Abstract
The use of silica nanoparticles for their cellular uptake capability opens up new fields in biomedical research. Among the toxicological effects associated with their internalization, silica nanoparticles induce apoptosis that has been recently reported as a biochemical cue required for muscle regeneration. To assess whether silica nanoparticles could affect muscle regeneration, we used the C2C12 muscle cell line to study the uptake of fluorescently labeled NPs and their cellular trafficking over a long period. Using inhibitors of endocytosis, we determined that the NP uptake was an energy-dependent process mainly involving macropinocytosis and clathrin-mediated pathway. NPs were eventually clustered in lysosomal structures. Myoblasts containing NPs were capable of differentiation into myotubes, and after 7 days, electron microscopy revealed that the NPs remained primarily within lysosomes. The presence of NPs stimulated the formation of myotubes in a dose-dependent manner. NP internalization induced an increase of apoptotic myoblasts required for myoblast fusion. At noncytotoxic doses, the NP uptake by skeletal muscle cells did not prevent their differentiation into myotubes but, instead, enhanced the cell fusion.
Keywords: silica, nanoparticle, muscle, cell encapsulation, transmission electron microscopy, apoptosis
Introduction
The increasing interest in nanoparticles is driven by the potential development of nanotechnology, biotechnology, and medicine, even though several nanoparticles such as metallic and carbon-based particles exhibit health risks associated with their use.1–4 A variety of nanoparticles including silica nanoparticles possess the ability to cross cell membranes. The cellular uptake of nanoparticles occurs either accidentally during environmental exposure or purposely for biomedical strategy. Toxicological effects related to cell internalization have been already identified and depend on several parameters, such as cell type and applied doses.5–7 The presence of nanoparticles induces oxidative stress and inflammation and affects degradation pathways with an appearance of autophagosomes and lysosomal dysfunctions, leading to autophagy and apoptosis.8–10 At biocompatible doses, their ability to easily enter the cell has been proposed as an asset for drug delivery,11–13 cell tracking, and cell therapy,14,15 with a particular focus on the delivery of anticancer therapeutics.16,17
Nanoparticle application has been recently evaluated in neuromuscular diseases such as Duchenne muscular dystrophy, which is caused by defects in the dystrophin gene. Duchenne muscular dystrophy muscles exhibit multiple cellular defects, including mitochondrial dysfunction and increased oxidative stress. Beneficial treatments targeting defective autophagy have been observed in mdx mice.18 Notably, rapamycin, an autophagy-inducing compound successfully delivered using nanoparticle formulation, enhanced physical performance.19 There is an emerging field of nanoparticle therapeutics for muscle disorders and, potentially, muscle repair. For instance, the differentiation of myoblasts has been stimulated with the use of silica nanoparticles loaded with γ-secretase inhibitors, blocking the Notch signaling pathway.20 Developments combining stem cells with nanoparticles provide an interesting strategy for cell therapy.21 Because muscles are scarcely exposed to nanoparticles, little attention has been given to how silica nanoparticles interact with muscle cells, unlike exposed tissues. Fundamental studies on the mechanisms of nanoparticle internalization and their cellular fate are required for providing a thorough view of their mechanism of action.
In muscle, the regeneration takes place with the activation of satellite cells, undifferentiated mononucleated muscle precursor cells (for reviews, see Rochlin et al22 and Abmayr et al23). After several cycles of proliferation, the majority of the cells fuse to repair damaged myofibers or to form new ones. The remaining cells become quiescent and restore the initial population of satellite cells. The myogenic differentiation is regulated by various transcription factors, including the myogenic regulatory factors Myf5, MyoD, myogenin, and MRF4.24 The in vitro differentiation of the C2C12 skeletal muscle cell line reproduces each step of myogenesis. Interestingly, a recent report showed that during myoblast fusion, some myoblasts exposed phosphatidylserine at their surface and underwent apoptosis.25
Phosphatidylserine and the receptor BAI1 induced a signal promoting the fusion of healthy myoblasts with the multinucleated myotubes. Thus, the presence of apoptotic cells and a receptor recognizing those phosphatidylserine-exposing cells plays a key role in myoblast fusion during muscle development, regeneration, and repair. Given that nanoparticles could induce apoptosis, it is of importance to study the internalization of bare silica nanoparticles in muscle cells and the consequences on the maintenance of their differentiation capacity.
In this study, we report the uptake of fluorescent silica nanoparticles (NPs) in C2C12 myoblasts. These myoblasts containing NPs were capable of differentiation into myotubes. After 7 days of differentiation, NPs were still present within the cytoplasm of myotubes. The presence of NPs promoted the formation of myotubes by enhancing myoblast fusion.
Material and methods
Nanoparticle synthesis
Fluorescein isothiocyanate (FITC) was incorporated inside the NP’s core during synthesis, leading to fluorescent 50 nm NPs, as previously described.26 Shortly, the synthesis was based on the method described by Van Blaaderen.27 In a first step, FITC (Thermo Fisher Scientific, Rockford, IL, USA) was covalently attached to a silane-coupling agent, (3-aminopropy1)triethoxysilane (APS), by the reaction of an amino group with an isothiocyanate group, leading to a thiourea link. The reaction was performed in the dark to avoid photobleaching and under anhydrous conditions to prevent hydrolysis of APS. Typically, an amount of 5 mg FITC was dissolved in 5 mL of 42.7 mM of APS in ethanol. After 12 hours of stirring, the fluorescent silane was added in a 500 mL two-neck flask immersed in a 50°C oil bath and containing 250 mL ethanol, 5 mL tetraethoxysilane, 7.6 mL ammonium hydroxide (28%), and 10.9 mL water. The reaction was allowed for 12 hours in the dark, under magnetic stirring. The so-prepared particles have an average diameter of about 30 nm. A seed-growth procedure was used to increase the NP size to 50 nm. The entire mixture was poured in a 1 L round-bottom flask containing 500 mL ethanol, 160 mL water, and 9.9 mL ammonium hydroxide (28%). Then 18.2 mL tetraethoxysilane was added at two times, spaced for 12 hours to prevent secondary nucleation. After this synthesis step, ammonia and ethanol were removed from the medium at 40°C, using a rotary evaporator. The fluorescent particles were extensively washed by centrifugation against ultrapure water (18 MΩ) at 13,000× g for 15 minutes until the disappearance of fluorescence in the supernatant. An estimation of silica concentration in the dispersion was carried out by inductively coupled plasma optical emission spectrometry and the gravimetric method. Physicochemical characteristics of NPs are accessible in the Supplementary materials (Table S1).
Cell culture
All cell culture reagents were purchased from Life Technologies unless otherwise specified. The C2C12 mouse myoblast cells were obtained from the American Type Culture Collection (Rockville, MD, USA). They were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (penicillin/streptomycin) in a humidified atmosphere with 5% CO2 at 37°C. All the experiments were performed with cells from passages 7–9. When necessary, myogenic differentiation was induced on 70% confluent cells cultured in gelatin-coated dishes by changing the growth medium to a differentiation medium (2% horse serum [HS] in DMEM). A serum deprivation triggered myoblast fusion into myotubes. Differentiation medium was changed every 3 or 4 days.
Cell treatment
NP dispersions were prepared by diluting the concentrated NP stock solution (19.6 mg/mL in water) into DMEM without phenol red at room temperature, immediately before the experiments, and were vortexed for 1 minute. Before NP exposure, cells were rinsed once with DMEM, and all treatments were performed in the absence of serum. For all experiments, cells were incubated with NPs at 36 μg/mL or 9 μg/cm2 for the required times with the exception of flow cytometry experiments of NP uptake, performed at 3 μg/cm2. After treatment, cells were washed twice with phosphate-buffered saline (PBS) to remove the unbound NPs. For the energy-dependent NP uptake, cells were preincubated for 30 minutes at 4°C or with sodium azide (100 mM; Sigma) and 2-deoxy D-glucose (200 mM; Sigma) in PBS before exposure. Endocytosis inhibitors (Merck), that is, chlorpromazine (15 μM), genistein (200 μM), and cytochalasin D (5 μM), were incubated with cells for 30 minutes before treatment with NPs. Then NPs were applied for 30 minutes in the presence of the inhibitors.
For the study of the NP effect on muscle cell differentiation and the long-term fate of NPs, 70% confluent myoblasts were washed and treated with NPs at 9 μg/cm2 for 1 hour just before inducing differentiation.
Cell viability
Myoblasts were seeded on 48 well plates at a density of 25,000 cells/cm2 in proliferation medium and allowed to adhere during the 24 hours before NP exposure. The WST-1 reagent (Roche) was used according to the manufacturer’s instructions to estimate the cell viability. The WST-1 reagent was added 16 hours after NP treatment to the cells and blanks and incubated for 4 hours. Absorbance was measured at 450 nm.
Fluorescence microscopy
Cells were cultured on coverslips and fixed with 4% formaldehyde in PBS for 20 minutes and then permeabilized with 1% Triton X-100 in PBS for 5 minutes at room temperature. Cells were blocked with 3% bovine serum albumin (BSA) for 2 hours at room temperature. Cells were then incubated for 3 hours with mouse antibodies to skeletal heavy chain myosin (M4276; Sigma) or to sarcomeric alpha actinin (A7811; Sigma) diluted 1:300 and 1:500, respectively, in PBS plus 1% BSA. After three washes in PBS, cells were then incubated with Alexa Fluor 594-conjugated donkey anti-mouse IgG antibody diluted 1:1,000 (Molecular Probes), diluted in PBS plus 1% BSA for 1 hour at room temperature. After washes in PBS, cells were incubated with Hoechst 34580 at 5 ng/mL (Invitrogen) for nuclei staining and mounted in low-fluorescence mounting medium (Dako) and examined by epifluorescence (an inverted Leica DMI 6000 microscope equipped with a Leica DFC 350FX camera). For the confocal spinning disk experiment, cells were incubated for 30 minutes with tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin (1 ng/mL in PBS; Sigma), and then nuclei were stained using Hoechst 34580. Coverslips were mounted in a low-fluorescence mounting medium (Dako). Images were acquired with a Leica DMI-6000 microscope equipped with a Yokogawa CSU-X1 confocal spinning disk.
Transmission electron microscopy preparation
Cells were fixed using 1.6% (v/v) glutaraldehyde in phosphate buffer 0.1 M at pH 7.4 overnight at 4°C. They were postfixed using 1% (v/v) osmium tetroxide for 90 minutes and dehydrated in ascending series of ethanol dilutions. They were then treated with propylene oxide, impregnated in ascending dilutions of resin in propylene oxide, left in pure resin overnight (Epon; Inland Europe), embedded, and polymerized at 60°C for 48 hours. Ultrathin sections (70 nm) performed on an ultramicrotome (RMC, powertome PC) were collected on butvar-coated single-slot copper grids and were stained with 2% (v/v) uranyl acetate for 30 minutes and with lead citrate for 2 minutes. For NP characterization, solution at 40 μg/mL was deposited on a carbon-coated grid previously submitted to a glow discharge (Elmo, Cordouan Technologies). Grids were examined by transmission electron microscopy (TEM; CM120; FEI), and the images were acquired using a digital 2×2 k Gatan camera.
Flow cytometry analysis of NP uptake
Cells were seeded at a density of 15,000 cells/cm2 in 12 well plates and allowed to adhere for 24 hours in growth medium before exposure to 1 mL NPs at 3 μg/cm2 in the different conditions. After treatment and washing, cells were trypsinized and resuspended in cold 10% fetal bovine serum (FBS)-supplemented medium without phenol red. Cells were stored on ice and analyzed within 1 hour. Cells were preincubated with 0.25% Trypan blue for 1 minute just before analysis to quench the fluorescence of NPs absorbed on the cell surface. Samples were analyzed on a BD Biosciences FACS Canto flow cytometer, and NPs were excited by a 488 nm laser. The analysis was performed using Diva software. Cell debris was excluded, based on the forward and side scatter characteristics of cell populations. The results are expressed as the percentage of the mean of fluorescence intensity. Ten thousand cells were analyzed in the gate.
Measurement of annexin V, 7-amino-actinomycin D vital dye, binding, and cleaved caspase-3
Phosphatidylserine exposure of floating and adherent myoblasts was measured using the annexin V phycoerythrin-7-amino-actinomycin D vital dye (7AAD) apoptosis detection kit (BD Biosciences). Annexin V has a high affinity for phosphatidylserine (PS) in the presence of calcium. In apoptotic cells, PS translocated to the outer leaflet of the plasma membrane can be detected by fluorescent annexin V conjugates. C2C12 cells exposed to NPs for 1 hour were incubated with differentiation or growth medium for the indicated times, washed with PBS two times, and trypsinized. Cell pellets were resuspended in binding buffer (10 mM Hepes, 140 mM NaCl, 25 mM CaCl2 at pH 7.4) at 1×106 cells/mL. Next, 150 μL were incubated with annexin V-PE (phycoerythrin) and 7AAD for 10 minutes at room temperature and were immediately analyzed by flow cytometry. The apoptotic cell population was defined as PS-positive and 7AAD-negative. Negative controls were performed in the free-calcium binding buffer.
For cleaved caspase labeling, floating and adherent C2C12 cells were fixed and permeabilized, using the Cytofix/Cytoperm solution (BD Biosciences). They were then stained with an anticleaved caspase-3 antibody PE conjugated (D3E9; Cell Signaling) and diluted 1:50 in washing buffer (0.1% saponin and 1% FBS in PBS at pH 7.2) overnight at 4°C. Cells were washed using the same washing buffer and resuspended in PBS containing 0.5% BSA (Sigma) before analysis. Samples were analyzed on a BD Biosciences FACS LSRFortessa flow cytometer. FITC and PE were excited by a 488 nm laser and a 561 nm laser, respectively. The analysis was performed using the Diva software. Cell debris was excluded on the basis of the forward and side scatter characteristics of cell populations. The results are expressed as the percentage of positive cells. Ten thousand cells were analyzed in the gate.
Quantitative analysis of fusion index
The evaluation of myoblast fusion was determined as previously described.28 At various times after inducing differentiation, cells were fixed, permeabilized, and stained for myosin, as described earlier, and nuclei were stained with Hoechst. The extent of fusion was determined as follows: fusion (%) = (number of nuclei in myotubes)/(total number of nuclei in myoblasts and myotubes) × 100. At least 10 random fields (100× final magnification) were analyzed per experiment, using the ImageJ software. The number of myosin positive myotubes per field with five or more nuclei was also quantified.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blot analysis
Cell pellets were dissolved in PBS containing 1% triton X-100 and 1× protease inhibitor cocktail and then sonicated briefly and lysed for 30 minutes at 4°C. The protein concentration of each cell lysate was determined by the Bradford method (Bradford Reagent; Sigma). After treatment with 5× sodium dodecyl sulfate loading buffer, 50 μg protein was resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Immobilon-P, Millipore) by the semidry transfer method. The membrane was blocked for 2 hours at room temperature with 5% (w/v) skimmed milk in Tris-buffered saline plus 0.1% Tween 20 (TBST). The membrane was then incubated for 2 hours at room temperature in TBST plus 1% skimmed milk containing the following primary antibodies: mouse anti-skeletal myosin diluted 1:500, anti-myogenin (sc-576; Santa Cruz) diluted 1:250, anti-alpha-actinin (sarcomeric) diluted 1:2,000, and anti-GAPDH (sc-25778; Santa Cruz) diluted 1:1,000. The membrane was next washed three times in TBST, and horseradish peroxidase conjugated secondary antibody (Sigma) diluted 1:50,000 was applied. Proteins were detected by incubating the membrane with an enhanced chemiluminescence system (ECL Prime Western blotting detection reagent; GE Healthcare) according to the manufacturer’s instructions, using the Gene Gnome system (Syngene). The obtained signals were quantified by using the ImageJ software.
Statistical analysis
Data were collected from at least three separate experiments and expressed as mean ± standard error of the mean. The statistical difference was analyzed by Student unpaired t-test when two groups were compared. For analysis of statistical difference between three or more groups, a one-way analysis of variance followed by a Bonferroni’s multiple comparison test was applied. P-values below 0.05 were considered to indicate statistical differences.
Results
Interactions of NPs with C2C12 cells
C2C12 myoblasts at 70% confluence were treated for 1 hour with a 9 μg/cm2 NP concentration compatible with the WST-1 proliferation and viability assay (Figure S1). Using confocal microscopy, aggregates of NPs were observed at the cell surface, and smaller fluorescent spots were detectable inside the cell, as revealed by orthogonal projections (white arrow, Figure 1A). More descriptive information about the NP distribution was provided by TEM observations of adherent cells sectioned perpendicularly to the support. Isolated NPs or small aggregates were observed attached to the cell surface (Figure 1B). According to zeta potential analysis and TEM observation, it is likely that these aggregates were formed in solution (Figures S2 and S3 and Table S1). Within cells, vesicles containing one or several NPs were present, suggesting NPs pass the plasma membrane via endocytic pathways (Figure 1C and D). In addition, few events were encountered, revealing isolated NPs engulfed by the plasma membrane (black arrows in Figure 1E) and within the cytoplasm without being surrounded by an apparent lipid membrane (Figure 1F). A similar distribution of silica nanoparticles has been already reported by Shapero et al in lung epithelial cell.29
Figure 1.
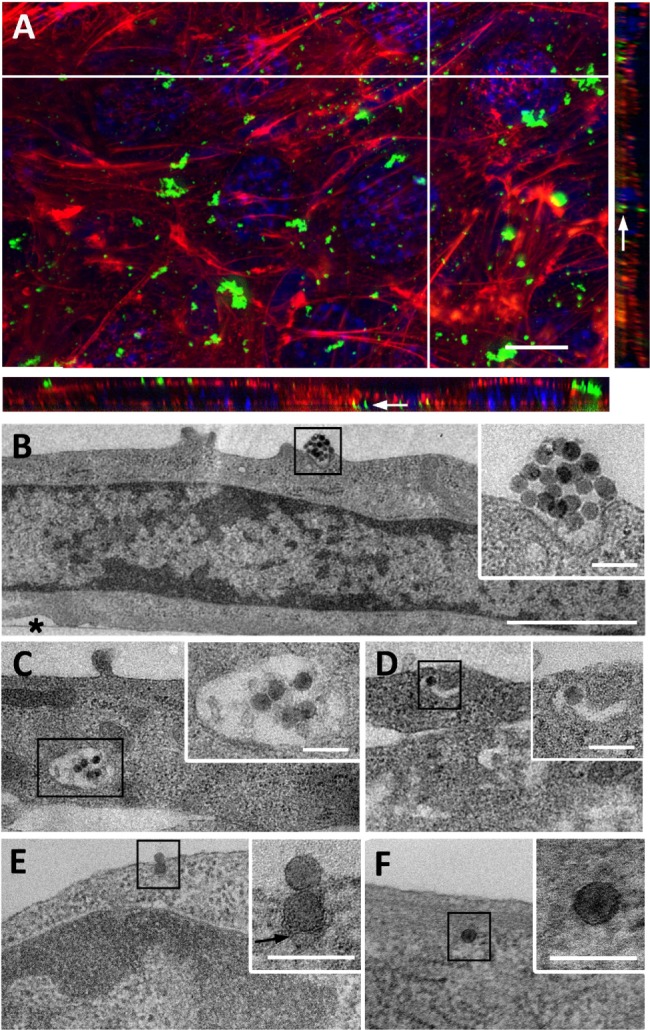
Uptake of NPs by C2C12 myoblasts.
Notes: (A) Z-projection of confocal images and xz and yz-slices (orthogonal views along white lines) of NPs interacting with cells for 1 hour. Blue, Hoescht-stained nuclei; red, TRITC-phalloidin-stained actin filaments; green, fluorescein isothiocyanate-labeled NPs. White arrow marks fluorescent spot inside the cell. Scale bar, 10 μm. (B–F) Transmission electron microscopy images of NPs interacting at cell surface (B) and NPs within cytoplasm (C–F). Insets are enlarged areas marked with black squares in (B–F). Black arrows indicate plasma membrane surrounding NP. Asterisk marks the cell side facing the support. Scale bars, 10 μm (A), 1 μm (B–F), and 100 nm (insets).
Abbreviations: NP, silica nanoparticle; TRITC, tetramethylrhodamine B isothiocyanate.
Note that the NP internalization decreased with the increase of cell density (Figure S4). For all experiments, the cell density has been set at 15,000 cells/cm2 and is estimated to reach 70% confluence 24 hours after seeding, which corresponds to conventional culture conditions amenable to myotube formation.
Internalization pathways of NPs within C2C12 cells
The involvement of different endocytic pathways has been investigated using several inhibitors of endocytosis: chlorpromazine for clathrin-mediated endocytosis, genistein for caveolin-mediated endocytosis, and cytochalasin D for actin-dependent macropinocytosis (Figure 2). As shown by flow cytometry, all inhibitors reduced NP uptake. Macropinocytosis and clathrin-mediated endocytosis were more efficient than caveolin-mediated endocytosis. When ATP production was impaired with 2-deoxyglucose and sodium azide or when the experiment was performed at 4°C, the fluorescence of cells exposed to NPs considerably decreased, corresponding to a reduced uptake of NPs. These results indicated that the uptake of NPs in muscle cells was mainly an energy-dependent process, as observed in various cell types, including pancreatic, hepatic, and epithelial cells.26,29–31 In addition, even though ATP depletion considerably reduced NP uptake, it is not impossible that NPs enter cells via ATP-independent processes, as already proposed.32,33 Accordingly, a physicochemical mechanism involving adhesion and membrane spreading around the NP surface has already been observed, with large unilamellar liposomes and red blood cells both lacking endocytic capabilities.34,35
Figure 2.
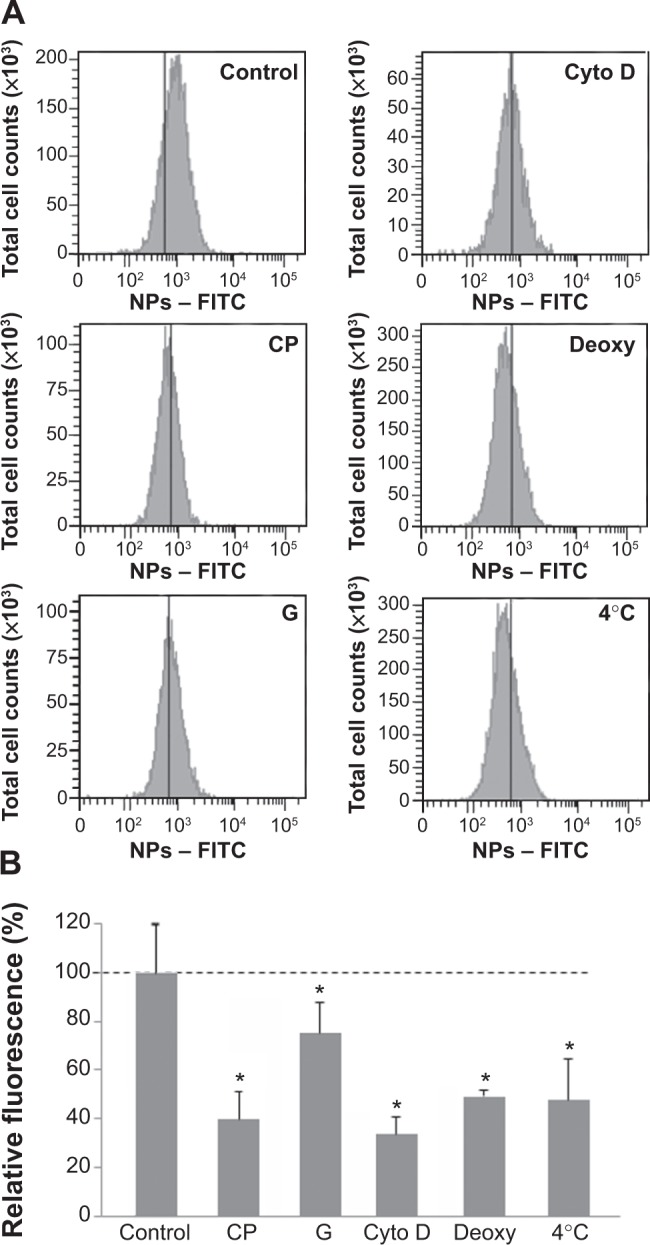
Effect of endocytosis inhibitors on cellular uptake of NPs.
Notes: Cells were pretreated at 37°C with chlorpromazine (CP), genistein (G), cytochalasin D (cyto D), sodium azide, and 2-deoxy-D-glucose (deoxy) for 30 minutes and then exposed to NPs (3 μg/cm2) and inhibitors for 30 minutes. Cells maintained at 4°C were exposed to NPs for 30 minutes after 30 minutes of preincubation at 4°C. Quantification of internalization was performed by flow cytometry, after addition of trypan blue. (A) Flow cytometry analysis of FITC-NPs uptake by treated cells. (B) Percentage of uptake relative to untreated cells exposed to NPs. This was calculated by dividing the mean cell fluorescence intensity after NP uptake in drug-treated cells by that of untreated cells. Untreated cells at 37°C ± standard error of the mean. Bars on the graph represent the standard of the mean. *Significantly different from the control (P≤0.05); n=3.
Abbreviation: NP, silica nanoparticle.
Fate of NPs within myoblasts and myotubes
To get further insight on the fate of NP after internalization, myoblasts were cultured in growth medium (FBS) for 4 hours after NP exposure. The intracellular distribution of NPs analyzed by TEM revealed the presence of large amounts of NPs within the endosomal vesicles. NPs were clustered in large vesicles (Figure 3A) and also in lipid membrane-containing vesicles that closely resembled lysosomes containing mitochondria undergoing degradation (Figure 3B). The presence of such multilamellar vesicles may be associated with cellular defense mechanisms to sequester NPs such as autophagy, as reported in the literature.8
Figure 3.
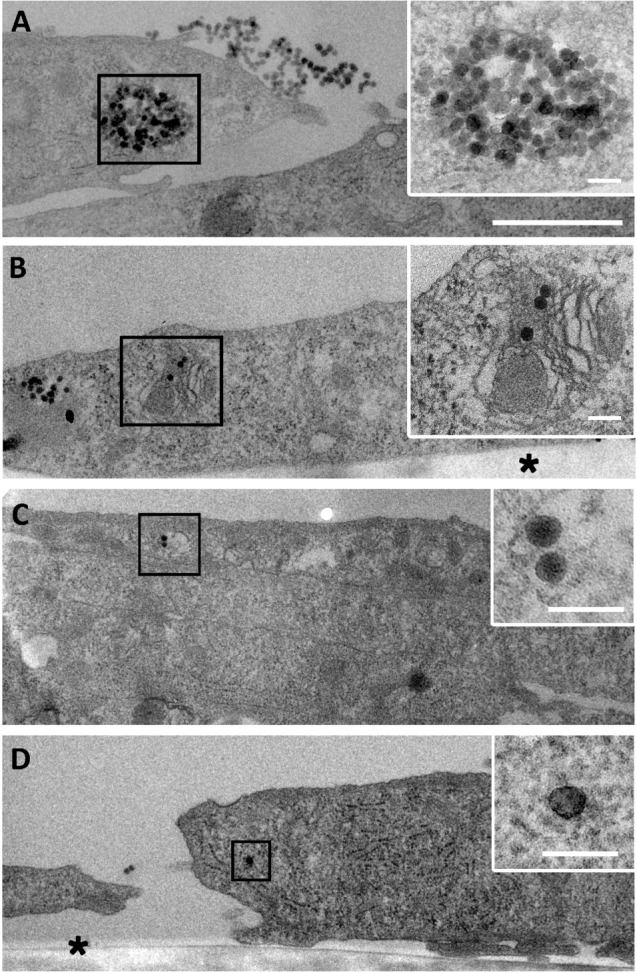
Fate of NP uptake by myoblasts after 4 hours.
Notes: Transmission electron microscopy images of a large amount of NPs accumulated in vesicles (A) or in organelles undergoing degradation (B). Few NPs in vesicle (C) and isolated NP in cytoplasm (D) were visible, as observed after 1 hour of exposure. Insets are enlarged areas marked with black squares. Asterisk marks the cell side facing the support. Scale bars, 1 μm (A–D) and 100 nm (insets).
Abbreviation: NP, silica nanoparticle.
It is important to note that NPs were still attached to the cell surface, even a few hours after removing the excess NP solution. Furthermore, NPs were observed to be isolated or in small vesicles corresponding to similar events encountered after 1 hour uptake, suggesting that the process of NP uptake lasted several hours after exposure (Figure 3C and D).
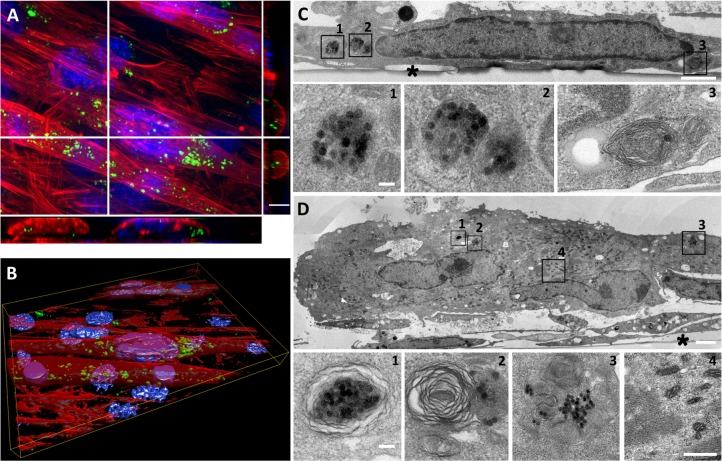
Then, the fate of NPs internalized into C2C12 myoblasts has been investigated in the context of the formation of myotubes to evaluate the effect of NPs on this differentiation process. Indeed, myoblast fusion into myotubes was induced by the use of a low percentage of HS. In these conditions, the cell cycle was arrested and C2C12 myoblasts fused to form polynucleated myotubes. C2C12 myoblasts were exposed to NPs just before serum deprivation and were cultivated over the course of 7 days. First myotubes appeared on day 2 and grew to day 7. These myotubes contained fluorescent NPs (Figure 4A and B). TEM observations revealed that NPs were mainly packed into multilamellar lysosomal structures (Figure 4D). Aside from myotubes, there were mononucleated myoblasts that still contained NPs clustered into unilamellar and multilamellar vesicles (Figure 4C).
Figure 4.
Long-term fate of NP uptake 7 days after initiating the differentiation process.
Notes: (A) Z-projection of confocal images and xz and yz slices (orthogonal views along white lines) of NPs within myotubes and myoblasts. Blue, Hoescht-stained nuclei; red, TRITC-phalloidin-stained actin filaments; green, fluorescein isothiocyanate-labeled NPs. Scale bar, 10 μm. (B) Three-dimensional view of the field shown in A. (C) TEM images of NPs accumulated in myoblasts. Typical vesicles containing NPs (1–3) are enlarged areas marked with black squares in C. (D) TEM images of NPs accumulated in polynucleated myotube. 1–3 are enlarged areas (marked with black squares in D) of typical vesicles containing NPs similar to those in C. Image 4 shows typical sarcomeric structures and numerous mitochondria characteristic of myotubes. Asterisks mark the cell side facing the support. Scale bars, 1 μm (C, D) and 100 nm (enlarged images).
Abbreviations: NP, silica nanoparticle; TEM, transmission electron microscopy.
Promyogenic effect of NPs
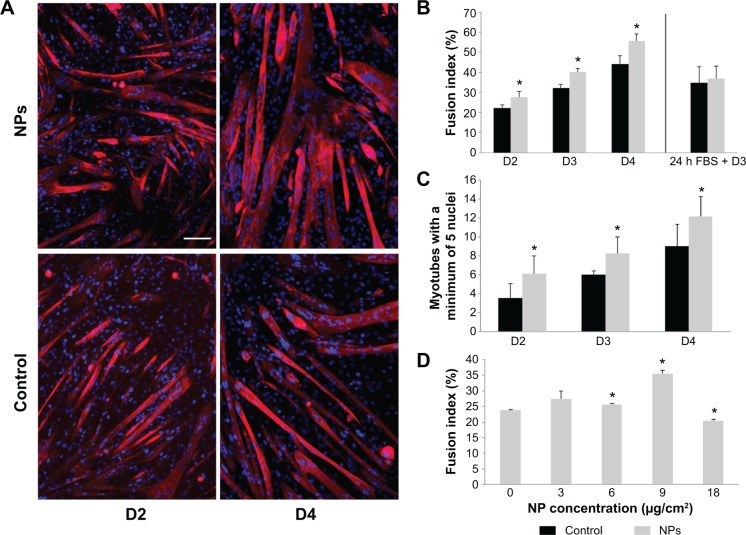
As NPs were present all along the differentiation process, we investigated the potential effect of NPs on myotube formation. Their morphologic features, characterized by a striated pattern of alpha-actinin (an early marker of sarcomere assembly), were similar to those of myotubes formed from untreated myoblasts (Figure 5A), as observed by TEM (Figure 4D4). The measurement of sarcomeric alpha-actinin and myosin expression levels in NP-containing myotubes at day 7 revealed a 2.5-fold and a 1.5-fold increase, respectively, whereas the expression of the myogenin transcription factor involved in the regulation of muscle-specific genes was not modified, suggesting that myogenesis was enhanced without activating the myogenin pathway (Figure 5B and C). The rate of fusion characterized by the fusion index (ratio of nuclei contained within myotubes) is a criterion indicative of myogenesis activity. Using immunolabeling against myosin, a specific marker of myotubes (Figure 6A), we measured the fusion index of NP-containing cells and showed it was significantly increased by 5.4%, 7.8%, and 11.4% at days 2, 3, and 4, respectively (Figure 6B). Accordingly, the myotubes containing at least five nuclei (another way to quantify the fusion velocity) were significantly more abundant in the presence of NPs (Figure 6C). Thus, NPs had a promyogenic effect by enhancing the formation of myotubes. In our experimental conditions, an exposure of NPs to myoblasts did not impair myogenesis but, instead, promoted cell differentiation into myotubes.
Figure 5.
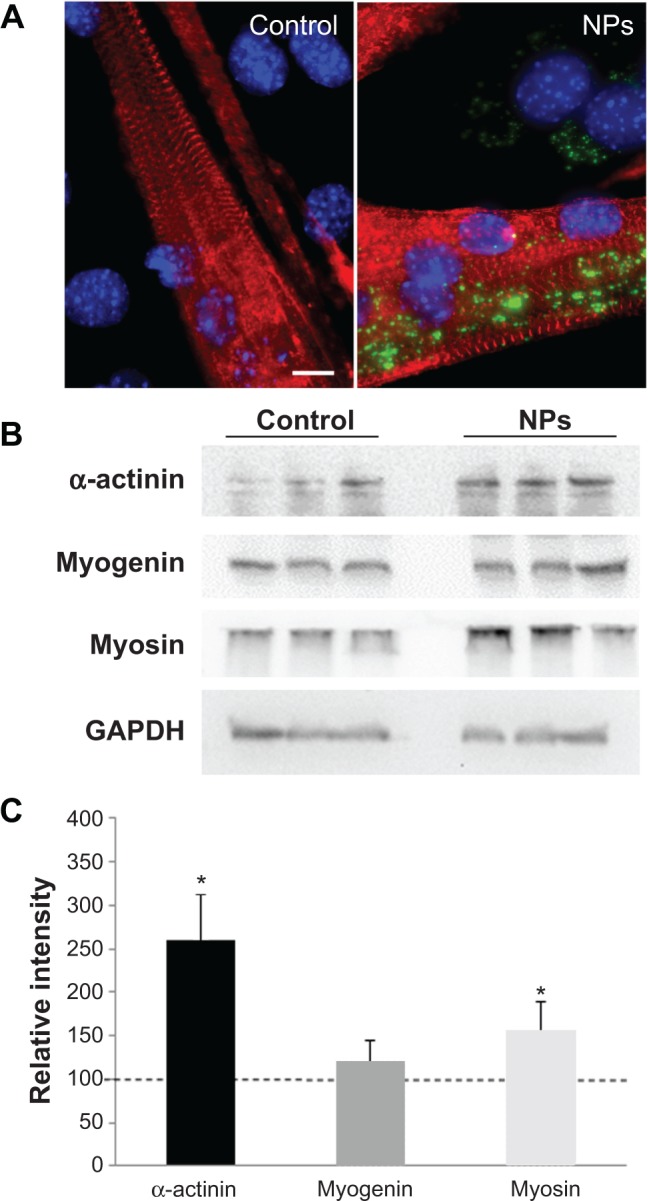
Effect of NPs on the expression of muscle specific markers in myotubes.
Notes: (A) Fluorescence images of myotubes after incubation with and without NPs. Red, staining of alpha-actinin characteristic of the sarcomeric organization of myofibers; blue, Hoechst stained-nuclei. Scale bar, 1 μm. (B) Expression of muscle-specific markers. After myoblast treatment with NPs, proteins were extracted from myotubes at day 7 of differentiation and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis. Immunodetection was carried out using specific antibodies against alpha-actinin, myosin heavy chain, and myogenin. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. (C) The level of the different proteins was quantified and expressed as a percentage of the level obtained in the untreated control cells. Bars on the graph represent the standard error of the mean. *Significantly different from the control (P≤0.05).
Abbreviation: NP, silica nanoparticle.
Figure 6.
Myotube formation promoted by NP uptake.
Notes: Myotube fusion was analyzed at days 2, 3, and 4 of differentiation when myoblasts were incubated or not (control) with NPs. (A) Fluorescent images of myoblasts and nascent myotubes at days 2 and 4 of differentiation. Myotubes arising from NP-treated myoblasts appeared larger. Nuclei (Hoechst, blue) and myotubes (anti-myosin antibody, red) were labeled. Scale bar, 100 μm. (B) Quantitative analysis of the fusion index. (C) Quantification of myotubes containing at least five nuclei. Their number increased significantly at the early days of differentiation. (D) Quantitative analysis of fusion index for several NP concentrations at day 3. Data are expressed as mean ± standard error of the mean; n=3. *Significantly different from the control (P≤0.05).
Abbreviation: NP, silica nanoparticle; FBS, fetal bovine serum.
The enhancement of myoblast fusion has been assessed for several NP concentrations (Figure 6D). A 3 μg/cm2 NP concentration did not significantly increase myoblast fusion, whereas the concentrations of 6 and 9 μg/cm2 induced a progressive promyogenic effect. A 18 μg/cm2 toxic concentration had a detrimental effect on fusion. Thus, this promyogenic effect seemed to be NP dose-dependent, with an optimal concentration close to the toxicity threshold.
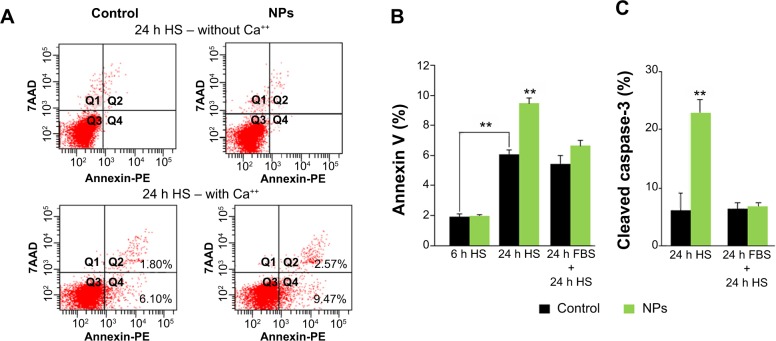
NPs increased apoptosis of myoblasts
It has been recently shown that after 24 hours in differentiation medium (ie, with a low percentage of HS), myoblasts exposed PS and underwent apoptosis, representing a signal for myoblast fusion.25 Therefore, we assessed two apoptosis markers: annexin V binding to PS exposed at the cell surface and the expression of cleaved caspase-3 (Figure 7). A significant increase of cells positive for annexin V binding (9.6% versus 6.1% of cells in control) and for cleaved caspase-3 (22.9% versus 6.2% of cells in control) was observed 24 hours after NP treatment, indicative of an apoptosis enhancement. Interestingly, this enhancement was measured only when NP treatment was concomitant to serum deprivation. Indeed, when differentiation process was delayed by 24 hours after NP exposure (24 hours FBS + 24 hours HS), neither annexin V binding nor cleaved caspase-3 were enhanced in the presence of NPs. The fusion index in this condition was similar to that of the control (Figure 6B). Altogether, these results indicated that NP entry transitorily enhanced apoptosis during myoblast differentiation.
Figure 7.
NP uptake enhances apoptosis required for myoblast fusion.
Notes: (A) Representative annexin V-PE/7AAD labeling pattern of myoblast populations in HS medium 24 hours (h) after NP exposure or not (control), in the absence and the presence of calcium. The population positive for annexin V and negative for 7AAD (Q4) was used to quantify PS exposure at cell surface. (B) Exposed myoblast population positive for annexin V analyzed after 6 h in HS, 24 h in HS, and 24 h in FBS, followed by 24 h in HS. (C) Percentage of cells positive for cleaved caspase-3 analyzed after 24 h in HS and 24 h in FBS, followed by 24 h in HS. Data are expressed as mean ± standard error of the mean from three independent experiments (**P<0.01).
Abbreviations: NP, silica nanoparticle; HS, horse serum; FBS, fetal bovine serum; 7AAD, 7-amino-actinomycin D; PE, phycoerythrin; PS, phosphatidylserine.
Discussion
The main concerns raised by the development of nanoparticles are cell toxicity, internalization pathways, nanomaterial biopersistence, and the effect on cell functionalities. The internalization of silica nanoparticles has been studied in cell types in relation with exposed tissues, mainly lung, gut, and skin.36–38 It has been proven that these nanoparticles may have deleterious effects and jeopardized cell survival above exposure thresholds. Below the toxic threshold, silica nanoparticles show potential development in biomedical research for bioimaging, diagnosis, drug delivery, and tissue engineering, including muscle regeneration therapy.20,39,40 We have studied the internalization of silica NPs in myoblasts and characterized their biopersistence during in vitro cell differentiation, representing an excellent model for muscle regeneration. Interestingly, we showed that at low doses, NP uptake was able to increase the formation of myotubes by enhancing myoblast fusion.
Our results on NP internalization show that cellular uptake is based on energy-dependent mechanisms, with macropinocytosis and the clathrin-mediated pathway being the predominant endocytic pathways. The uptake of the same NP by NCl-H292 lung adenocarcinoma cells exhibits similar routes.26 After a rapid internalization into myoblasts, our TEM observations show that NPs are clustered into vesicles. Moreover, NPs are still present within these vesicles over several days of differentiation and are also transferred into myotubes during myoblast fusion. The NP feature, in terms of size and shape, remains unchanged, as observed for other silica nanoparticles.41 The load of NPs sequestered into these multilamellar structures does not prevent myoblast differentiation into myotubes or their further growth.
The internalization of NPs and their persistence within myoblasts have an unexpected effect on cell differentiation. Our results show that myotube growth is enhanced in the presence of NPs and raises the question of by which mechanism NPs enhance this process. It has been shown that apoptotic myoblasts were required for inducing myotube formation.25,42 The apoptotic myoblasts would interact with healthy myoblasts via their receptor BAI1. After its recognition of PS lipids on apoptotic myoblasts, BAI1 signaling through the ELMO/Dock180/Rac1 pathway would then promote the fusion of healthy myoblasts.25 In agreement with these data, our results show a significant increase of apoptotic cells after 24 hours in differentiation medium. Interestingly, cell incubation with NPs before switching to differentiation medium enhances PS exposure and cleaved caspase-3, indicative of an increase of apoptotic cells. Apoptosis induced by silica nanoparticles has already been measured on several cell types, including macrophages and hepatic and endothelial cells.43–45 Thus, according to the mechanism described by Hochreiter-Hufford et al25 NP uptake could very likely enhance myoblast fusion via an apoptotic induction that acts in synergy with the apoptotic process triggered by differentiation medium.
Among the four myogenic regulatory factors, MyoD, myogenin, and Myf-5 regulate the cell cycle, whereas MRF4 is involved in terminal differentiation. Myogenin is an early marker for the entry of myoblasts into the differentiation pathway and possesses an antiproliferative activity.46 Therefore, we have analyzed the myogenin expression as an early marker of differentiation and showed that NP treatment did not modify the level of myogenin expression, indicating that the effect of NPs is not directly related to the myogenin expression. Likewise, Hochreiter-Hufford et al25 who reported on the promoting role of apoptosis on myoblast fusion, did not observe a modification of myogenin expression, suggesting that apoptosis is required either downstream or parallel to myogenin.
The effect of NPs on myotube growth has been observed when differentiation was initiated after NP incubation. As we have observed that NP internalization and clustering into vesicles was an ongoing process a few hours after rinsing, the internalization process occurs concomitantly with the differentiation process. To assess whether this internalization process is important for myotube formation, the initiation of differentiation has been postponed for 24 hours, corresponding to a time course for a complete NP internalization.26 In these conditions, the percentage of apoptotic cells is not significantly higher than that of untreated cells (Figure 7). In addition, the morphology of myotubes and the fusion index are similar to those obtained in untreated myoblasts, meaning that NP load has no or few effects on the myotube growth. Altogether, these results indicate that NP internalization and the related subsequent cellular events are able to promote apoptosis that enhances myoblast fusion when combined with differentiation medium.
Although preliminary, our results could be useful for the potential development of stem cells to restore muscle structure and function in pathological conditions such as congenital skeletal muscle defects.18,47 Several limitations concerned the survival of implanted cells and nonspecific cell delivery, impairing clinical applications.48 The use of fluorescent silica nanoparticles as a cell tracker and/or drug delivery vector for myogenic inducer could facilitate this strategy.21 In a recent report, silica nanoparticles loaded with Notch signaling modulators were used to induce efficient differentiation of myoblasts into myotubes.20 C2C12 cells have been grown on a film of silica nanoparticles. After their cellular uptake, these nanoparticles delivered the Notch inhibitor, suggesting that a surface or a three-dimensional scaffold impregnated with nanoparticles can be used for cell-targeted delivery. Our study may help in the choice of nanomaterial in the perspective of muscle regeneration therapy.
Conclusion
We have studied in muscle cells the internalization of 50 nm fluorescent NPs and their effect on myoblast fusion. These NPs were internalized in an energy-dependent process. We showed that the differentiation of myoblasts into myotubes could be enhanced after NP exposure. NP internalization induced an increase of apoptotic myoblasts and exposed phosphatidylserine essential for myoblast differentiation. Thus, low doses of NPs could have a beneficial effect on muscle growth and are worth attention in the field of regenerative medicine and muscle repair.
Supplementary materials
Cytotoxicity of NPs in C2C12 myoblasts.
Notes: Twenty-four hours after seeding at 15,000 cells/cm2, cells were exposed to various doses of NPs for 1 hour in Dulbecco’s Modified Eagle’s Medium. Cell viability was measured 16 hours after treatment, using the WST-1 assay. The toxicity threshold is observed for a NP concentration of 9 μg/cm2. Data are the percentages of surviving cells relative to the control reported as the mean ± standard error of the mean of three independent experiments.
Abbreviation: NP, silica nanoparticle.
Transmission electron microscopy characterization of 50 nm NPs.
Notes: Transmission electron microscopy images of NPs suspended in water (40 μg/mL). Scale bar, 100 nm. A histogram showed the size distribution of 100 NPs measured using the ImageJ software. The mean diameter was 49.5±3.9 nm (standard error of the mean).
Abbreviation: NP, silica nanoparticle.
Dynamic light scattering curves of silica nanoparticles in water and in Dulbecco’s Modified Eagle’s Medium.
Abbreviations: DMEM, Dulbecco’s Modified Eagle’s Medium; NPs, silica nanoparticles.
NP internalization as a function of cell confluence.
Notes: Cells were treated with NPs during 30 minutes at 3 μg/cm2 in Dulbecco’s Modified Eagle’s Medium, 24 hours after seeding at three concentrations: 2,000 (C1), 15,000 (C2), and 50,000 (C3) cells/cm2, corresponding to about 50%, 70%, and ~100% confluence, respectively. Quantification of NP internalization was performed by flow cytometry after the addition of Trypan blue. Results are expressed as mean cell fluorescence intensity (arbitrary unit) ± standard error of the mean; n=3.
Abbreviation: NP, silica nanoparticle.
Table S1.
Physicochemical characteristics of NPs
| Characteristic | H2O | Dulbecco’s Modified Eagle’s Medium |
|---|---|---|
| Zeta potential, mV | −35.6 | −31.2 |
| Hydrodynamic radius, nm | 81.8 | 122.9 |
Notes: NPs were characterized for their hydrodynamic radius and zeta potential suspended in water and Dulbecco’s Modified Eagle’s Medium. Dynamic light scattering and zeta potential values were measured by a zetasiser 300 HS (Malvern) and Vasco granulometer (CordouanTechnologies). Size estimation using the cumulant method shows a slight destabilization of NPs in Dulbecco’s Modified Eagle’s Medium (122.9 nm) compared with H2O (81.8 nm).
Abbreviation: NP, silica nanoparticle.
Acknowledgments
This work was supported in part by TiSiTrans (PNR-EST-2010/2/79), ANSES (Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travai) grants, and Conseil Régional d’Aquitaine (20121301007) grants. We acknowledge the Bordeaux Imaging Center for use of the confocal spinning disk system and V Pitard at the cytometry platform at University Bordeaux Segalen. We thank Cordouan Technologies for the use of their apparatus and J-C Taveau for help with image analysis. We also thank N Arraud for the annexin V binding experiment and A Baeza-Squiban for critical reading and comments.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Biju V. Chemical modifications and bioconjugate reactions of nano-materials for sensing, imaging, drug delivery and therapy. Chem Soc Rev. 2014;43(3):744–764. doi: 10.1039/c3cs60273g. [DOI] [PubMed] [Google Scholar]
- 2.Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nano-level. Science. 2006;311(5761):622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 3.Oberdörster G, Oberdörster E, Oberdörster J. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005;113(7):823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Napierska D, Thomassen LCJ, Lison D, Martens JA, Hoet PH. The nanosilica hazard: another variable entity. Part Fibre Toxicol. 2010;7(1):39. doi: 10.1186/1743-8977-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu Q, Hondow NS, Krzemiński L, Brown AP, Jeuken LJC, Routledge MN. Mechanism of cellular uptake of genotoxic silica nanoparticles. Part Fibre Toxicol. 2012;9(1):29. doi: 10.1186/1743-8977-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer AT, Strozyk EA, Gorzelanny C, et al. Cytotoxicity of silica nanoparticles through exocytosis of von Willebrand factor and necrotic cell death in primary human endothelial cells. Biomaterials. 2011;32(33):8385–8393. doi: 10.1016/j.biomaterials.2011.07.078. [DOI] [PubMed] [Google Scholar]
- 7.Uboldi C, Giudetti G, Broggi F, Gilliland D, Ponti J, Rossi F. Amorphous silica nanoparticles do not induce cytotoxicity, cell transformation or genotoxicity in Balb/3T3 mouse fibroblasts. Mutat Res. 2012;745(1–2):11–20. doi: 10.1016/j.mrgentox.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Stern ST, Adiseshaiah PP, Crist RM. Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity. Part Fibre Toxicol. 2012;9(1):20. doi: 10.1186/1743-8977-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy J, Inkielewicz-Stępniak I, Corbalan JJ, Radomski MW. Mechanisms of toxicity of amorphous silica nanoparticles on human lung submucosal cells in vitro: protective effects of fisetin. Chem Res Toxicol. 2012;25(10):2227–2235. doi: 10.1021/tx3002884. [DOI] [PubMed] [Google Scholar]
- 10.Corbalan JJ, Medina C, Jacoby A, Malinski T, Radomski MW. Amorphous silica nanoparticles trigger nitric oxide/peroxynitrite imbalance in human endothelial cells: inflammatory and cytotoxic effects. Int J Nanomedicine. 2011;6:2821–2835. doi: 10.2147/IJN.S25071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slowing II, Vivero-Escoto JL, Wu C-W, Lin VS-Y. Mesoporous silica nanoparticles as controlled release drug delivery and gene transfection carriers. Adv Drug Deliv Rev. 2008;60(11):1278–1288. doi: 10.1016/j.addr.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Vivero-Escoto JL, Slowing II, Trewyn BG, Lin VS-Y. Mesoporous silica nanoparticles for intracellular controlled drug delivery. Small. 2010;6(18):1952–1967. doi: 10.1002/smll.200901789. [DOI] [PubMed] [Google Scholar]
- 13.Bharali DJ, Klejbor I, Stachowiak EK, et al. Organically modified silica nanoparticles: a nonviral vector for in vivo gene delivery and expression in the brain. Proc Natl Acad Sci U S A. 2005;102(32):11539–11544. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Xu C, Ow H. Commercial nanoparticles for stem cell labeling and tracking. Theranostics. 2013;3(8):544–560. doi: 10.7150/thno.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zielinska M, Sawosz E, Grodzik M, et al. Effect of heparan sulfate and gold nanoparticles on muscle development during embryogenesis. Int J Nanomedicine. 2011;6:3163–3172. doi: 10.2147/IJN.S26070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7(9):771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 17.Benezra M, Penate-Medina O, Zanzonico PB, et al. Multimodal silica nanoparticles are effective cancer-targeted probes in a model of human melanoma. J Clin Invest. 2011;121(7):2768–2780. doi: 10.1172/JCI45600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauly M, Daussin F, Burelle Y, et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol. 2012;181(2):583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Bibee KP, Cheng Y-J, Ching JK, et al. Rapamycin nanoparticles target defective autophagy in muscular dystrophy to enhance both strength and cardiac function. FASEB J. 2014;28(5):2047–2061. doi: 10.1096/fj.13-237388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böcking D, Wiltschka O, Niinimäki J, et al. Mesoporous silica nanoparticle-based substrates for cell directed delivery of Notch signalling modulators to control myoblast differentiation. Nanoscale. 2014;6(3):1490–1498. doi: 10.1039/c3nr04022d. [DOI] [PubMed] [Google Scholar]
- 21.Rivière C, Lecoeur C, Wilhelm C, et al. The MRI assessment of intraurethrally-delivered muscle precursor cells using anionic magnetic nanoparticles. Biomaterials. 2009;30(36):6920–6928. doi: 10.1016/j.biomaterials.2009.08.056. [DOI] [PubMed] [Google Scholar]
- 22.Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2010;341(1):66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139(4):641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132(12):2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 25.Hochreiter-Hufford AE, Lee CS, Kinchen JM, et al. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497(7448):263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vranic S, Boggetto N, Contremoulins V, et al. Deciphering the mechanisms of cellular uptake of engineered nanoparticles by accurate evaluation of internalization using imaging flow cytometry. Part Fibre Toxicol. 2013;10(1):2. doi: 10.1186/1743-8977-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Blaaderen A, Vrij A. Synthesis and characterization of colloidal dispersions of fluorescent, monodisperse silica spheres. Langmuir. 1992;8(12):2921–2931. [Google Scholar]
- 28.Pajcini KV, Pomerantz JH, Alkan O, Doyonnas R, Blau HM. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. J Cell Biol. 2008;180(5):1005–1019. doi: 10.1083/jcb.200707191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapero K, Fenaroli F, Lynch I, Cottell DC, Salvati A, Dawson KA. Time and space resolved uptake study of silica nanoparticles by human cells. Mol Biosyst. 2011;7(2):371–378. doi: 10.1039/c0mb00109k. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Liong M, Sherman S, et al. Mesoporous Silica Nanoparticles for Cancer Therapy: Energy-Dependent Cellular Uptake and Delivery of Paclitaxel to Cancer Cells. NanoBiotechnology. 2007;3(2):89–95. doi: 10.1007/s12030-008-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing X, He X, Peng J, Wang K, Tan W. Uptake of silica-coated nanoparticles by HeLa cells. J Nanosci Nanotechnol. 2005;5(10):1688–1693. doi: 10.1166/jnn.2005.199. [DOI] [PubMed] [Google Scholar]
- 32.Geiser M, Rothen-Rutishauser B, Kapp N, et al. Ultrafine particles cross cellular membranes by nonphagocytic mechanisms in lungs and in cultured cells. Environ Health Perspect. 2005;113(11):1555–1560. doi: 10.1289/ehp.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothen-Rutishauser BM, Schürch S, Haenni B, Kapp N, Gehr P. Interaction of fine particles and nanoparticles with red blood cells visualized with advanced microscopic techniques. Environ Sci Technol. 2006;40(14):4353–4359. doi: 10.1021/es0522635. [DOI] [PubMed] [Google Scholar]
- 34.Le Bihan O, Bonnafous P, Marak L, et al. Cryo-electron tomography of nanoparticle transmigration into liposome. J Struct Biol. 2009;168(3):419–425. doi: 10.1016/j.jsb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Sun X, Zhang G, Trewyn BG, Slowing II, Lin VS-Y. Interaction of mesoporous silica nanoparticles with human red blood cell membranes: size and surface effects. ACS Nano. 2011;5(2):1366–1375. doi: 10.1021/nn103077k. [DOI] [PubMed] [Google Scholar]
- 36.Geiser M, Kreyling WG. Deposition and biokinetics of inhaled nanoparticles. Part Fibre Toxicol. 2010;7(1):2. doi: 10.1186/1743-8977-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rancan F, Gao Q, Graf C, et al. Skin penetration and cellular uptake of amorphous silica nanoparticles with variable size, surface functionalization, and colloidal stability. ACS Nano. 2012;6(8):6829–6842. doi: 10.1021/nn301622h. [DOI] [PubMed] [Google Scholar]
- 38.Bergin IL, Witzmann FA. Nanoparticle toxicity by the gastrointestinal route: evidence and knowledge gaps. Int J Biomed Nanosci Nanotechnol. 2013;3:1–2. doi: 10.1504/IJBNN.2013.054515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jokerst JV, Thangaraj M, Kempen PJ, Sinclair R, Gambhir SS. Photoacoustic imaging of mesenchymal stem cells in living mice via silica-coated gold nanorods. ACS Nano. 2012;6(7):5920–5930. doi: 10.1021/nn302042y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose S, Prevoteau A, Elzière P, Hourdet D, Marcellan A, Leibler L. Nanoparticle solutions as adhesives for gels and biological tissues. Nature. 2014;505(7483):382–385. doi: 10.1038/nature12806. [DOI] [PubMed] [Google Scholar]
- 41.Besic Gyenge E, Darphin X, Wirth A, et al. Uptake and fate of surface modified silica nanoparticles in head and neck squamous cell carcinoma. J Nanobiotechnology. 2011;9(1):32. doi: 10.1186/1477-3155-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt LC, Upadhyay A, Jazayeri JA, Tudor EM, White JD. Caspase-3, myogenic transcription factors and cell cycle inhibitors are regulated by leukemia inhibitory factor to mediate inhibition of myogenic differentiation. Skelet Muscle. 2011;1(1):17. doi: 10.1186/2044-5040-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, Yun H-S, Kim S-H. The comparative effects of mesoporous silica nanoparticles and colloidal silica on inflammation and apoptosis. Biomaterials. 2011;32(35):9434–9443. doi: 10.1016/j.biomaterials.2011.08.042. [DOI] [PubMed] [Google Scholar]
- 44.Lu X, Qian J, Zhou H, et al. In vitro cytotoxicity and induction of apoptosis by silica nanoparticles in human HepG2 hepatoma cells. Int J Nanomedicine. 2011;6:1889–1901. doi: 10.2147/IJN.S24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan J, Yu Y, Li Y, Yu Y, Sun Z. Cardiovascular toxicity evaluation of silica nanoparticles in endothelial cells and zebrafish model. Biomaterials. 2013;34(23):5853–5862. doi: 10.1016/j.biomaterials.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 46.Singh K, Dilworth FJ. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013;280(17):3991–4003. doi: 10.1111/febs.12188. [DOI] [PubMed] [Google Scholar]
- 47.Krampera M, Pizzolo G, Aprili G, Franchini M. Mesenchymal stem cells for bone, cartilage, tendon and skeletal muscle repair. Bone. 2006;39(4):678–683. doi: 10.1016/j.bone.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 48.Meregalli M, Farini A, Belicchi M, et al. Perspectives of stem cell therapy in Duchenne muscular dystrophy. FEBS J. 2013;280(17):4251–4262. doi: 10.1111/febs.12083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cytotoxicity of NPs in C2C12 myoblasts.
Notes: Twenty-four hours after seeding at 15,000 cells/cm2, cells were exposed to various doses of NPs for 1 hour in Dulbecco’s Modified Eagle’s Medium. Cell viability was measured 16 hours after treatment, using the WST-1 assay. The toxicity threshold is observed for a NP concentration of 9 μg/cm2. Data are the percentages of surviving cells relative to the control reported as the mean ± standard error of the mean of three independent experiments.
Abbreviation: NP, silica nanoparticle.
Transmission electron microscopy characterization of 50 nm NPs.
Notes: Transmission electron microscopy images of NPs suspended in water (40 μg/mL). Scale bar, 100 nm. A histogram showed the size distribution of 100 NPs measured using the ImageJ software. The mean diameter was 49.5±3.9 nm (standard error of the mean).
Abbreviation: NP, silica nanoparticle.
Dynamic light scattering curves of silica nanoparticles in water and in Dulbecco’s Modified Eagle’s Medium.
Abbreviations: DMEM, Dulbecco’s Modified Eagle’s Medium; NPs, silica nanoparticles.
NP internalization as a function of cell confluence.
Notes: Cells were treated with NPs during 30 minutes at 3 μg/cm2 in Dulbecco’s Modified Eagle’s Medium, 24 hours after seeding at three concentrations: 2,000 (C1), 15,000 (C2), and 50,000 (C3) cells/cm2, corresponding to about 50%, 70%, and ~100% confluence, respectively. Quantification of NP internalization was performed by flow cytometry after the addition of Trypan blue. Results are expressed as mean cell fluorescence intensity (arbitrary unit) ± standard error of the mean; n=3.
Abbreviation: NP, silica nanoparticle.
Table S1.
Physicochemical characteristics of NPs
| Characteristic | H2O | Dulbecco’s Modified Eagle’s Medium |
|---|---|---|
| Zeta potential, mV | −35.6 | −31.2 |
| Hydrodynamic radius, nm | 81.8 | 122.9 |
Notes: NPs were characterized for their hydrodynamic radius and zeta potential suspended in water and Dulbecco’s Modified Eagle’s Medium. Dynamic light scattering and zeta potential values were measured by a zetasiser 300 HS (Malvern) and Vasco granulometer (CordouanTechnologies). Size estimation using the cumulant method shows a slight destabilization of NPs in Dulbecco’s Modified Eagle’s Medium (122.9 nm) compared with H2O (81.8 nm).
Abbreviation: NP, silica nanoparticle.