sherman B. T., Lempicki R. A. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang da W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 44.Subramaniam S., Hsiao G. 2012. Gene expression measurement: variance-modeling considerations for robust data analysis. Nat. Immunol. 13: 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pico A. R., Kelder T., van Iersel M. P., Hanspers K., Conclin B. R., Evelo C. 2008. WikiPathways: pathway editing for the people. PLoS Biol. 6: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population- based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorden D. L., Ivanova P. T., Myers D. S., McIntyre J. O., VanSaun M. N., Wright J. K., Matrisian L. M., Brown H. A. 2011. Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS ONE. 6: e22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Cañaveras J. C., Donato M. T., Castell J. V., Lahoz A. 2011. A comprehensive untargeted metabolomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J. Proteome Res. 10: 4825–4834. [DOI] [PubMed] [Google Scholar]
- 49.Lake A. D., Novak P., Fisher C. D., Jackson J. P., Hardwick R. N., Billheimer D. D., Klimetcki W. T., Cherrington N. J. 2011. Analysis of global and absorption, distribution, metabolism, and eliminaton gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab. Dispos. 39: 1954–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lake A. D., Novak P., Hardwick R. N., Flores-Keown B., Zhao F., Klimecki W. T., Cherrington N. J. 2014. The adaptive endoplasmic reticulum stress response to lipotoxicity in progressive human nonalcoholic fatty liver disease. Toxicol. Sci. 137: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Younossi Z. M., Gorreta F., Ong J. P., Schlauch K., Del Giacco L., Elariny H., VanMeter A., Younoszai A., Goodman Z., Baranova A., et al. 2005. Hepatic gene expression in patients with obesity-related non-alcoholic steatohepatitis. Liver Int. 25: 760–771. [DOI] [PubMed] [Google Scholar]
- 52.Younossi Z. M., Baranova A., Ziegler K., Del Giacco L., Schlauch K., Born T. L., Elariny H., Gorreta F., VanMeter A., Younoszai A., et al. 2005. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 42: 665–674. [DOI] [PubMed] [Google Scholar]
- 53.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., Hobbs H. H. 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40: 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merrill A. H., Jr 2011. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111: 6387–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuellig R. A., Hornemann T., Othman A., Hehl A. B., Bode H., Güntert T., Ogunshola O. O., Saponara E., Grabliauskaite K., Jang J. H., et al. 2014. Deoxysphingolipids, novel biomarkers for type 2 diabetes, are cytotoxic for insulin-producing cells. Diabetes. 63: 1326–1339. [DOI] [PubMed] [Google Scholar]
- 56.Jiménez-Rojo N., Sot J., Busto J. V., Shaw W. A., Duan J., Merrill A. H., Jr, Alonso A., Goñi F. M. 2014. Biophysical properties of novel 1-deoxy-(dihydro)ceramides occurring in mammalian cells. Biophys. J. 107: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watt M. J., Barnett A. C., Bruce C. R., Schenk S., Horowitz J. F., Hoy A. J. 2012. Regulation of plasma ceramide levels with fatty acid oversupply: evidence that the liver detects and secretes de novo synthesized ceramide. Diabetologia. 55: 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merrill A. H., Jr, Lingrell S., Wang E., Nikolova-Karakashian M., Vales T. R., Vance D. E. 1995. Sphingolipid biosynthesis de novo by rat hepatocytes in culture. Ceramide and sphingomyelin are associated with, but not required for, very low density lipoprotein secretion. J. Biol. Chem. 270: 13834–13841. [DOI] [PubMed] [Google Scholar]
- 59.McGettrick A. F., O’Neill L. A. 2013. How metabolism generates signals during innate immunity and inflammation. J. Biol. Chem. 288: 22893–22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schleicher J., Gutke R., Dahmen U., Dirsch O., Holzhuetter H. G., Schuster S. 2014. A theoretical study of lipid accumulation in the liver-implications for nonalcoholic fatty liver disease. Biochim. Biophys. Acta. 1841: 62–69. [DOI] [PubMed] [Google Scholar]
- 61.Mantena S. K., Vaughn D. P., Andringa K. K., Eccleston H. B., King A. L., Abrams G. A., Doeller J. E., Kraus D. W., Darley-Usmar V. M., Bailey S. M. 2009. High fat diet induces dysregulation of hepatic oxygen gradients and mitochondrial function in vivo. Biochem. J. 417: 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qu A., Taylor M., Xue X., Matsubara T., Metzger D., Chambon P., Gonzalez F. J., Shah Y. M. 2011. Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology. 54: 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong R. J., Cheung R., Ahmed A. 2014. Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the US. Hepatology. 59: 2188–2195. [DOI] [PubMed] [Google Scholar]
- 64.Ridgeway N. D. 2013. The role of phosphatidylcholine and choline metabolites to cell proliferation and survival. Crit. Rev. Biochem. Mol. Biol. 48: 20–38. [DOI] [PubMed] [Google Scholar]
- 65.Ide Y., Waki M., Hayasaka T., Nishio T., Morita Y., Tanaka H., Sasaki T., Koizumi K., Matsunuma R., Hosokawa Y., et al. 2013. Human breast cancer tissues contain abundant phosphatidylcholine (36:1) with high stearoyl-CoA desaturase-1 expression. PLoS ONE. 8: e61204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy E. P. 1987. Metabolism and function of membrane lipids. Klin. Wochenschr. 65: 205–212. [DOI] [PubMed] [Google Scholar]
- 67.Smith R. E., Lespi P., Di Luca M., Bustos C., Marra F. A., de Alaniz M. J., Marra C. A. 2008. A reliable biomarker derived from plasmalogens to evaluate malignancy and metastatic capacity of human cancers. Lipids. 43: 79–89. [DOI] [PubMed] [Google Scholar]
- 68.Wree A., McGeough M. D., Peña C. A., Schlattjan M., Li H., Inzaugarat M. E., Messer K., Canbay A., Hoffman H. M., Feldstein A. E. 2014. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J. Mol. Med. (Berl). 92: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedman S. L. 2013. Liver fibrosis in 2012: Convergent pathways that cause hepatic fibrosis in NASH. Nat. Rev. Gastroenterol. Hepatol. 10: 71–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.