Background: The mechanism of gene regulation by the H2A deubiquitinase BAP1 is unclear.
Results: BAP1 loss increases FoxK2 target gene expression but not in the absence of the H2A ubiquitin ligase complex Ring1B-Bmi1.
Conclusion: BAP1 counteracts Ring1B-Bmi1-dependent activation of FoxK2 target genes.
Significance: BAP1 deficiency in cancer cells might cause up-regulation of target genes in a Ring1B-Bmi1-dependent manner.
Keywords: Deubiquitylation (Deubiquitination), Gene Expression, Phosphorylation, Tumor Suppressor Gene, Ubiquitin
Abstract
BRCA1-associated protein 1 (BAP1), which is frequently mutated in cancer, functions as a deubiquitinase (DUB) for histone H2A. Although BAP1 interacts with a transcriptional regulator, HCF-1, and transcription factors FoxK1 and FoxK2, how BAP1 controls gene expression remains unclear. This study investigates the importance of BAP1 DUB activity and the interactions with FoxK2 and HCF-1 in the regulation of FoxK2 target genes. We show that FoxK2 recruits BAP1 to the target genes through the forkhead-associated domain, which interacts with Thr(P)-493 on BAP1. BAP1, in turn, recruits HCF-1, thereby forming a ternary complex in which BAP1 bridges FoxK2 and HCF-1. BAP1 represses FoxK2 target genes, and this effect requires BAP1 DUB activity but not interaction with HCF-1. Importantly, BAP1 depletion causes up-regulation of FoxK2 target genes only in the presence of the Ring1B-Bmi1 complex, an E3 ubiquitin ligase for histone H2A, indicating an antagonizing role of BAP1 against Ring1B-Bmi1. Our findings suggest that BAP1 deficiency causes increased expression of target genes in a Ring1B-Bmi1-dependent manner.
Introduction
BRCA1-associated protein 1 (BAP1) is a tumor suppressor gene that is somatically mutated in a variety of cancer types, including mesothelioma, uveal melanoma, renal cell carcinoma, and cholangiocarcinomas (1–8). In addition, heterozygous germ line mutations in the BAP1 gene cause a hereditary cancer predisposition syndrome that is characterized by high incidences of mesothelioma, uveal melanoma, renal cell carcinoma, atypical melanocytic tumors, and others (1, 8–11). BAP1 is also a tumor suppressor in experimental systems. Restoration of BAP1 in a BAP1-null cell line, H226, suppresses its proliferation and tumor formation in a xenograft model (12), whereas conditional knockout of BAP1 in adult mice causes myeloid transformation (13). However, despite the mounting evidence linking BAP1 deficiency with cancer, how loss of BAP1 drives tumor formation is less understood.
BAP1 is a deubiquitinase (DUB)2 in the ubiquitin C terminus hydrolase family. On one hand, BAP1 has been shown to deubiquitinate two BAP1-interacting proteins, HCF-1 (a transcription coregulator) and O-linked N-acetylglucosamine transferase (13–15). However, the biological significance of this deubiquitination has not been established. On the other hand, in Drosophila, the BAP1 homolog Calypso and a polycomb protein, Additional sex comb (ASX), form the Polycomb repressive (PR)-DUB complex, which deubiquitinates histone H2A (16). Similarly, this H2A DUB activity is conserved in human PR-DUB, a complex of BAP1 and ASXL1 (16). Monoubiquitinated H2A (H2Aub) is implicated in gene silencing, and the majority of H2A ubiquitination in mammalian cells is catalyzed by polycomb repressive complex 1 (PRC1), in which a heterodimer of Ring1A or Ring1B and Bmi1 (also known as Polycomb group RING finger 4 (PCGF4)) or one of five other PCGF proteins serves as a catalytic core (17–20).
The study in Drosophila demonstrated that Calypso (a BAP1 homolog) represses Hox genes in a DUB activity-dependent manner (16). Because both the ubiquitin E3 ligase PRC1 and the deubiquitinase PR-DUB are required for repression of Hox genes in Drosophila (16, 21), it has been suggested that dynamic balancing of H2Aub levels by PRC1 and PR-DUB is necessary for proper repression. However, whether the human PR-DUB plays a similar repressive role in gene expression is currently unclear. In fact, a study in human cells has shown that BAP1 activates YY1-regulated genes in a DUB-dependent manner (22). In addition, genome-wide gene expression analyses showed that many genes are up- or down-regulated by BAP1 knockdown or knockout, indicating that BAP1 can be a corepressor or coactivator (13, 22, 23). Although these data suggest a role for BAP1 as a regulator of gene expression, further studies are needed to understand which genes are regulated by BAP1, how BAP1 is recruited to the target genes, and how BAP1 controls gene expression. In particular, assessing the role of DUB activity and the requirement of the interactions with coregulators such as HCF-1 are necessary to figure out the complex mechanism of gene regulation by BAP1.
The BAP1-interacting proteins FoxK1 and FoxK2 are members of the Fox transcription factors, which are characterized by the forkhead DNA-binding domains (24, 25). FoxK1 and FoxK2 are distinguished from other Fox transcription factors by the presence of the forkhead-associated (FHA) domains, which recognize phosphothreonine-containing peptides (26, 27). Functionally, FoxK1 has been implicated in the proper control of cell proliferation and the cell cycle and plays a critical role in myogenic progenitor cells in mice (28–30). Little is known about the physiological roles for FoxK2, but its function has been linked to cell cycle regulation because it is phosphorylated by cyclin-dependent kinases (31). Moreover, FoxK2 colocalizes with AP-1 and promotes AP-1-mediated transcriptional activation (32).
It has been reported recently that FoxK2 functions as a chromatin-targeting factor for BAP1 (33). The present study utilizes FoxK2-mediated gene regulation to dissect the role of BAP1 DUB activity and the importance of the interactions with HCF-1 and FoxK2. We demonstrate that FoxK2 recruits BAP1 via its FHA domain in a phosphorylation-dependent manner. BAP1 functions as a corepressor at FoxK2 target genes, and this function is dependent on DUB activity but not HCF-1 interaction. Our data indicate that BAP1 plays an antagonizing role against Ring1B in gene expression, raising the interesting possibility that Ring1B can be an activator in the absence of BAP1.
EXPERIMENTAL PROCEDURES
Plasmids
Full-length BAP1 cDNA (15) was subcloned into a mammalian expression vector, pEFF-C (C-terminal FLAG tag), pEFF-C/2NLS (C-terminal FLAG tag and two SV40 nuclear localization signals), and a retrovirus vector, pMX-Neo. Full-length FoxK1 and FoxK2 cDNAs were generated from the breast epithelial cell line MCF10A by reverse transcription PCR using LA Taq (Takara) and cloned into pEFF-N (N-terminal FLAG tag) and pEFM-N (N-terminal Myc tag). The cDNAs encoding the N-terminal portions of FoxK1 (1–234) and FoxK2 (8–190) were amplified by PCR and cloned into pGEX-5X-1. Site-directed mutagenesis was performed using pfu Turbo (Agilent Technologies), and all introduced mutations were confirmed by sequencing.
Cell Culture
The human embryonic kidney cell line 293T and the human lung cancer cell lines H1299 and H226 were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. For transient protein expression, 293T cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen). Retroviruses and lentiviruses were produced by transfecting viral and packaging plasmids in 293T cells as described previously (15). Cells were infected with virus-containing media in the presence of 4 μg/ml Polybrene (Sigma) and selected with 2 μg/ml puromycin (Sigma) for 2–5 days. Infected cells were cultured in the absence of puromycin for at least 24 h before subsequent assays.
RNA Interference
Transfection of siRNAs was performed using Lipofectamine RNAi Max (Invitrogen), and cells were analyzed after 48 h. For stable knockdown using shRNA, cells were infected with pLKO.1-based lentiviral vectors and selected with 2 μg/ml puromycin (Sigma) for 4 days. The target sequences of siRNA and shRNAs were as follows: siControl, 5′-CGUACGCGGAAUACUUCGA-3′; siBAP1.214, 5′-CGACCUUCAGAGCAAAUGU-3′; siBAP1.2132, 5′-GAGUUCAUCUGCACCUUUA-3′; siFoxK2.1734, 5′-CAGGGAAGUCAAAGUGAAA-3′; siFoxK2.2287, 5′-GCUGGCCUUAACACUCCUU-3′; shControl, 5′-CAACAAGAUGAAGAGCACCAA-3′; shBAP1.321, 5′-CGUCCGUGAUUGAUGAUGAUA-3′; shBAP1.2119, 5′-CCACAACUACGAUGAGUUCAU-3′; shFoxK2.2287, 5′-GCUGGCCUUAACACUCCUUAA-3′; shRing1B, 5′-GCCAGGAUCAACAAGCACAAU-3′; and shBmi1, 5′-CCAGACCACUACUGAAUAUAA-3′.
Immunoprecipitation and Western Blotting
Cells were lysed in Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.1% Nonidet P-40, 5 mm EDTA, 50 mm NaF, 1 mm Na2VO4, and 10% glycerol) containing a protease inhibitor mixture (Sigma). For immunoprecipitation, cell lysates containing 1–2 mg of proteins were mixed with antibodies overnight at 4 °C. Immune complexes were then captured using protein A- or protein G-Sepharose beads (GE Healthcare). For precipitation of FLAG-tagged proteins, anti-FLAG M2 affinity beads (Sigma) were mixed with lysates for 4 h at 4 °C. Beads were then washed five times with Nonidet P-40 lysis buffer, and precipitated proteins were eluted in 2× SDS-PAGE sample buffer by boiling. The following antibodies were used for Western blotting: anti-BAP1 and anti-GST (Santa Cruz Biotechnology), anti-FoxK1 and anti-FoxK2 (Bethyl Laboratory), anti-Ring1B and anti-FoxO3a (Cell Signaling Technology), anti-FLAG and anti-β-actin (Sigma), anti-GAPDH (GeneTex), and anti-Bmi1 (Millipore). Anti-Myc (9E10) ascites fluid was produced at Cocalico Biologicals. Anti-HCF-1-N antibodies have been described previously (15). Anti-HCF1-C and anti-KAP1 antibodies were obtained from Winship Herr and Zhenkun Lou, respectively.
Recombinant Proteins and Pulldown Assays
GST fusion proteins were expressed in Escherichia coli BL21(DE3), purified using glutathione-agarose beads (Sigma), and desalted using spin columns (Thermo Scientific). For GST pulldown assays, 5 μg of GST fusion proteins was immobilized on glutathione-agarose beads and incubated with 293T lysates containing 1 mg of proteins at 4 °C for 4 h. Where indicated, 293T cell lysates were treated with λ protein phosphatase (New England Biolabs) for 30 min at 30 °C in the presence or absence of phosphatase inhibitors (5 mm NaF, 1 mm Na3VO3, and 2 mm β-glycerophosphate). The beads were washed five times with GST binding buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1.0% Nonidet P-40, and 25 mg/ml bovine serum albumin) by rotating for 5 min at 4 °C. Samples were then boiled in 2× SDS-PAGE sample buffer and subjected to Western blotting. GST proteins were visualized with Coomassie Brilliant Blue. For peptide pulldown assays, 15 μg of synthetic peptides corresponding to amino acids 487–499 of human BAP1 with or without phosphorylation at Thr-493 (CTPSNESpTDTASEI or CTPSNESTDTASEI) was covalently attached to Sulfolink resin (Thermo Scientific). Beads were incubated with 0.5 μg of GST-FoxK1-N or GST-FoxK2-N proteins in PBS containing 0.1% Triton X-100 for 2 h at 4 °C and washed five times with PBS containing 0.1% Triton X-100 for 5 min at 4 °C. Proteins were eluted by boiling beads in 2× SDS-PAGE sample buffer and analyzed by Western blotting.
ChIP Assays
Cells (1 × 107) were fixed with 1% formaldehyde for 10 min and quenched with 125 mm glycine. Cells were lysed in ChIP lysis buffer (50 mm HEPES-NaOH (pH 7.5), 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate) a containing protease inhibitor mixture (Sigma). The cell lysates were sonicated by a sonic dismembrator (Fisher Scientific, model 500) to fragment the genomic DNA to ∼200–400 bp. Immunoprecipitation was performed by incubating lysates with 2 μg of anti-BAP1 antibodies (Santa Cruz Biotechnology) or 5 μl of rabbit anti-HCF-1-N antiserum (15) overnight at 4 °C, followed by incubation with protein G or protein A beads for 2 h. The beads were washed twice with ChIP lysis buffer for 5 min at 4 °C, twice with Tris/LiCl buffer (10 mm Tris-HCl (pH 8.0), 0.25 m LiCl, 1 mm EDTA, 0.5% Nonidet P-40, and 0.5% sodium deoxycholate) for 5 min at 4 °C, and twice with 2× Tris/EDTA buffer (20 mm Tris-HCl (pH 8.0) and 2 mm EDTA) for 5 min at 4 °C and then incubated in 1× Tris/EDTA buffer overnight at 65 °C for de-cross-linking. Proteins were digested with proteinase K at 55 °C for 30 min, and DNA was purified by phenol/chloroform/isoamyl extraction and ethanol precipitation. Precipitated DNA was analyzed by quantitative PCR (qPCR) using iQ SYBR Green PCR Master Mix (Bio-Rad). Primer pairs for ChIP assays were as follows: MCM3, 5′-ATCAGAACTGCCCTCCAGTG-3′ and 5′-CGGCGATGTCTTTATGGAGT-3′; CDC14A, 5′-ACCACCACAACAAACAGCAA-3′ and 5′-TACATGCACTGCCCTGTACC-3′; CDKN1B, 5′-GGCCTCAGAAGACGACGTCAAAC-3′ and 5′-AGCCTTCCCCATTGCTACTT-3′; and SRF int3, 5′-GCCACAGGGCAGTAGATGTT-3′ and 5′-GCCACAGGGCAGTAGATGTT-3′ as reported previously (32).
For ChIP assays using anti-Ring1B or anti-H2Aub antibodies, cells were lysed in extraction buffer (50 mm Tris-HCl (pH 7.5), 150 mm KCl, 1% Nonidet P-40, 0.04% sodium dodecyl sulfate, and 0.2% sodium deoxycholate) containing a protease inhibitor mixture. Immunoprecipitation was performed with protein A beads prebound with 2 μg of anti-Ring1B (Cell Signaling Technology) or anti-H2Aub (Cell Signaling Technology) antibodies overnight at 4 °C. Beads were washed five times with radioimmune precipitation assay wash buffer (50 mm Tris-HCl (pH 7.5), 150 mm KCl, 1% Nonidet-P-40, and 0.25% sodium deoxycholate) for 5 min at 4 °C and twice with TE wash buffer (10 mm Tris-HCl (pH 7.5) and 1 mm EDTA).
Quantitative RT-PCR
Total RNA was purified using the RNeasy mini kit (Qiagen), and cDNA was generated using Superscript III reverse transcriptase (Invitrogen). qPCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) and the following primer pairs: MCM3, 5′-CGCAGTGTCCACTACTGTCC-3′ and 5′-CTCCTGGATGGTGATGGTCT-3′; CDC14A, 5′-CATCGATGAGGAGCTGGTCT-3′ and 5′-TTTGCTCTTTTCCGTTGGTC-3′; CDKN1B, 5′-CATTTGGTGGACCCAAAGAC-3′ and 5′-CTTCTGAGGCCAGGCTTCTT-3′; and GAPDH, 5′-CAATGACCCCTTCATTGACC-3′ and 5′-GACAAGCTTCCCGTTCTCAG-3′.
RESULTS
BAP1 Forms Ternary Complexes Containing FoxK1/K2 and HCF-1
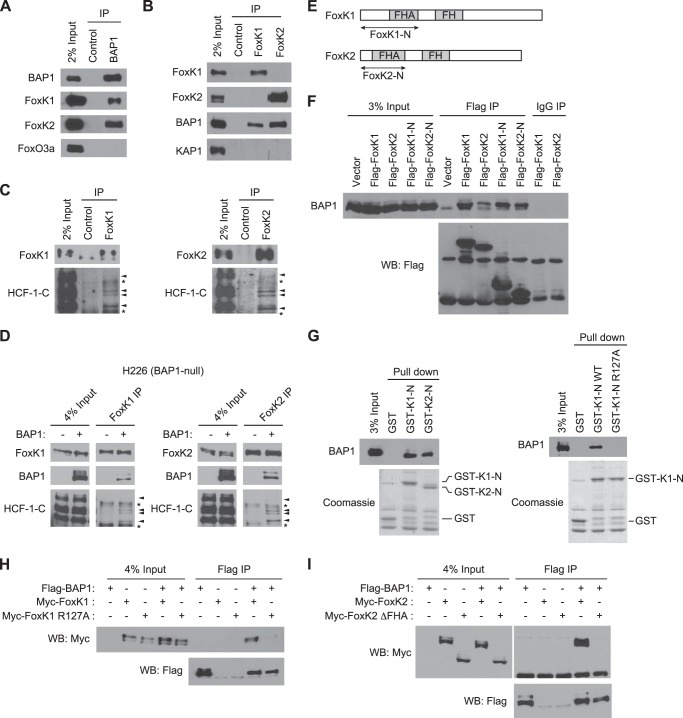
FoxK1 and FoxK2 (hereafter FoxK1/K2) have been identified as BAP1-interacting proteins by mass spectrometry analyses of BAP1-containing complexes (13, 15, 22, 34). We confirmed the interactions between BAP1 and FoxK1/K2 at the endogenous levels by coimmunoprecipitation. Both FoxK1 and FoxK2, but not FoxO3a, were coprecipitated with BAP1 (Fig. 1A). Reciprocally, BAP1 was recovered in FoxK1 and FoxK2 immunoprecipitations whereas KAP1 was not (Fig. 1B). As shown in Fig. 1C, FoxK1/K2 immunoprecipitations also contained a known BAP1-interacting protein, HCF-1 (13, 15, 22), suggesting that FoxK1/K2 and HCF-1 are present in the same complexes. To determine the role of BAP1 in the interactions of FoxK1/K2 and HCF-1, we performed coimmunoprecipitation experiments in a BAP1-deficient cell line, H226, which has a homozygous deletion of the BAP1 gene (35). The interactions between FoxK1/K2 and HCF-1 were undetectable in H226 cells but were restored when BAP1 was expressed exogenously (Fig. 1D). These results suggest that BAP1, FoxK1/K2, and HCF-1 form a ternary complex and that BAP1 is required for the interaction between FoxK1/K2 and HCF-1.
FIGURE 1.
FoxK1 and FoxK2 form ternary complexes with BAP1 and HCF-1. A, interaction of endogenous FoxK1/K2 with BAP1 in vivo. BAP1 was immunoprecipitated (IP) with specific antibodies, and the indicated proteins were detected by Western blotting. FoxO3a is shown as a negative control. B, association of FoxK1/K2 with BAP1 in vivo. FoxK1 or FoxK2 was immunoprecipitated, and the indicated proteins were detected by Western blotting. KAP1 is shown as a negative control. C, complex formation of FoxK1/K2 with HCF-1. FoxK1 or FoxK2 was immunoprecipitated, and the indicated proteins were detected by Western blotting. D, in vivo association of FoxK1/K2 with BAP1 and HCF-1 in H226 BAP1-deficient cells. BAP1 was expressed in H226 cells, and FoxK1 or FoxK2 was immunoprecipitated with specific antibodies. The indicated proteins were detected by Western blotting. The arrowheads and asterisks indicate HCF-1 bands and nonspecific bands, respectively. E, schematic of FoxK1 and FoxK2 proteins. F, interactions between the N-terminal regions of FoxK1/K2 and BAP1. Full-length or N-terminal regions (indicated in E) of FoxK1 and FoxK2 were expressed with a FLAG tag and immunoprecipitated with anti-FLAG antibodies or control IgG. Coprecipitated endogenous BAP1 was detected by Western blotting (WB). G, in vitro interactions between BAP1 and the N-terminal regions of FoxK1/K2. GST pulldown assays were performed with the indicated GST fusion proteins mixed with 293T cell lysates. GST fusion proteins in the precipitates were visualized by Coomassie staining, and coprecipitated BAP1 was detected by Western blotting. H, coimmunoprecipitation experiments of BAP1 and the FoxK1 R127A mutant. The indicated proteins were coexpressed in 293T cells, and FLAG-BAP1 was precipitated with anti-FLAG antibodies. Myc-FoxK1 was detected by anti-Myc Western blotting. I, coimmunoprecipitation experiments of BAP1 and the FHA deletion mutant (ΔFHA) of FoxK2.
The FHA Domains of FoxK1 and FoxK2 Are Necessary to Bind BAP1
To gain more insight into the interaction between BAP1 and FoxK1/K2, we next determined the BAP1-interacting regions of FoxK1/K2. Coimmunoprecipitation experiments revealed that the FoxK1/K2 N-terminal regions (FoxK1-N and FoxK2-N), which contain the FHA domains (Fig. 1E), are sufficient to interact with BAP1 in vivo (Fig. 1F). In the in vitro pulldown assays, GST fusion proteins of the FoxK1/K2 N-terminal regions efficiently pulled down BAP1 from cell lysates (Fig. 1G, left panel). Taken together, these experiments show that the N-terminal regions of FoxK1 and FoxK2, which contain the FHA domain, are sufficient to interact with BAP1.
Next we tested whether the FHA domains of FoxK1/K2 are required for the interaction with BAP1. Previous studies have shown that the conserved Arg residue in the FHA domain plays a key role in recognizing phosphothreonine and that mutation of the Arg residue diminishes interactions with phosphopeptides (27). We introduced the equivalent mutation at Arg-127 (R127A) in the FoxK1 FHA domain and examined its effect on the interaction between FoxK1 and BAP1. This mutation diminished the interaction of GST-FoxK1-N and BAP1 in the in vitro pulldown assays (Fig. 1G, right panel) and also disrupted the interaction between full-length FoxK1 and BAP1 in vivo (Fig. 1H). Similarly, we found that a FoxK2 mutant lacking the FHA domain (ΔFHA) was incapable of interacting with BAP1 in the coimmunoprecipitation experiments (Fig. 1I), confirming the requirement of the FHA domain for the interaction with BAP1 (33). Collectively, these data show that FoxK1 and FoxK2 interact with BAP1 through their FHA domains.
Identification of the FoxK1/K2-interacting Region of BAP1
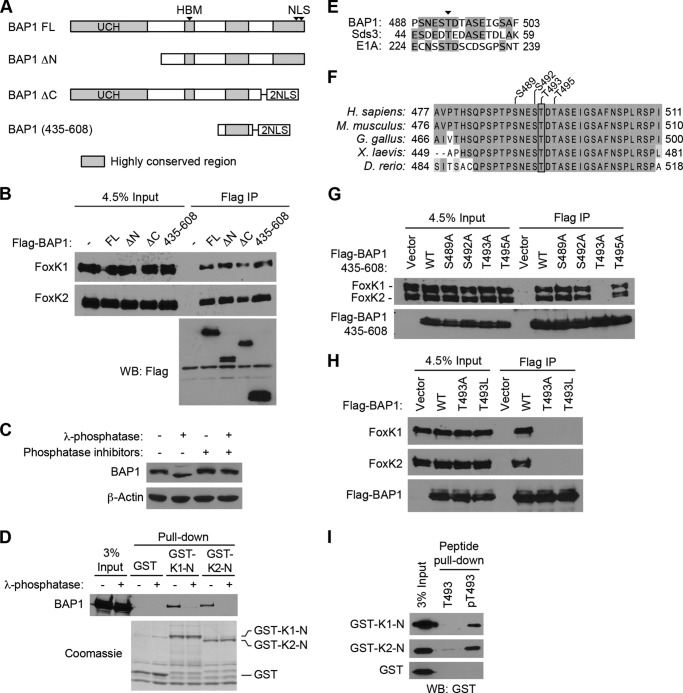
To better understand the mechanism of interaction between BAP1 and FoxK1/K2, we next determined the FoxK1/K2-interacting region of BAP1. As shown in Fig. 2A, we generated BAP1 deletion mutants and determined their ability to interact with FoxK1/K2 by immunoprecipitation. It has been reported recently that deletion of the N-terminal ubiquitin C terminus hydrolase domain or the C-terminal domain from BAP1 reduced its interaction with FoxK2 (33). In our experiments, however, deletion of the N-terminal or C-terminal region of BAP1 did not affect the interaction between the deletion fragments and endogenous FoxK1/K2 (Fig. 2B), suggesting that the central region of BAP1, which contains an HCF-1-binding motif (HBM) and a highly conserved region (477–526) (Fig. 2A), might interact with FoxK1/K2. Indeed, a BAP1 fragment containing this highly conserved region precipitated FoxK1/K2 as efficiently as full-length BAP1 (Fig. 2B), indicating that this conserved region of BAP1 is sufficient to interact with FoxK1/K2.
FIGURE 2.
Phosphorylation-dependent interactions between BAP1 and FoxK1/K2. A, schematic of BAP1 and its truncation mutants. Regions highly conserved between species are shown in gray. FL, full-length; UCH, ubiquitin C-terminal hydrolase; NLS, nuclear localization signal. B, mapping of the FoxK1/K2-interacting domains of BAP1. Full-length BAP1 and its truncation mutants, depicted in A, were expressed with a FLAG tag and precipitated with anti-FLAG antibodies. Coprecipitated endogenous FoxK1 and FoxK2 proteins were detected by Western blotting (WB). IP, immunoprecipitation. C, effect of λ-phosphatase treatments on BAP1 gel mobility. Cell lysates were treated with λ-phosphatase in the presence or absence of phosphatase inhibitors, and the indicated proteins were assessed by Western blotting. D, in vitro interaction between BAP1 and the N-terminal region of FoxK1/FoxK2. GST pulldown assays were performed using the indicated GST fusion proteins, and 293T cell lysates were treated with or λ-phosphatase or left untreated. Precipitated GST fusion proteins were visualized by Coomassie staining, and coprecipitated BAP1 was detected by Western blotting. E, alignment of BAP1, Sds3, and E1A sequences surrounding the conserved Thr residue (arrowhead), which is phosphorylated in Sds3. Identical amino acids are highlighted. F, alignment of the FoxK1/K2-interacting region of BAP1 from various species. Identical and similar amino acids are highlighted with different contrasts. G, in vivo interactions between BAP1 (435–608) and FoxK1/K2. FLAG-tagged BAP1 (435–608) with various point mutations was expressed in 293T cells and immunoprecipitated with anti-FLAG antibodies. Coprecipitated endogenous FoxK1 and FoxK2 were detected by Western blotting. H, in vivo interactions between FoxK1/K2 and full-length BAP1. FLAG-tagged BAP1 proteins (wild-type, T493A, and T493L) were expressed and immunoprecipitated with anti-FLAG antibodies. Coprecipitated endogenous FoxK1/K2 was detected by Western blotting. I, peptide pulldown assays. Peptides corresponding to the 487–499 amino acid region of BAP1 were synthesized with or without phosphorylation at Thr-493 and conjugated to beads. Pulldown assays were performed by mixing beads and recombinant GST-FoxK1-N or GST-FoxK2-N proteins, and precipitated GST-fusion proteins were detected by Western blotting using anti-GST antibodies. T493, non-phosphorylated peptide; pT493, peptide phosphorylated at Thr-493.
Phosphorylation of BAP1 Is Required for Interaction with FoxK1/K2
Because FHA domains are known to interact with phosphothreonine peptides, we wondered whether the interactions between BAP1 and FoxK1/K2 might involve phosphorylation of BAP1. Indeed, a gel mobility shift of BAP1 was observed when cell lysates were treated with λ-phosphatase (Fig. 2C), suggesting that BAP1 is a phosphoprotein. We therefore examined the possibility that FoxK1/K2 interacts with BAP1 in a phosphorylation-dependent manner by in vitro GST pulldown assays using cell lysates treated with λ-phosphatase. Interactions of GST-FoxK1-N and GST-FoxK2-N with BAP1 in cell lysates were greatly reduced by λ-phosphatase treatment (Fig. 2D), indicating that the interactions between BAP1 and FoxK1/K2 are dependent on phosphorylation.
We next sought to identify the BAP1 phosphorylation site required for the FoxK1/K2 interaction. Previous studies showed that the FHA domain of FoxK1 interacted with Thr(P)-493 in Sds3 (36). Examination of the FoxK1/K2-interacting region of BAP1 (amino acids 435–608) revealed a segment that resembles the sequence of Sds3 surrounding Thr-49 (Fig. 2E). Furthermore, these sequences have a similarity to the FoxK1/K2-interacting domain of adenovirus E1A (Fig. 2E), another protein known to interact with FoxK1 and FoxK2 in a phosphorylation-dependent manner (37). In addition, Thr-493 in human BAP1 is highly conserved among BAP1 homologs (Fig. 2F). We therefore hypothesized that Thr-493 phosphorylation is necessary for the interaction with FoxK1/K2. To test this possibility, we first substituted Thr-493 with alanine (T493A) and performed immunoprecipitation assays. The T493A mutation disrupted the interaction with endogenous FoxK1/K2 proteins, whereas the point mutations at Ser or Thr residues in the surrounding region did not affect the interaction (Fig. 2G). Similarly, full-length BAP1 harboring the T493A or T493L mutation failed to interact with FoxK1/K2 in the coimmunoprecipitation experiments (Fig. 2H). These results indicate that Thr-493 of BAP1 is required for the interaction with FoxK1/K2.
We next determined the effect of Thr-493 phosphorylation on the interactions with FoxK1/K2. We prepared two synthetic peptides corresponding to the sequences surrounding Thr-493 of BAP1: one containing non-phosphorylated Thr-493 and the other containing Thr(P)-493. These synthetic peptides were immobilized on beads, and the interaction with the N-terminal regions of FoxK1/K2 produced in E. coli was examined by in vitro pulldown assays (Fig. 2I). The Thr(P)-493 peptides pulled down GST-FoxK1-N and GST-FoxK2-N efficiently whereas non-phosphorylated peptide did not. Altogether, these data suggest that phosphorylation of BAP1 at Thr-493 is important for the interactions with FoxK1/K2 FHA domains.
FoxK2 Recruits BAP1 and HCF-1 to FoxK2 Target Genes
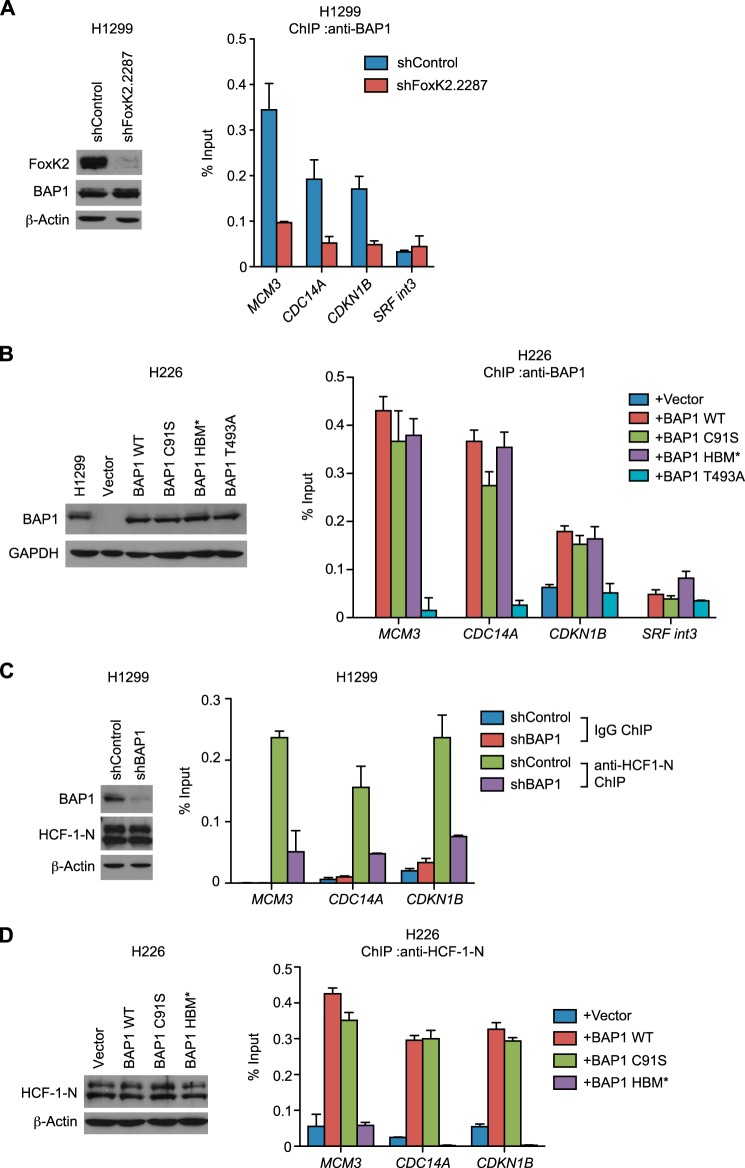
We next determined whether the ternary complex of FoxK2, BAP1, and HCF-1 is formed on the FoxK2-binding regions on the genome. ChIP assays showed that BAP1 was recruited to the previously reported FoxK2-binding regions of MCM3, CDC14A, and CDKN1B (32) but not to a negative control region (SRF int3) (Fig. 3A). Importantly, BAP1 recruitment was greatly reduced in FoxK2-depleted cells, suggesting that recruitment of BAP1 to FoxK2 targets is dependent on FoxK2. These results are consistent with a recent report that concluded that FoxK2 recruits BAP1 to its binding regions (33).
FIGURE 3.
Recruitment of BAP1 and HCF-1 to FoxK2 binding regions. A, recruitment of BAP1 to FoxK2 target genes. ChIP using anti-BAP1 antibodies was performed with control and FoxK2-depleted H1299 cells. Expression levels of FoxK2 and BAP1 assessed by Western blotting are shown in the left panel. B, recruitment of various BAP1 mutants to FoxK2 target genes. BAP1 mutants were expressed at endogenous levels in the H226 BAP1-deficient cell line, and ChIP was performed with anti-BAP1 antibodies. Expression levels of BAP1 assessed by Western blotting are shown in the left panel. H1299 is shown as a control for the endogenous BAP1 level. C, recruitment of HCF-1 to FoxK2 target genes. ChIP using anti-HCF-1-N antiserum was performed with control and BAP1-depleted H1299 cells. Expression levels of BAP1 and HCF-1-N assessed by Western blotting are shown in the left panel. D, ChIP analyses of HCF-1 recruitment in the H226 BAP1-deficient cell line expressing various BAP1 mutants. BAP1 mutants were expressed as in B, and ChIP was performed with anti-HCF-1-N antiserum. Expression levels of HCF-1-N were assessed by Western blotting and are shown in the left panel. β-actin and GAPDH served as loading controls in Western blotting. qPCR was done in triplicate, and mean ± S.D. is shown. In ChIP experiments, the amount of precipitated DNA at each region was expressed as a percentage of input.
To gain more insight into BAP1 recruitment to FoxK2 binding regions, we expressed various BAP1 mutants in H226 BAP1-null cells and examined their recruitment to FoxK2 binding regions (Fig. 3B). As expected, exogenously expressed wild-type BAP1 (BAP1 WT) was recruited to FoxK2 binding regions but not to a control region. A catalytically inactive mutant of BAP1 (C91S) was also recruited to FoxK2-binding regions, indicating that the BAP1 recruitment does not require its DUB activity. Similarly, the interaction with HCF-1 is not necessary for the localization of BAP1 to FoxK2-binding regions because the recruitment was not affected by the HBM* mutation (the HBM sequence (NHNY) was replaced with AAAA), which disrupts interaction with HCF-1 (15). On the other hand, a point mutation at Thr-493 (T493A) phosphorylation site, which impairs interaction with FoxK2 (Fig. 2H), diminished BAP1 recruitment to FoxK2 target genes, supporting our model that FoxK2 recruits phosphorylated BAP1 to FoxK2 target genes.
Although it has been shown recently that HCF-1 localizes to FoxK2-binding regions (33), it remains to be determined whether FoxK2 directly recruits HCF-1 or does so indirectly through BAP1 recruitment. To address this point, we first tested whether BAP1 is necessary for HCF-1 recruitment. Knockdown of BAP1 in BAP1-proficient cells reduced HCF-1 recruitment to FoxK2 target genes (Fig. 3C), whereas exogenous expression of BAP1 in a BAP-deficient cell line restored HCF-1 recruitment (Fig. 3D), suggesting that HCF-1 recruitment is dependent on BAP1. BAP1 could recruit HCF-1 through the direct protein-protein interaction via its HBM, or BAP1 might induce HCF-1 recruitment indirectly through modulation of the chromatin structure via H2A deubiquitination. To address these points, we expressed BAP1 mutants that are deficient in catalytic activity (C91S) or HCF-1 interaction (HBM*) in the BAP1-deficient cell line and examined HCF-1 recruitment. The catalytically inactive mutant recruited HCF-1 as efficiently as the wild type, whereas the HBM* mutant exhibited greatly diminished HCF-1 recruitment (Fig. 3D). These data indicate that BAP1 recruits HCF-1 through direct interaction but not through DUB activity. Collectively, these data suggest that FoxK2 recruits BAP1 through recognition of Thr(P)-493, and BAP1, in turn, recruits HCF-1 via HBM.
BAP1 Represses Expression of FoxK2 Target Genes
Having established the recruitment of BAP1 and HCF-1 to the FoxK2 target genes, we examined their effects on gene expression. BAP1 depletion in BAP1-proficient cells increased expression of the FoxK2-target genes MCM3, CDC14A, and CDKN1B (Fig. 4A), whereas restoration of BAP1 in a BAP1-negative cell line repressed the expression levels (Fig. 4B), demonstrating that BAP1 negatively regulates the expression of FoxK2 target genes. Further studies showed that the DUB activity of BAP1 is necessary for this repression because the inactive mutant of BAP1 did not reduce gene expression (Fig. 4B). Interestingly, the HBM* mutant of BAP1 reduced target gene expression as efficiently as the wild type, suggesting that HCF-1 interaction is not necessary for the repression of gene expression. On the other hand, the T493A mutant of BAP1, which is deficient in interacting with FoxK2 (Fig. 2H), exhibited diminished abilities to repress target genes. This result suggests that down-regulation of BAP-regulated gene expression requires its local recruitment to target genes by FoxK2. Consistent with the notion that FoxK2 recruits BAP1 to repress target genes, knockdown of FoxK2 in the BAP1-proficient cell line increased target gene expression (Fig. 4C), mimicking the effect of BAP1 depletion.
FIGURE 4.
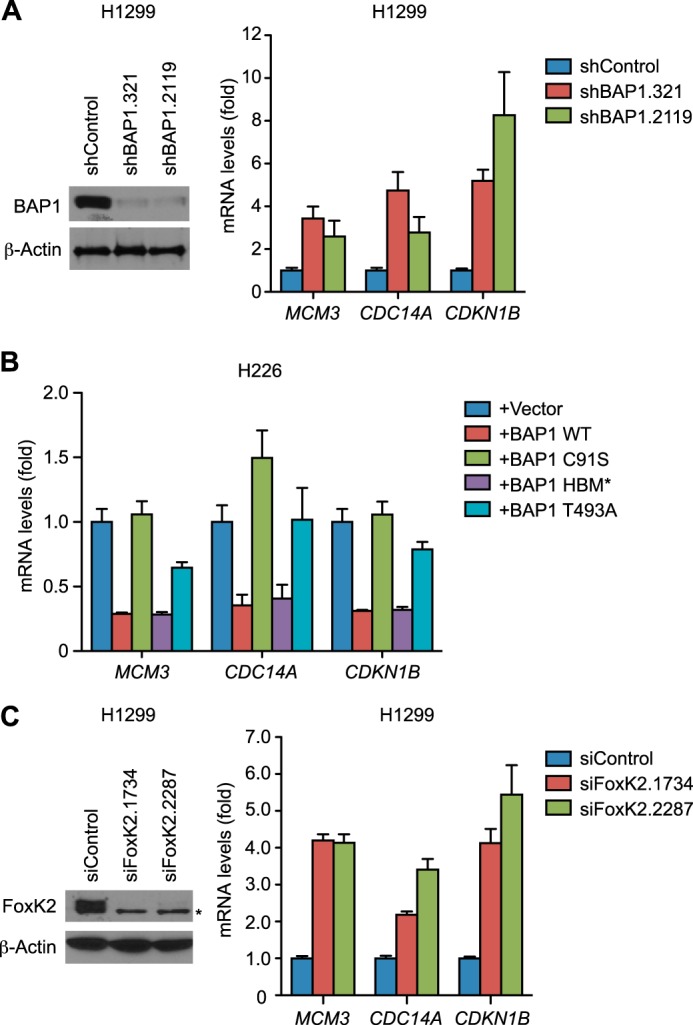
BAP1 down-regulates FoxK2 target genes. A, expression levels of FoxK2 target genes in BAP1-depleted H1299 cells. mRNA levels of the indicated FoxK2 target genes were measured by RT-PCR in control and BAP1-depleted H1299 cells (right panel). Knockdown of BAP1 was assessed by Western blotting (left panel). β-Actin was used as a loading control. B, effects of various mutations in BAP1 on expression of FoxK2 target genes. The indicated BAP1 mutants were expressed in the H226 BAP1-deficient cell line as in Fig. 3B, and expression of the indicated genes was quantitated by RT-PCR. C, expression of FoxK2 target genes in FoxK2-depleted H1299 cells. FoxK2 was depleted with the indicated siRNAs, and FoxK2 protein levels were assessed by Western blotting (left panel). An asterisk indicates a nonspecific band. mRNA levels of the indicated genes were measured by RT-PCR (right panel). qPCR was done in triplicate, and mean ± S.D. is shown. Values were normalized to GAPDH and are shown relative to control samples.
BAP1 Loss Causes PRC1-mediated Up-regulation of FoxK2 Target Genes
Because repression of the FoxK2 target genes requires BAP1 DUB activity (Fig. 4B), we next explored possible BAP1 substrates involved in this regulation. Although HCF-1 can be deubiquitinated by BAP1 (14, 15), we ruled out HCF-1 as a relevant substrate because mutation of the HBM (HBM*), which recruits HCF-1, did not affect the repression by BAP1 (Fig. 4B). Histone H2A is also a known substrate of BAP1 (16), and we confirmed that H2Aub levels increased at FoxK2-binding regions following BAP1 depletion but not at a negative control region (SRF int3) (Fig. 5A). A reciprocal experiment also showed that BAP1 restoration in a BAP1-deficient cell line reduced H2Aub at the FoxK2-binding regions but not at the negative control region (Fig. 5B). The inactive mutant of BAP1 (C91S) and the T493A mutant did not reduce H2Aub levels at these loci, suggesting that BAP1 directly deubiquitinates H2A at the FoxK2-binding regions.
FIGURE 5.
Regulation of H2Aub at FoxK2 target genes by BAP1. A and B, effect of BAP1 on H2Aub levels at the FoxK2-binding regions. ChIP assays were performed using anti-H2Aub antibodies in H1299 with control or BAP1 knockdown (A) and the H226 BAP1-deficient cell line expressing the wild type or the inactive BAP1 mutant (C91S) (B). C and D, recruitment of Ring1B to FoxK2 binding regions. ChIP assays were performed using control IgG or anti-Ring1B antibodies in H1299 with control or BAP1 knockdown (C) and the H226 BAP1-deficient cell line with or without BAP1 expression (D). qPCR was done in triplicate, and mean ± S.D. is shown.
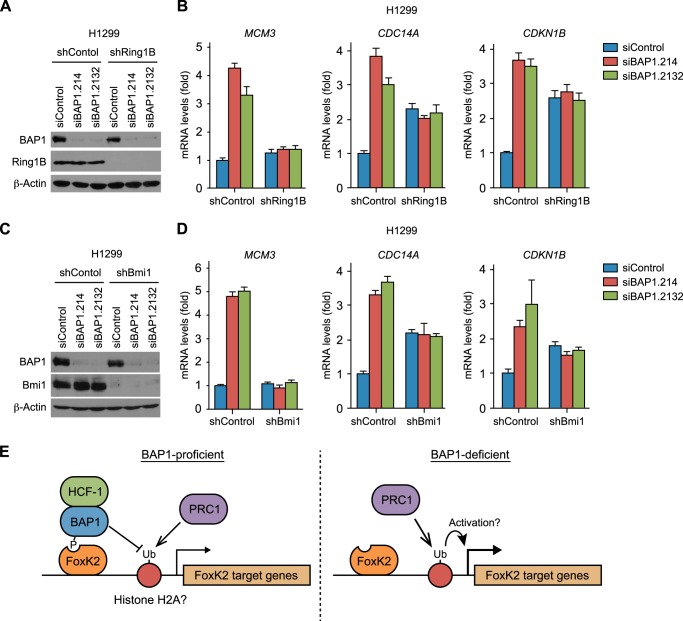
Histone H2A is ubiquitinated by Ring1B, a ubiquitin E3 ligase and the catalytic subunit of PRC1. Accordingly, we next addressed the interplay between BAP1 and Ring1B on FoxK2-regulated gene expression. ChIP experiments confirmed that Ring1B was detectable at the FoxK2-binding regions of MCM3, CDC14A, and CDKN1B (Fig. 5, C and D). The amount of Ring1B at these regions was unaffected by BAP1 depletion in BAP1-proficient cells (Fig. 5C) or by BAP1 restoration in BAP1-deficient cells (Fig. 5D), indicating that the down-regulation of H2Aub by BAP1 is not due to reduction in Ring1B recruitment. To assess the functional relationship between BAP1 and Ring1B in the repression of the FoxK2 target genes, we codepleted Ring1B and BAP1 (Fig. 6A). Consistent with the experiments shown above (Fig. 4A), BAP1 depletion up-regulated the FoxK2 target genes MCM3, CDC14A, and CDKN1B in the control experiments. In striking contrast, BAP1 depletion in the Ring1B-depleted cells did not up-regulate these genes, indicating that up-regulation of FoxK2 target genes upon BAP1 requires Ring1B. This was most obvious in the MCM3 gene, in which Ring1B depletion by itself minimally affected MCM3 expression, and, yet, it diminished up-regulation after BAP1 knockdown (Fig. 6B, left panel). Interpretation of the data was slightly complicated at CDC14A and CDKN1B (Fig. 6B, center and right panels) because Ring1B depletion by itself up-regulated these genes, a phenomenon most likely reflecting the loss of Ring1B-mediated silencing. Nonetheless, BAP1 knockdown did not increase gene expression under the Ring1B-depleted condition, suggesting a role of BAP1 as a suppressor of Ring1B-dependent activation of FoxK2 target genes.
FIGURE 6.
BAP1 suppresses PRC1-mediated activation of FoxK2 target genes. A–D, codepletion of BAP1 with Ring1B or Bmi1. BAP1 and Ring1B (A and B) or Bmi1 (C and D) were depleted individually or in combination. Verification of knockdown by Western blotting is shown in A and C. β-actin served as a loading control. Expression of the indicated FoxK2 target genes was assessed by RT-PCR and is shown in B and D. Values were normalized to GAPDH and are shown relative to the control RNAi sample. qPCR was done in triplicate, and mean ± S.D. is shown. E, model of BAP1-mediated regulation of FoxK2 target genes. See “Discussion” for details.
Because Ring1B dimerizes with Bmi1 to form an active E3 ligase in PRC1 (19), we tested whether Bmi1 also has an antagonizing relationship with BAP1. Like Ring1B depletion, knockdown of Bmi1 diminished up-regulation of FoxK2 target genes after BAP1 depletion (Fig. 6, C and D). Together, these results suggest that BAP1 loss causes the PRC1-dependent induction of FoxK2 target gene expression.
DISCUSSION
In this study, we investigated the molecular basis of the interaction between BAP1 and FoxK1/K2 and examined the role of BAP1 in the regulation of FoxK2 target genes. On the basis of our data, we propose that the FHA domain of FoxK2 recruits BAP1 phosphorylated at Thr-493. BAP1, in turn, brings HCF-1 to FoxK2 target genes via direct interaction at HBM (Fig. 6E). Our study found that BAP1 represses FoxK2-target genes but that this repression is mainly mediated by suppressing PRC1-dependent activation through BAP1 DUB activity rather than by HCF-1 recruitment. The data presented here are consistent with the model that BAP1 represses gene expression by deubiquitination of H2A, although it remains possible that additional substrates play a role (Fig. 6E).
Our study showed that the interactions between FoxK1/K2 and BAP1 are phosphorylation-dependent. On the contrary, Ji et al. (33) reported recently that FoxK2 interacts with BAP1 in a phosphorylation-independent manner. This discrepancy might come from the methods used in the experiments. In our experiments, we performed GST pulldown assays using cell lysates pretreated with λ-phosphatase. This may have allowed λ-phosphatase to remove phosphate groups from BAP1 before it interacted with the FHA domains in the subsequent GST pulldown assays. In contrast, Ji et al. (33) performed immunoprecipitation from the phosphatase-treated cell lysates in which FoxK2-bound BAP1 might have been protected from dephosphorylation at Thr-493 because of interactions with the FHA domain. Supporting our phosphorylation dependence model, the peptide pulldown assays showed a clear dependence on phosphorylation at Thr-493 (Fig. 2I). Given that the FHA domain of FoxK1 interacts with Sds3 in a phosphorylation-dependent manner (36), our data suggest that the FHA domains of FoxK1 and FoxK2 share a common interaction mechanism in which the interaction is greatly enhanced by threonine phosphorylation.
We also showed that FoxK1/K2, BAP1, and HCF-1 form a ternary complex in which BAP1 bridges FoxK1/K2 and HCF-1. It is interesting that FoxK2 recruits HCF-1 through BAP1, whereas some other transcription factors, such as E2F, recruit HCF-1 directly (38). It is possible that recruitment through BAP1 allows another layer of regulation for HCF-1. For example, HCF-1 is known to interact with a number of histone-modifying enzymes (39), so deubiquitination of histone H2A by BAP1 could create a different environment for these chromatin regulators. Alternatively, given that BAP1 can deubiquitinate HCF-1 (14, 15), it is possible that BAP1 remodels HCF-1 complexes, potentially regulating recruitment of histone-modifying enzymes. Although this study did not find a critical function of HCF-1 at the FoxK2 target genes, further studies will be needed to fully understand the role of the BAP1·HCF-1 complex in gene regulation.
Our finding that BAP1 represses gene expression is consistent with a previous report showing that Drosophila PR-DUB, a complex of the BAP1 ortholog Calypso and the Polycomb group protein Asx, repressed Hox genes (16). However, unlike our study, codepletion of the Ring1B ortholog dRing in Drosophila did not correct Hox gene misexpression caused by PR-DUB depletion (through an Asx mutation). Instead, the codepletion resulted in an even more rapid loss of Hox gene silencing than individual depletion, suggesting that both ubiquitination and deubiquitination of histone H2A are important for gene silencing (16, 21). Therefore, one might ask whether the up-regulation of target genes following BAP1 depletion in our study is simply due to loss of the Ring1B-mediated silencing rather than activation by Ring1B. We believe that our results cannot be explained by this model for three reasons. First, such a model does not explain the effect on MCM3, where BAP1 depletion up-regulated MCM3 but Ring1B depletion had only a minor effect (Fig. 6B, left panel). Second, the model predicts that BAP1 depletion in the Ring1B-depleted background would increase gene expression at least to the same levels as in control. However, the increased levels after BAP1 knockdown were lower in Ring1B-depleted cells than in the control in our experiments (Fig. 6B). Third, we observed both effects (silencing and activation) of Ring1B at CDKN1B and CDC14A. At these loci, Ring1B is necessary to repress basal level expression, whereas it up-regulates gene expression when BAP1 is depleted (Fig. 6B). Therefore, it appears that Ring1B has two opposing effects on CDC14A and CDKN1B: one is repression, possibly mediated by the silencing function of Ring1B, and the other is activation, which is normally suppressed by BAP1. Depletion of Bmi1, a dimerizing partner of Ring1B, has the same effect as Ring1B knockdown, suggesting that activation of FoxK2 target genes in BAP1-deficient cells is mediated by PRC1. On the basis of these data, we propose a model in which BAP1 suppresses PRC1-dependent activation of FoxK2 target genes.
Importantly, our model predicts that PRC1 can be a positive factor for gene expression in the absence of BAP1 (Fig. 6E). Although this seems surprising given that Ring1B and H2A ubiquitination are generally associated with gene silencing, gene activation by Ring1B has been reported in quiescent lymphocytes (40). It should be noted, however, that the activating effect of PRC1 becomes detectable only after BAP1 depletion in our experiments. At this point, it is not clear whether this PRC1-dependent activation reflects a normal function of PRC1 or an abnormal condition that occurs only when BAP1 is absent. In either case, our findings might have an important implication in the understanding of BAP1-mutated cancer. Loss of BAP1 in cancer cells might create an unusual condition in which PRC1 elevates expression of tumor-promoting factors.
In conclusion, this study demonstrates an antagonizing effect of BAP1 against PRC1 on FoxK2-regulated genes and uncovers a potential role of PRC1 as a coactivator in the absence of BAP1. Further studies will determine the mechanism of the activation and its implication in the pathogenesis of BAP1-mutated cancer.
Acknowledgment
We thank Larry M. Karnitz for critical reading of the manuscript.
This work was supported by the Minnesota Partnership for Biotechnology and Medical Genomics (to Y. J. M.) and Eagles Funds for Cancer Research (Y. J. M.).
- DUB
- deubiquitinase
- FHA
- forkhead-associated
- qPCR
- quantitative PCR
- HBM
- HCF-1-binding motif
- FH
- forkhead
- ASX
- Additional sex comb
- PR
- Polycomb repressive
- PCGF
- Polycomb group RING finger.
REFERENCES
- 1. Testa J. R., Cheung M., Pei J., Below J. E., Tan Y., Sementino E., Cox N. J., Dogan A. U., Pass H. I., Trusa S., Hesdorffer M., Nasu M., Powers A., Rivera Z., Comertpay S., Tanji M., Gaudino G., Yang H., Carbone M. (2011) Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 43, 1022–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bott M., Brevet M., Taylor B. S., Shimizu S., Ito T., Wang L., Creaney J., Lake R. A., Zakowski M. F., Reva B., Sander C., Delsite R., Powell S., Zhou Q., Shen R., Olshen A., Rusch V., Ladanyi M. (2011) The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 43, 668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harbour J. W., Onken M. D., Roberson E. D., Duan S., Cao L., Worley L. A., Council M. L., Matatall K. A., Helms C., Bowcock A. M. (2010) Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peña-Llopis S., Vega-Rubín-de-Celis S., Liao A., Leng N., Pavía-Jiménez A., Wang S., Yamasaki T., Zhrebker L., Sivanand S., Spence P., Kinch L., Hambuch T., Jain S., Lotan Y., Margulis V., Sagalowsky A. I., Summerour P. B., Kabbani W., Wong S. W., Grishin N., Laurent M., Xie X. J., Haudenschild C. D., Ross M. T., Bentley D. R., Kapur P., Brugarolas J. (2012) BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 44, 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo G., Gui Y., Gao S., Tang A., Hu X., Huang Y., Jia W., Li Z., He M., Sun L., Song P., Sun X., Zhao X., Yang S., Liang C., Wan S., Zhou F., Chen C., Zhu J., Li X., Jian M., Zhou L., Ye R., Huang P., Chen J., Jiang T., Liu X., Wang Y., Zou J., Jiang Z., Wu R., Wu S., Fan F., Zhang Z., Liu L., Yang R., Liu X., Wu H., Yin W., Zhao X., Liu Y., Peng H., Jiang B., Feng Q., Li C., Xie J., Lu J., Kristiansen K., Li Y., Zhang X., Li S., Wang J., Yang H., Cai Z., Wang J. (2012) Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat. Genet. 44, 17–19 [DOI] [PubMed] [Google Scholar]
- 6. Chan-On W., Nairismägi M. L., Ong C. K., Lim W. K., Dima S., Pairojkul C., Lim K. H., McPherson J. R., Cutcutache I., Heng H. L., Ooi L., Chung A., Chow P., Cheow P. C., Lee S. Y., Choo S. P., Tan I. B., Duda D., Nastase A., Myint S. S., Wong B. H., Gan A., Rajasegaran V., Ng C. C., Nagarajan S., Jusakul A., Zhang S., Vohra P., Yu W., Huang D., Sithithaworn P., Yongvanit P., Wongkham S., Khuntikeo N., Bhudhisawasdi V., Popescu I., Rozen S. G., Tan P., Teh B. T. (2013) Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 45, 1474–1478 [DOI] [PubMed] [Google Scholar]
- 7. Jiao Y., Pawlik T. M., Anders R. A., Selaru F. M., Streppel M. M., Lucas D. J., Niknafs N., Guthrie V. B., Maitra A., Argani P., Offerhaus G. J., Roa J. C., Roberts L. R., Gores G. J., Popescu I., Alexandrescu S. T., Dima S., Fassan M., Simbolo M., Mafficini A., Capelli P., Lawlor R. T., Ruzzenente A., Guglielmi A., Tortora G., de Braud F., Scarpa A., Jarnagin W., Klimstra D., Karchin R., Velculescu V. E., Hruban R. H., Vogelstein B., Kinzler K. W., Papadopoulos N., Wood L. D. (2013) Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 45, 1470–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiesner T., Obenauf A. C., Murali R., Fried I., Griewank K. G., Ulz P., Windpassinger C., Wackernagel W., Loy S., Wolf I., Viale A., Lash A. E., Pirun M., Socci N. D., Rütten A., Palmedo G., Abramson D., Offit K., Ott A., Becker J. C., Cerroni L., Kutzner H., Bastian B. C., Speicher M. R. (2011) Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 43, 1018–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdel-Rahman M. H., Pilarski R., Cebulla C. M., Massengill J. B., Christopher B. N., Boru G., Hovland P., Davidorf F. H. (2011) Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J. Med. Genet. 48, 856–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Popova T., Hebert L., Jacquemin V., Gad S., Caux-Moncoutier V., Dubois-d'Enghien C., Richaudeau B., Renaudin X., Sellers J., Nicolas A., Sastre-Garau X., Desjardins L., Gyapay G., Raynal V., Sinilnikova O. M., Andrieu N., Manié E., de Pauw A., Gesta P., Bonadona V., Maugard C. M., Penet C., Avril M. F., Barillot E., Cabaret O., Delattre O., Richard S., Caron O., Benfodda M., Hu H. H., Soufir N., Bressac-de Paillerets B., Stoppa-Lyonnet D., Stern M. H. (2013) Germline BAP1 mutations predispose to renal cell carcinomas. Am. J. Hum. Genet. 92, 974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farley M. N., Schmidt L. S., Mester J. L., Peña-Llopis S., Pavia-Jimenez A., Christie A., Vocke C. D., Ricketts C. J., Peterson J., Middelton L., Kinch L., Grishin N., Merino M. J., Metwalli A. R., Xing C., Xie X. J., Dahia P. L., Eng C., Linehan W. M., Brugarolas J. (2013) A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol. Cancer Res. 11, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ventii K. H., Devi N. S., Friedrich K. L., Chernova T. A., Tighiouart M., Van Meir E. G., Wilkinson K. D. (2008) BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 68, 6953–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., Modrusan Z., Seshagiri S., Dong K., Lin Z., Balazs M., Suriben R., Newton K., Hymowitz S., Garcia-Manero G., Martin F., Levine R. L., Dixit V. M. (2012) Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Misaghi S., Ottosen S., Izrael-Tomasevic A., Arnott D., Lamkanfi M., Lee J., Liu J., O'Rourke K., Dixit V. M., Wilson A. C. (2009) Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 29, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machida Y. J., Machida Y., Vashisht A. A., Wohlschlegel J. A., Dutta A. (2009) The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 284, 34179–34188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scheuermann J. C., de Ayala Alonso A. G., Oktaba K., Ly-Hartig N., McGinty R. K., Fraterman S., Wilm M., Muir T. W., Müller J. (2010) Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 18. de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., Brockdorff N. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676 [DOI] [PubMed] [Google Scholar]
- 19. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., Reinberg D. (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gutiérrez L., Oktaba K., Scheuermann J. C., Gambetta M. C., Ly-Hartig N., Müller J. (2012) The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development 139, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu H., Mashtalir N., Daou S., Hammond-Martel I., Ross J., Sui G., Hart G. W., Rauscher F. J., 3rd, Drobetsky E., Milot E., Shi Y., Affar el B. (2010) The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 30, 5071–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matatall K. A., Agapova O. A., Onken M. D., Worley L. A., Bowcock A. M., Harbour J. W. (2013) BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer 13, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hannenhalli S., Kaestner K. H. (2009) The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lam E. W., Brosens J. J., Gomes A. R., Koo C. Y. (2013) Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 26. Mahajan A., Yuan C., Lee H., Chen E. S., Wu P. Y., Tsai M. D. (2008) Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal 1, re12. [DOI] [PubMed] [Google Scholar]
- 27. Durocher D., Jackson S. P. (2002) The FHA domain. FEBS Lett. 513, 58–66 [DOI] [PubMed] [Google Scholar]
- 28. Garry D. J., Meeson A., Elterman J., Zhao Y., Yang P., Bassel-Duby R., Williams R. S. (2000) Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. U.S.A. 97, 5416–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hawke T. J., Jiang N., Garry D. J. (2003) Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J. Biol. Chem. 278, 4015–4020 [DOI] [PubMed] [Google Scholar]
- 30. Grant G. D., Gamsby J., Martyanov V., Brooks L., 3rd, George L. K., Mahoney J. M., Loros J. J., Dunlap J. C., Whitfield M. L. (2012) Live-cell monitoring of periodic gene expression in synchronous human cells identifies Forkhead genes involved in cell cycle control. Mol. Biol. Cell 23, 3079–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marais A., Ji Z., Child E. S., Krause E., Mann D. J., Sharrocks A. D. (2010) Cell cycle-dependent regulation of the forkhead transcription factor FOXK2 by CDK.cyclin complexes. J. Biol. Chem. 285, 35728–35739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji Z., Donaldson I. J., Liu J., Hayes A., Zeef L. A., Sharrocks A. D. (2012) The forkhead transcription factor FOXK2 promotes AP-1-mediated transcriptional regulation. Mol. Cell. Biol. 32, 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji Z., Mohammed H., Webber A., Ridsdale J., Han N., Carroll J. S., Sharrocks A. D. (2014) The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res. 42, 6232–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jensen D. E., Proctor M., Marquis S. T., Gardner H. P., Ha S. I., Chodosh L. A., Ishov A. M., Tommerup N., Vissing H., Sekido Y., Minna J., Borodovsky A., Schultz D. C., Wilkinson K. D., Maul G. G., Barlev N., Berger S. L., Prendergast G. C., Rauscher F. J., 3rd. (1998) BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16, 1097–1112 [DOI] [PubMed] [Google Scholar]
- 36. Shi X., Seldin D. C., Garry D. J. (2012) Foxk1 recruits the Sds3 complex and represses gene expression in myogenic progenitors. Biochem. J. 446, 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Komorek J., Kuppuswamy M., Subramanian T., Vijayalingam S., Lomonosova E., Zhao L. J., Mymryk J. S., Schmitt K., Chinnadurai G. (2010) Adenovirus type 5 E1A and E6 proteins of low-risk cutaneous β-human papillomaviruses suppress cell transformation through interaction with FOXK1/K2 transcription factors. J. Virol. 84, 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tyagi S., Chabes A. L., Wysocka J., Herr W. (2007) E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 39. Wysocka J., Myers M. P., Laherty C. D., Eisenman R. N., Herr W. (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17, 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frangini A., Sjöberg M., Roman-Trufero M., Dharmalingam G., Haberle V., Bartke T., Lenhard B., Malumbres M., Vidal M., Dillon N. (2013) The aurora B kinase and the polycomb protein ring1B combine to regulate active promoters in quiescent lymphocytes. Mol. Cell 51, 647–661 [DOI] [PubMed] [Google Scholar]