Background: Miniature chromosome maintenance 7 (MCM7) is a pivotal DNA replication licensing factor.
Results: MCM7 interacts with splicing factor 3B, subunit 3 (SF3B3). SF3B3/MCM7 binding is required for RNA splicing of several growth factor receptors and reporter gene oxytocin-enhanced green fluorescence protein.
Conclusion: MCM7 is an essential component for RNA splicing.
Significance: MCM7 plays significant role in RNA splicing through its interaction with SF3B3.
Keywords: DNA Replication, Epigenetics, Oncogene, RNA, Spliceosome
Abstract
Miniature chromosome maintenance 7 (MCM7) is an essential component of DNA replication licensing complex. Recent studies indicate that MCM7 is amplified and overexpressed in a variety of human malignancies. In this report, we show that MCM7 binds SF3B3. The binding motif is located in the N terminus (amino acids 221–248) of MCM7. Knockdown of MCM7 or SF3B3 significantly increased unspliced RNA of epidermal growth factor receptor, platelet-derived growth factor receptor, and c-Met. A dramatic drop of reporter gene expression of the oxytocin exon 1-intron-exon 2-EGFP construct was also identified in SF3B3 and MCM7 knockdown PC3 and DU145 cells. The MCM7 or SF3B3 depleted cell extract failed to splice reporter RNA in in vitro RNA splicing analyses. Knockdown of SF3B3 and MCM7 leads to an increase of cell death of both PC3 and DU145 cells. Such cell death induction is partially rescued by expressing spliced c-Met. To our knowledge, this is the first report suggesting that MCM7 is a critical RNA splicing factor, thus giving significant new insight into the oncogenic activity of this protein.
Introduction
Miniature chromosome maintenance (MCM)2 proteins were initially identified from autonomously replicating sequence in Saccharomyces cerevisiae. Mutations of some of these proteins such as MCM7 or MCM3 in yeast result in loss of the large chunk of yeast chromosomes. MCM7 cDNA encodes a 543-amino acid protein and is ubiquitously expressed in all tissues. Initiation of DNA replication is a complex process involving the concerted action of many proteins. A large body of studies indicate that MCM7 is a critical component of DNA replication licensing complex in the yeast and Xenopus (1–4). Some studies suggest that the MCM4, MCM6, and MCM7 complex contains DNA helicase activity (5, 6). DNA replication licensing complex is multimeric and phase-specific. In the yeast, DNA replication licensing proteins such as MCM2–7 and several replication origin binding proteins such as CDC6, germinin, and CDT1 form a DNA replication licensing complex in G1 phase to enable DNA replication and to promote cell cycle entry into the S phase. Such a complex, however, dissipates in the S, G2, and M phases to prevent refiring of DNA replication and thus protect the integrity of genomes. There is little interest in the MCM complex as target for oncogenic or tumor suppressor pathway until the links of MCM7 overexpression and amplification to several human malignancies were found (7–10).
A recent study has suggested that MCM7 is overexpressed and amplified in a variety of human malignancies (11). MCM7 genome sequence contains a cluster of microRNA that has been shown to down-regulate the expression of several tumor suppressors including p21, E2F1, BIM, and pTEN (10–12). The oncogenic potential of both MCM7 and its embedded microRNA had been demonstrated vigorously in in vitro experiments and in animal models and appears to cooperate with each other in initiating cancers (13, 14). Interestingly, MCM7 protein also serves as a critical target for oncogenic signaling pathways such androgen receptor signaling or cell metabolism or tumor suppressor pathways such as integrin α7, protein kinase C, or Rb signaling (15–18). A recent study suggests that MCM7 serves as a co-transcription factor for androgen receptor because androgen receptor transcription activity is MCM7-dependent (7). In this study, we showed that MCM7 binds splicing factor SF3B3 and is essential for splicing of epidermal growth factor receptor (EGFR), c-Met, and platelet-derived growth factor (FDGFR). Thus, MCM7 appears to be a versatile molecule involving in multiple critical functions for cell growth and survival.
MATERIALS AND METHODS
Plasmid Construction and Quantitative RT-PCR
For construction of pCDNA4-MCM7, primers (TGCATAAGCTTACGTTTCGCGCCAATTTCGGTT/TAGTTCTAGAGACAAAAGTGATCCGTGTCCGGGA) corresponding to the sequence encompassing the full-length MCM7 were used in a PCR using template of pCR-MCM7 vector. The PCR product was restricted with HindIII and XbaI and ligated into a similarly restricted pCDNA4-TO vector. To construct mutant of MCM7 that contains amino acids 1 and 221–248, mutagenesis PCR was performed using primers GACTCAGATGCTTAAGGCGCGGCCGCACGGCCCTCGGCAGCGATGGCATATCTGCAGACACGGGGCTCC/TATCTGCAGACACGGGGCTCCAGATTCATCAAATTCCAGGAGATGAAGATGCAAGAACATAGTGATCAGGTGCCTGTGGGAAAT. The PCR product was then restricted with AflII and XbaI and ligated into similarly restricted pCDNA4-TO vector to produce pCDNA4-ΔMCM7aa1,221–248. For construction of pET28a-SF3B3 vector, a mutagenic primer set (AACTGCTAGCATGACTGAGCAGATGACCCT and AACTGCGGCCGCTCTAGCGTGTGCCAATGGTCA) was designed to create two restriction sites (NheI and NotI). A PCR was performed on pCMV-SF3B3. The PCR product was restricted with NheI and NotI and ligated into a similarly restricted pET28a vector to generate a His tag-SF3B3 (aa 2–317). To construct pCMV-SF3B3, PCR product on SF3B3 cDNA using primers cagtgccggatccctcgtcgctgcagcgacacac/ttatttgggtaccctctgccataaacttctagcgtgtg was digested with BamHI and KpnI. The digested product was ligated into similarly digested pCMVscript. pSG-c-Met construction was previously described (26). For quantitative RT-PCR, 1 μg of total RNA treated with DNase I was reversed transcribed by SuperscriptII using random hexamer to synthesize the first strand cDNA following manufacturer's manual. PCR was then performed by the primers listed in Table 1 using following conditions: 94 °C for 30 s and then 30 cycles of 94 °C for 5 s, 61 °C for 10 s, and 72 °C for 1 min. The real time PCR was carried out in Eppendorf Realplex2 master cycler.
TABLE 1.
Primer sequences for real time RT-PCR
| Gene | Intron/exon | Primer name | Sequence |
|---|---|---|---|
| EGFR | Intron 4 | EGFR intronF | GCTGCCCTAGGAGGATATTTG |
| EGFR | Intron 4 | EGFR intronR | TTAAAGGGCCATGTTCCCTGG |
| EGFR | Exon 3 | EGFR splicingF | TGTGCATTTGCTGTGGGTTCC |
| EGFR | Exon 4 | EGFR splicingR | TGATGCCTTCCTCTTCTTGCC |
| PDGFR | Intron 12 | PDGFR intronF | AGAAGGGAGTGCCCAAGTCTG |
| PDGFR | Intron 12 | PDGFR intronR | ACTGTGAGGATCACATGAGC |
| PDGFR | Exon 11 | PDGFR splicingF | AGCTGATCCGTGCTAAGGAAG |
| PDGFR | Exon 12 | PDGFR splicingR | TGAGCCATGGTGATCATCGAC |
| c-Met | Intron 11 | c-Met intronF | GGCCAGAAATGGGAGTTTCTC |
| c-Met | Intron 11 | c-Met intronR | GAGAGGATCCATGCTGAGCTG |
| c-Met | Exon 8 | c-Met splicingF | CTCCTTGGAAATGAGAGCTGC |
| c-Met | Exon 10 | c-Met splicingR | CTGTATTGTGTTGTCCCGTGG |
Construction of Inducible MCM7 and ΔMCM7 Expression in PC-3 Cell Line
The plasmid pCDNA4-ΔMCM7aa1,221–248 and pCDNA6 were then co-transfected into PC3 cells with pcDNA6. Transfected cells were selected with Blasticidin (500 μg/ml) and Zeocin (1 μg/ml) (Invitrogen). Clones were expanded and tested for inducible MCM7 expression by exposure to 5 μg/ml tetracycline and by Western blot analysis with antibody specific for MCM7 and β-actin. For SF3B3 knockdown assays, siRNA specific for SF3B3 (5′-rCrArC rCrArU rUrUrC rCrUrC rUrGrC rCrArU rCrUrG rCrUrG rCrUrU-3′/5′-rGrCrA rGrCrA rGrArU rGrGrC rArGrA rGrGrA rArArU rGrGT G-3′) or for siMCM7 (5′-rUrUrU rArUrU rUrArC rCrArC rUrUrC rCrCrU rCrUrC rCrUrU rGrUrA-3′/5′-rCrArA rGrGrA rGrArG rGrGrA rArGrU rGrGrU rArArA rUrAA A-3′) or scramble siRNA (5′-UAA UGUAUUGGAACGCAUAUU-3′/5′-UAUGCGUUCCAAUACAUUA-3′) was transfected into cultured cells using Lipofectamine 2000 (Invitrogen). The detailed procedure followed the manufacturer's manual.
Yeast Two-hybrid Analysis
The yeast-competent cell preparation was previously described (15, 21, 27–30). One hundred microliters of freshly prepared competent AH109 cells were mixed with plasmid DNA (0.25–0.50 μg) plus 0.5 μg of DNA from prostate yeast two-hybrid cDNA library constructed in pACT2 in 0.5 ml of polyethylene glycol/LiAc, incubated at 30 °C for 30 min. Following this initial incubation with plasmid DNA, the cell solution was combined with 20 μl of DMSO and subjected to 15 min incubation at 42 °C. The cells were pelleted, resuspended in 1 ml of YPD medium, and shaken at 30 °C for 40 min. The transformed cells were then pelleted, resuspended in 0.5 ml of 0.9% NaCl, and plated onto the appropriate S.D. agar plate. The transformants were first plated on low and medium stringency plates of SD−Leu/−Trp and SD−Leu/−Trp/−His, respectively. The grown colonies were subjected to the β-galactosidase assay as previously described (27) and allowed to grow further in the high stringency plate (SD−Ade/−His/−Leu/−Trp).
Immunoprecipitation
Protein extracts of PC3 cells or DU145 cells were incubated with MCM7 (mouse monoclonal, 1:400) or SF3B3 (1:500, goat polyclonal) antibodies for 16 h and then with protein G-Sepharose for 3 h. The complex was washed five times with radioimmune precipitation assay buffer, and the bound proteins were eluted with SDS-PAGE sample buffer. The bound SF3B3 or MCM7 was electrophoresed in 10% SDS-PAGE and immunoblotted with anti-SF3B3 antibodies or MCM7 antibodies (1:2000).
GST Fusion Proteins Pull Down to Examine SF3B3/MCM7 Binding
The Escherichia coli cells harboring pGST-MCM7 mutants or pGST were grown in 100 ml of Luria-Bertani medium supplemented with ampicillin (100 μg/ml) overnight and induced by isopropyl-l-thio-β-d-galactopyranoside (final concentration of 1 mm) for 3 h. The cells were then pelleted, resuspended in 1× PBS, and sonicated for 2 min. The proteins were solubilized in 1% Triton X-100. The supernatant was collected after centrifugation at 15,000 × g for 5 min. The GST, GST-MCM7c, GST-MCM7m, GST-MCM7n, and other MCM7 mutant fusion proteins were purified through a glutathione-Sepharose 4B column (Amersham Bioscience). The E. coli-produced His tag-SF3B3 was purified through a histidine column. The purified His tag-SF3B3 was then incubated with GST fusion protein-packed glutathione-Sepharose 4B at 4 °C for 2 h. The column was spun at 3,000 × g at room temperature for 1 min and further washed twice with PBS. The proteins were eluted from the column with 40 μl of SDS-PAGE gel sample loading dye. SDS-PAGE and Western blot analyses were subsequently conducted.
In Vitro Splicing Assay
PCR was performed using primers 5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGATAAAAAGGCCAGGCCGGAG-3′/5′-GCTGCAGCAGAGGCCCAAGAC-3′, to generate a OXT exon 1-intron-exon 2 DNA template with T7 promoter attaching to the 5′ end (T7-OXTex1-int1-ex2). In vitro transcription was performed with T7-OXTex1-int1-ex2 template using Megascript kit from Ambion, Inc. (Grand island, NY). The IVT product was then purified using RNeasy column. In vitro splicing reactions were carried out as described previously (31). Briefly, 100 ng of RNA was incubated in 80 mm KCl, KAc, or KGlu; 2 mm MgCl2 or MgAc2; 1 mm ATP; 5 mm creatine phosphate; and 0.2 nm T7-OXTex1-int1-ex2 pre-mRNA template, with a volume of 40% cell nucleus extract at 30 °C for 30 min. The reactions were quenched with 10 volumes of a splicing stop buffer (100 mm Tris, pH 7.5, 10 mm EDTA, 1% SDS, 150 mm NaCl, 300 mm NaAc). RNeasy column (Qiagen) were used to purify the RNAs. Spliced RNA was visualized in an Agilent 2100 Bioanalyzer using an RNA chip.
RESULTS
To investigate what proteins regulate the function of MCM7 and how such interaction impacts the function of MCM7, we performed a yeast two-hybrid screening using GAL4 DNA-binding domain-MCM7 fusion proteins, utilizing MATCHMAKER system 3 from CLONETECH, INC. Three BD-MCM7s were constructed (Fig. 1A). All were demonstrated with proper expression in the yeast (data not shown). Using pBD-MCM7, we have identified 24 positive colonies after three rounds of metabolic screening of a prostate yeast two-hybrid cDNA library. These colonies were subsequently isolated. After several restriction enzyme digestions, several redundant clones were eliminated. Three unique clones were identified and sequenced. One of these clones contains cDNA encoding splicing factor 3B3.
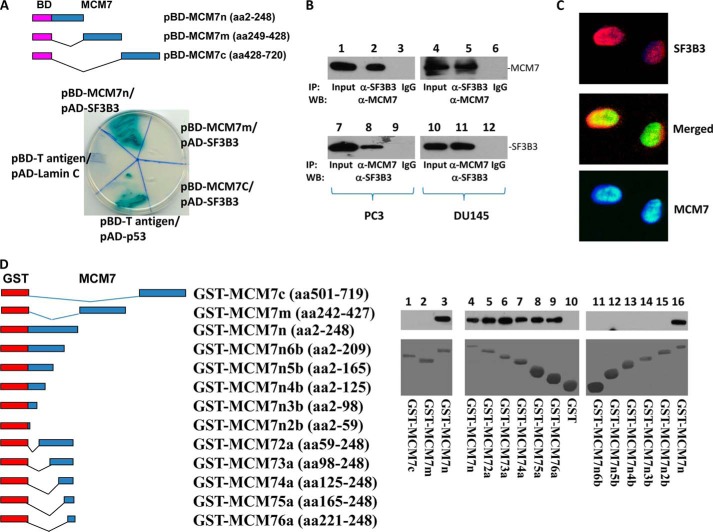
FIGURE 1.
MCM7 interacts with SF3B3. A, yeast two-hybrid analysis of MCM7 N terminus binding with SF3B3. Top panel, schematic diagram of DNA-binding domain (BD)-MCM7 fragment constructs. Bottom panel, binding of BD-MCM7 fragments with activation domain (AD)-SF3B3. Co-transfection of pBD-T-antigen and pAD-Lamin C is negative control, whereas co-transfection of pBD-T-antigen and pAD-p53 is a positive control. B, co-immunoprecipitation of MCM7 and SF3B3 proteins. Protein extracts from PC3 and DU145 cells were immunoprecipitated by the indicated antibodies and immunoblotted with antibodies specific for MCM7 (top panel) or SF3B3 (bottom panel). C, SF3B3 co-localized with MCM7. PC3 cells were stained with antibodies against MCM7 (mouse monoclonal and FITC-conjugated donkey anti-mouse antibodies) and SF3B3 (rabbit polyclonal and Cy5-conjugated donkey anti-rabbit antibodies). D, MCM7 binds with SF3B3 in cell free system. Left panel, schematic diagram of GST-MCM7 constructs. Right panel, binding of GST-MCM7 fragments with His tag-SF3B3 generated from E. coli. IP, immunoprecipitation; WB, Western blot.
To validate the yeast two-hybrid screening results, pAD-SF3B3 and pBD-MCM7 were co-transfected into yeast AH109 cells, grown in high stringency medium and tested for α-galactosidase activity. pBD-MCM7n (N terminus) showed positive galactosidase activity, whereas the C terminus and midsegment of MCM7 were negative, suggesting that the SF3B3 binding activity is mediated by a region located in the N terminus of MCM7 (Fig. 1A). SF3B3 is abundantly expressed in PC3 and DU145 cell lines (Fig. 1B). To verify MCM7/SF3B3 interaction, an in vivo MCM7-SF3B3 binding analysis was performed using protein extracts of PC3 and DU145 cells. As shown in Fig. 1B, co-immunoprecipitation of MCM7 and SF3B3 was readily apparent in both cell lines. To visualize whether MCM7 and SF3B3 co-localize in these cells, double immunofluorescence staining using antibodies against MCM7 and SF3B3 were performed in PC3 cells. As demonstrated in Fig. 1C, MCM7 and a significant amount of SF3B3 were co-localized in the nuclei of PC3 cells. Similar co-localization results were obtained with DU145 cells (data not shown).
To validate the interaction between MCM7 N terminus and SF3B3 in vitro, a fragment of 247 amino acids from the N terminus of MCM7 was constructed into pGEX-5T to create a GST-MCM7n fusion protein. The results of the in vitro binding assays with recombinant His tag-SF3B3 indicate that GST-MCM7n but not GST-MCM7m nor GST-MCM7c binds with SF3B3 in cell free system (Fig. 1D). These results indicate that the interaction between MCM7 and SF3B3 is direct and does not require “bridge protein” in their interaction. A series of deletion mutants of GST-MCM7n were constructed to identify the motifs that are required to interact with SF3B3. A 30-amino acid sequence corresponds to positions 221–248 of MCM7 was found crucial for MCM7 binding with SF3B3, because the fusion proteins deleted of this sequence did not bind with SF3B3, whereas all proteins containing this sequence bound with SF3B3 (Fig. 1D).
SF3B3 is a critical splicing factor that converts pre-mRNA to mRNA by forming a spliceosome with several other factors. Binding of MCM7 with SF3B3 suggests that MCM7 is a part of the spliceosome. To investigate the impact of MCM7 on splicing activity of RNA, we chose to investigate splicing of several important growth factors that might associate with MCM7 oncogenic activity: EGFR, c-Met, and PDGFR, through quantitative RT-PCR of both pre-mRNA and mRNA of these genes. To analyze the splicing of EGFR, RT-PCR was performed using primers corresponding to exons 3 and 4 of EGFR to examine the spliced RNA and primers corresponding to intron 4 of this gene to examine the unspliced RNA. As shown in Fig. 2A, when SF3B3 was knocked down by siRNA, 2.6-fold increase (p < 0.05) of unspliced EGFR RNA in PC3 cells and 1.81-fold (p < 0.05) in DU145 cells were found. Knockdown of MCM7 produced 2.3-fold increase (p < 0.05) of unspliced EGFR in PC3 cells and 1.81-fold (p < 0.05) in DU145 cells, respectively, extremely similar to those identified in SF3B3 knockdown. When we examined PDGFR splicing with primers corresponding to exons 11 and 12 and intron 12 to detect spliced and unspliced RNA, respectively, knockdown of SF3B3 produced 2.4-fold increase (p < 0.05) of unspliced RNA in PC3 cells and 2-fold increase (p < 0.05) in DU145 cells in comparison with scramble controls. These are comparable with 2.1-fold (p < 0.05) and 1.9-fold (p < 0.05) increases of PDGFR unspliced RNA in MCM7 knockdown PC3 and DU145 cells, respectively. Knockdown of SF3B3 and MCM7 also produced similar increase of unspliced c-Met RNA in PC3 and DU145 cells: 2.3-fold (p < 0.05) for SF3B3 versus 1.8 (p < 0.05) for MCM7 in PC3 cells and 2.8-fold (p < 0.05) for SF3B3 versus 2.4 (p < 0.05) for MCM7. These results indicate that MCM7 is an important splicing factor for these growth factor receptors.
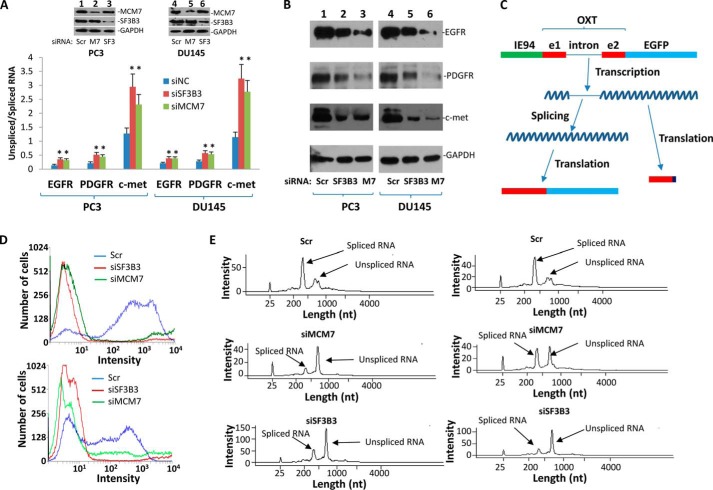
FIGURE 2.
MCM7 is required for efficient RNA splicing for EGFR, PDGFR, and c-Met. A, knockdown of MCM7 or SF3B3 increased unspliced RNA for EGFR, PDGFR, and c-Met. *, p < 0.05 in comparison with controls. PC3 or DU145 cells (1 × 106) were transfected with 80 pmol of siMCM7, siSF3B3, or siScramble (Scr). These cells were harvested for analysis 24 h after the transfection. B, knockdown of SF3B3 and MCM7 decreased protein expression of EGFR, PDGFR, and c-Met as of A. C, schematic diagram of oxytocin-EGFP reporter gene system. D, knockdown of SF3B3 or MCM7 drastically reduced oxytocin-EGFP reporter gene activity. Top panel, PC3 cells. Bottom panel, DU145 cells. E, MCM7 or SF3B3 is required for oxytocin-EGFP RNA splicing in vitro. RNA template of oxytocin-EGFP was generated by in vitro translation under T7 promoter from a PCR product. The transcription was then incubated with nuclear protein extracts of PC3 (left) or DU145 (right) cells treated with siRNA specific for MCM7, SF3B3, or scramble control. The spliced RNA was analyzed through Agilent bioanalyzer 2100.
To examine the impact of increased unspliced RNA on protein expression of these genes, immunoblot analyses were performed to identify the protein expression levels of EGFR, PDGFR, and c-Met upon treatment of PC3 and DU145 cells with siRNA specific for SF3B3 or MCM7. As shown in Fig. 2B, knockdown of MCM7 or SF3B3 had significantly reduced the protein levels of all three proteins examined, suggesting that reducing splicing of EGFR, PDGFR, and c-Met reduced proper translation of the RNA encoding for these proteins. To investigate whether the impact of MCM7 or SF3B3 on splicing also occur in on other genes, a reporter gene system utilizing oxytocin genome containing exon 1, intron 1, and exon 2 was constructed by ligating the genome sequence into pEGFP vector to create a fusion transcript that links EGFP coding region with a properly spliced oxytocin. If oxytocin pre-mRNA is not spliced, translation ribosome will migrate into intron 1 of oxytocin. A stop codon in the intron will terminate the translation. Thus, only spliced oxytocin-EGFP transcript would express the fusion protein that elicits green fluorescence (Fig. 2C). To establish the reporter system, this construct was transfected into PC3 and DU145 cells to produce stable cell lines expressing the fusion protein. As shown in Fig. 2D, when these cells were transfected with siRNA specific for SF3B3 or MCM7, a dramatic reduction of cells expressing green fluorescence signals was identified in both PC3 and DU145 cells, in comparison with cells treated with scramble siRNA. To verify whether reduction of green fluorescence signal is due to an increase of unspliced oxytocin-EGFP pre-mRNA, in vitro splicing assays were performed on an oxytocin-EGFP unspliced transcript generated from in vitro transcription under T7 promoter. Our results indicated that nuclear extracts from MCM7 or SF3B3 knockdown produced little splicing activity for oxytocin-EGFP in vitro transcribed RNA template, whereas nuclear extracts from scramble control spliced most of the pre-mRNA from the same transcription (Fig. 2E). These experiments clearly suggest that MCM7 is a co-splicing factor, and RNA splicing appears to be dependent on MCM7 protein.
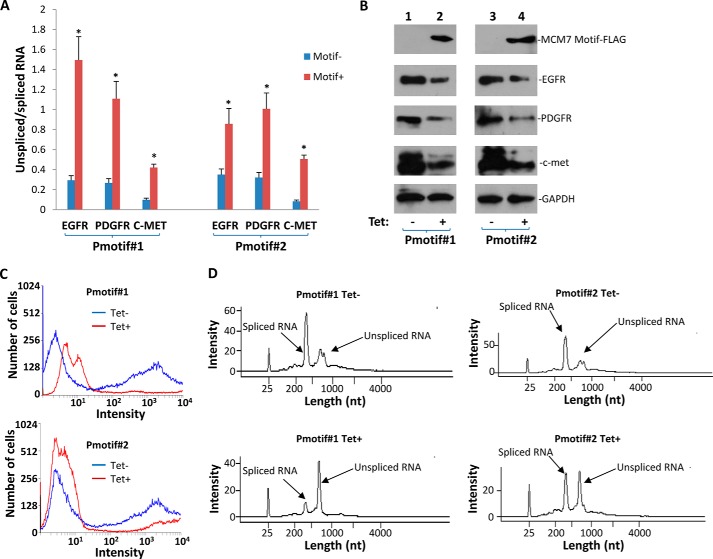
To investigate whether interaction between MCM7 and SF3B3 is essential for MCM7-mediated RNA splicing activity, a mutant MCM7 that contains only the binding motif for SF3B3 (aa 221–248) was constructed and ligated in frame with FLAG TAG into pCDNA4-FLAG vector to create pCDNA4-ΔMCM7aa1,221–248. This construct expresses an interference peptide that impedes the binding between wild type MCM7 and SF3B3. This construct was co-transfected with pCDNA6 into PC3 cells. Two clones that showed expression of ΔMCM7aa1,221–248 were selected for further analyses. As shown in Fig. 3A, expression of the binding interference peptide in both clones significantly increased the nonspliced RNAs for EGFR, PDGFR, and c-Met: 2.4–5.1-fold for EGFR, 3.2–4.3-fold for PDGFR, and 4.3–6.1-fold for c-Met. Significant decreases of EGFR, PDGFR, and c-Met protein levels were also detected in these cells (Fig. 3B). The binding interference between wild type MCM7 and SF3B3 also similarly blocked the splicing of reporter gene expression of oxytocin-EGFP (Fig. 3C). Such blockade was verified in in vitro splicing analyses (Fig. 3D). These results suggest that MCM7 splicing activity is dependent on its interaction with SF3B3.
FIGURE 3.
Efficient RNA splicing of RNA of EGFR, PDGFR, and c-Met requires interaction between MCM7 and SF3B3. A, two clones (Pmotif#1 and Pmotif#2) of PC3 cells transformed with pCDNA4-ΔMCM7aa1,221–248/pCDNA6 were induced to express the MCM7/SF3B3 binding interference peptide with 5 μg/ml tetracycline. Quantitative RT-PCR was performed to quantify the ratio of unspliced/spliced RNA of EGFR, PDGFR, and c-met 24 h after the induction. *, p < 0.05 in comparison with controls. B, expression of ΔMCM7aa1,221–248 decreased protein level of EGFR, PDGFR, and c-Met. C, expression of ΔMCM7aa1,221–248 decreased oxytocin-EGFP reporter gene expression. D, expression of ΔMCM7aa1,221–248 decreased splicing of oxytocin-EGFP RNA in vitro.
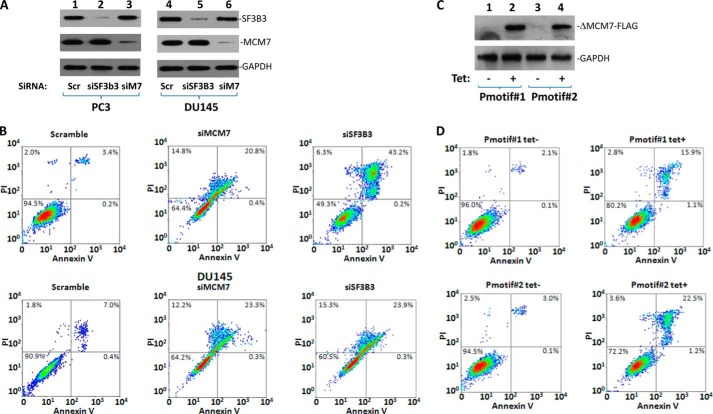
To investigate the impact of MCM7-mediated RNA splicing on cell survival, cell death analyses were performed on cells with knockdown MCM7. As shown in Fig. 4 and Table 2, knockdown of MCM7 through MCM7-specific siRNA produced a 6.4-fold (p < 0.01) increase of PC3 cell death and 3.9-fold (p < 0.01) DU145 cell death in comparison with scramble controls. Knockdown of SF3B3 generated 8.9-fold (p < 0.01) increase of PC3 cell death and 4.3-fold (p < 0.01) DU145 cell death. Because MCM7 is a versatile molecule performing multiple functions, it is necessary to distinguish the impact of MCM7-mediated RNA splicing from its other activities. To investigate the RNA splicing specific impact of MCM7, cell death analyses were performed on cell lines with inducible expression of SF3B3 binding interference peptide (Pmotif#1 and Pmotif#2). As shown in Fig. 4 (C and D), induced expression of MCM7/SF3B3 binding interference peptide produced an average 4.9-fold (p < 0.01) increase of cell death in two different clones, in comparison with the uninduced controls. These results suggest that most cell deaths generated by MCM7 knockdown are due to a decrease of RNA splicing. These experiments suggest that maintaining proper splicing of critical growth factor receptors is critically important for cell survival. The presence of MCM7 is essential for these activities.
FIGURE 4.
Knockdown of MCM7 or SF3B3 or interference of MCM7/SF3B3 interaction resulted in cell death. A, immunoblotting of SF3B3, MCM7, and GAPDH in PC3 or DU145 cells treated with indicated siRNA. PC3 or DU145 cells (1 × 106) were transfected with 80 pmol of siMCM7, siSF3B3, or siScramble (Scr). These cells were harvested for analysis 24 h after the transfection. B, annexin V and propidium iodide staining of PC3 or DU145 cells treated with siRNA as in A to detect apoptosis and necrotic cell death. C, immunoblotting of ΔMCM7aa1,221–248-FLAG and GAPDH in Pmotif#1 and Pmotif#2 cells induced with or without tetracycline (5 μg/ml) for 24 h using antibodies specific for FLAG and GAPDH. D, annexin V and propidium iodide staining of Pmotif#1 and Pmotif#2 cells as in C to detect apoptosis and necrotic cell death.
TABLE 2.
MCM7/SF3B3-mediated RNA splicing is essential for cell survival
| Cells | Treatment | Cell death |
|---|---|---|
| % | ||
| PC3 | Scr | 5.7 ± 0.3 |
| PC3 | siSF3B3 | 50.1 ± 1.9 |
| PC3 | siMCM7 | 37.9 ± 0.8 |
| DU145 | Scr | 9.1 ± 0.5 |
| DU145 | siSF3B3 | 40.1 ± 2.2 |
| DU145 | siMCM7 | 36.1 ± 1.4 |
| Pmotif#1 | Tet− | 4.4 ± 0.7 |
| Pmotif#1 | Tet+ | 20.5 ± 0.9 |
| Pmotif#2 | Tet− | 5.8 ± 0.5 |
| Pmotif#2 | Tet+ | 27.7 ± 1.1 |
DISCUSSION
MCM7 has been long considered an essential component of DNA replication licensing. Together with CDT1, Geminin, and MCM1–6 proteins, MCM7 facilitate forming the DNA replication licensing complex (6, 19, 20). Recent study also showed that MCM7 is a co-transcription factor for androgen receptor (21). Our analysis, however, suggest that MCM7 is an essential component for RNA splicing through its interaction with SF3B3. Several lines of evidence support that MCM7 binds SF3B3 and activates its RNA splicing activity. First, yeast two-hybrid DNA-binding domain and activation domain interaction demonstrated binding of MCM7 N terminus with SF3B3. Such interaction was confirmed in another type of eukaryotic cells (PC3 and DU145) using a co-immunoprecipitation approach. Furthermore, MCM7 and SF3B3 were found co-localized in both PC3 and DU145 cells. Second, in vitro cell free binding was identified using recombinant MCM7 and SF3B3 generated from E. coli. This finding indicates that the binding between MCM7 and SF3B3 is direct and requires no bridge protein. It also suggests that the binding between MCM7 and SF3B3 does not require post-translation modification of the proteins because there is minimal post-translational protein modification in E. coli. Third, a peptide that mimics the SF3B3 binding motif in MCM7 effectively blocked RNA splicing for EGFR, PDGFR, and c-Met, as well as reporter transcript oxytocin-EGFP. Thus, we conclude that MCM7 is an important splicing factor for RNA in eukaryotic cells. Its RNA splicing activity is mediated by its binding with SF3B3.
RNA splicing required multiple factors including SF3A, SF3B, and 12SRNA. These components form nuclear ribonucleoproteins complex (U2 snRNP) (22, 23). SF3B3 was also identified as a co-transcription factor and may be implicated in having a role in chromatin modification and DNA repair (24, 25). However, the exact mechanism of SF3B3-mediated RNA splicing is not clear. Our study suggests that MCM7 is essential for RNA splicing, because evidence of aborted RNA splicing when MCM7 was removed from the splicing complex. Because the magnitude of increase of unspliced RNAs is very similar between cells with MCM7 knockdown or SF3B3 knockdown, we speculate that the function of SF3B3 for RNA splicing is dependent on MCM7 binding. MCM7 functions to activate SF3B3 RNA splicing activity. Because MCM7 binds other DNA replication licensing factors in the chromatin, it is likely that the binding MCM7 with SF3B3 may also generate a large “DNA replication-RNA splicing” super complex that facilitates, multitasks, and integrates DNA replication, RNA transcription, and RNA splicing activities simultaneously. This hypothesis is also consistent with our previous finding that MCM7 binds with androgen receptor and facilitates its transcription activity and that transcription factor AR facilitates DNA replication licensing.
To our knowledge, this is the first report suggesting that a DNA replication licensing protein possesses RNA splicing activity. MCM7 genome amplification was documented in prostate cancer and gastric cancer (7, 10). Overexpression of MCM7 is associated with the high level of cell proliferation and the more aggressive behavior of a variety of human malignancies (11). The RNA splicing activity of MCM7, particularly its activity on several critical growth factor receptor RNAs, may play significant roles in producing the pro-growth phenotype that is associated with MCM7 amplification and overexpression. MCM7 appears a pivotal and an extremely versatile protein playing multiple roles in signal transduction, DNA replication, transcription, and RNA splicing. Thus, targeting at MCM7 in gene therapy may be a promising approach to treat human malignancies.
This work was supported by National Cancer Institute Grant RO1 CA098249 (to J.-H. L.) and American Cancer Society Grant RSG-08-137-01-CNE (to Y. P. Y.).
- MCM
- miniature chromosome maintenance
- aa
- amino acids
- EGFR
- epidermal growth factor receptor
- PDGFR
- platelet-derived growth factor receptor.
REFERENCES
- 1. Kearsey S. E., Maiorano D., Holmes E. C., Todorov I. T. (1996) The role of MCM proteins in the cell cycle control of genome duplication. Bioessays 18, 183–190 [DOI] [PubMed] [Google Scholar]
- 2. Chong J. P., Thömmes P., Blow J. J. (1996) The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem. Sci. 21, 102–106 [PubMed] [Google Scholar]
- 3. Coxon A., Maundrell K., Kearsey S. E. (1992) Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 20, 5571–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dalton S., Whitbread L. (1995) Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 92, 2514–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272, 24508–24513 [DOI] [PubMed] [Google Scholar]
- 6. You Z., Komamura Y., Ishimi Y. (1999) Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity. Mol. Cell. Biol. 19, 8003–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren B., Yu G., Tseng G. C., Cieply K., Gavel T., Nelson J., Michalopoulos G., Yu Y. P., Luo J. H. (2006) MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene 25, 1090–1098 [DOI] [PubMed] [Google Scholar]
- 8. Honeycutt K. A., Chen Z., Koster M. I., Miers M., Nuchtern J., Hicks J., Roop D. R., Shohet J. M. (2006) Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene 25, 4027–4032 [DOI] [PubMed] [Google Scholar]
- 9. Brake T., Connor J. P., Petereit D. G., Lambert P. F. (2003) Comparative analysis of cervical cancer in women and in a human papillomavirus-transgenic mouse model: identification of minichromosome maintenance protein 7 as an informative biomarker for human cervical cancer. Cancer Res. 63, 8173–8180 [PubMed] [Google Scholar]
- 10. Kan T., Sato F., Ito T., Matsumura N., David S., Cheng Y., Agarwal R., Paun B. C., Jin Z., Olaru A. V., Selaru F. M., Hamilton J. P., Yang J., Abraham J. M., Mori Y., Meltzer S. J. (2009) The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology 136, 1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo J. H. (2011) Oncogenic activity of MCM7 transforming cluster. World J. Clin. Oncol. 2, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petrocca F., Visone R., Onelli M. R., Shah M. H., Nicoloso M. S., de Martino I., Iliopoulos D., Pilozzi E., Liu C. G., Negrini M., Cavazzini L., Volinia S., Alder H., Ruco L. P., Baldassarre G., Croce C. M., Vecchione A. (2008) E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13, 272–286 [DOI] [PubMed] [Google Scholar]
- 13. Sikand K., Slane S. D., Shukla G. C. (2009) Intrinsic expression of host genes and intronic miRNAs in prostate carcinoma cells. Cancer Cell Int. 9, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poliseno L., Salmena L., Riccardi L., Fornari A., Song M. S., Hobbs R. M., Sportoletti P., Varmeh S., Egia A., Fedele G., Rameh L., Loda M., Pandolfi P. P. (2010) Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 3, ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han Y. C., Yu Y. P., Nelson J., Wu C., Wang H., Michalopoulos G. K., Luo J. H. (2010) Interaction of integrin-linked kinase and miniature chromosome maintenance 7-mediating integrin α7 induced cell growth suppression. Cancer Res. 70, 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi Y. K., Yu Y. P., Tseng G. C., Luo J. H. (2010) Inhibition of prostate cancer growth and metastasis using small interference RNA specific for minichromosome complex maintenance component 7. Cancer Gene Ther. 17, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gladden A. B., Diehl J. A. (2003) The cyclin D1-dependent kinase associates with the pre-replication complex and modulates RB. MCM7 binding. J. Biol. Chem. 278, 9754–9760 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X. Y., Tang L. Z., Ren B. G., Yu Y. P., Nelson J., Michalopoulos G., Luo J. H. (2013) Interaction of MCM7 and RACK1 for activation of MCM7 and cell growth. Am. J. Pathol. 182, 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romanowski P., Madine M. A., Laskey R. A. (1996) XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc. Natl. Acad. Sci. U.S.A. 93, 10189–10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blow J. J., Hodgson B. (2002) Replication licensing: defining the proliferative state? Trends Cell Biol. 12, 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Y. K., Yu Y. P., Zhu Z. H., Han Y. C., Ren B., Nelson J. B., Luo J. H. (2008) MCM7 interacts with androgen receptor. Am. J. Pathol. 173, 1758–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Golas M. M., Sander B., Will C. L., Lührmann R., Stark H. (2003) Molecular architecture of the multiprotein splicing factor SF3b. Science 300, 980–984 [DOI] [PubMed] [Google Scholar]
- 23. Das B. K., Xia L., Palandjian L., Gozani O., Chyung Y., Reed R. (1999) Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell. Biol. 19, 6796–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinez E., Palhan V. B., Tjernberg A., Lymar E. S., Gamper A. M., Kundu T. K., Chait B. T., Roeder R. G. (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21, 6782–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brand M., Moggs J. G., Oulad-Abdelghani M., Lejeune F., Dilworth F. J., Stevenin J., Almouzni G., Tora L. (2001) UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20, 3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu Y. P., Yu G., Tseng G., Cieply K., Nelson J., Defrances M., Zarnegar R., Michalopoulos G., Luo J. H. (2007) Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 67, 8043–8050 [DOI] [PubMed] [Google Scholar]
- 27. Yu Y. P., Luo J. H. (2006) Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 66, 7414–7419 [DOI] [PubMed] [Google Scholar]
- 28. Wang H., Luo K., Tan L. Z., Ren B. G., Gu L. Q., Michalopoulos G., Luo J. H., Yu Y. P. (2012) p53-induced gene 3 mediates cell death induced by glutathione peroxidase 3. J. Biol. Chem. 287, 16890–16902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Z. H., Yu Y. P., Zheng Z. L., Song Y., Xiang G. S., Nelson J., Michalopoulos G., Luo J. H. (2010) Integrin α7 interacts with high temperature requirement A2 (HtrA2) to induce prostate cancer cell death. Am. J. Pathol. 177, 1176–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu Z. H., Yu Y. P., Shi Y. K., Nelson J. B., Luo J. H. (2009) CSR1 induces cell death through inactivation of CPSF3. Oncogene 28, 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Masuhiro Y., Mezaki Y., Sakari M., Takeyama K., Yoshida T., Inoue K., Yanagisawa J., Hanazawa S., O'Malley B W., Kato S. (2005) Splicing potentiation by growth factor signals via estrogen receptor phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 102, 8126–8131 [DOI] [PMC free article] [PubMed] [Google Scholar]