Background: The terminal organelle is responsible for a new type of cell motility in mycoplasmas.
Results: In this work, the specific interaction between the terminal organelle proteins MG200 and MG491 was identified and characterized by SPR and NMR.
Conclusion: The MG200-MG491 interaction affects motility and cell morphology.
Significance: The MG200-MG491 interaction, critical for the stability of the terminal organelle, might be determinant for the infectiveness of M. genitalium.
Keywords: Cell Motility, Nuclear Magnetic Resonance (NMR), Protein Structure, Protein-Protein Interaction, Structural Biology
Abstract
Several mycoplasmas, such as the emergent human pathogen Mycoplasma genitalium, developed a complex polar structure, known as the terminal organelle (TO), responsible for a new type of cellular motility, which is involved in a variety of cell functions: cell division, adherence to host cells, and virulence. The TO cytoskeleton is organized as a multisubunit dynamic motor, including three main ultrastructures: the terminal button, the electrodense core, and the wheel complex. Here, we describe the interaction between MG200 and MG491, two of the main components of the TO wheel complex that connects the TO with the cell body and the cell membrane. The interaction between MG200 and MG491 has a KD in the 80 nm range, as determined by surface plasmon resonance. The interface between the two partners was confined to the “enriched in aromatic and glycine residues” (EAGR) box of MG200, previously described as a protein-protein interaction domain, and to a 25-residue-long peptide from the C-terminal region of MG491 by surface plasmon resonance and NMR spectroscopy studies. An atomic description of the MG200 EAGR box binding surface was also provided by solution NMR. An M. genitalium mutant lacking the MG491 segment corresponding to the peptide reveals specific alterations in cell motility and cell morphology indicating that the MG200-MG491 interaction plays a key role in the stability and functioning of the TO.
Introduction
Mycoplasmas are a prevalent group of prokaryotes that evolved from Gram-positive bacteria by reductive evolution, becoming obligatory parasites with small cell sizes and minimal genomes (1). Another interesting property of mycoplasmas is the absence of cell wall, which renders penicillin or other β-lactam-based antibiotics ineffective against these pathogens (2). Mycoplasma genitalium belongs to the pneumoniae cluster of mycoplasmas (3) and, with only 482 protein-coding genes, is considered to have one of the smallest genomes among cells capable of growing in axenic cultures (4). This microorganism is also a sexually transmitted human pathogen associated with non-gonoccocal, non-chlamidial urethritis in men and several inflammatory reproductive tract syndromes in women (5–7). Morphologically, M. genitalium presents a polar extension of the cell membrane, known as the terminal organelle (TO),6 that protrudes from the cell body and confers a classical flasklike shape to cells (8, 9). The TO is formed by a highly ordered cytoskeleton that mediates cell division, adherence to host cells, and motility (10–12). TO-containing mycoplasmas can move along solid surfaces by gliding motility (13) despite lacking classic motor proteins, suggesting the existence of a novel prokaryotic motility mechanism (14). The overall structural organization of mycoplasma TO has been described for M. pneumoniae thanks to complementary studies performed with electron microscopy and electron cryotomography (15–17). These studies revealed that the cytoskeleton of the TOs is organized as a multisubunit dynamic motor, which includes three main ultrastructures: a terminal button, a pair of electrodense rods, and a wheel complex. Moreover, the major adhesins of the cell also cluster around the membrane that connects with the TO.
In Mycoplasma pneumoniae, up to 11 proteins have been localized in the TO (10), which must interact to form the TO ultrastructures. Among these, at least four are components of the wheel complex that connects the TO with the cell body and the cell membrane: TopJ, P24, P200, and P41 (18–21). Their respective orthologs in M. genitalium (MG200, MG219, MG386, and MG491) are also thought to constitute the TO wheel complex in this microorganism. Cell and molecular biology studies on these proteins revealed that (i) P24 has an important role in the regulation of the TO formation and in gliding motility (20); (ii) P41 is required for positioning P24, to anchor the TO to the cell body and for the formation of new TOs at WT rates (20, 22); (iii) P200 (23) as well as its ortholog MG386 (24) were shown to be implicated in gliding motility but not in cytadherence; and (iv) TopJ is involved in gliding motility and cytadherence (25), whereas MG200 is involved only in gliding motility (24). P200, TopJ, MG386, and MG200 were found to share some features, such as the presence of unusual acidic and proline-rich domains (26) and also of “enriched in aromatic and glycine residues” (EAGR) boxes (27). The latter, which constitute well conserved domains found only in proteins from mycoplasma TOs, was proposed to participate in protein-protein interactions, and a variant lacking the MG200 EAGR box showed a specific contribution of the domain to cell motility (28).
The lack of detailed molecular information on mycoplasma motility machinery, which is primordial for cell infection and pathogenic processes, together with the increasing necessity to find new antibiotics or multivalent vaccines against the emergent human pathogen M. genitalium (29), led us to study the proteins that compose the TOs of mycoplasmas. In the present study, the interaction between the TO wheel complex proteins MG200 and MG491 was identified by SPR, and the EAGR box from MG200 was demonstrated to be essential for complex formation. Provided the backbone resonance assignment of the MG200 EAGR box, the residues involved in the binding to MG491 were identified through chemical shift perturbation (CSP) mapping studies. Moreover, further SPR and NMR experiments revealed that a 25-residue-long peptide at the C-terminal region of MG491 is responsible for the interaction with MG200. This information results in a model for the MG200-MG491 interaction that contributes to cellular motility in a very specific way, as confirmed by in vivo studies on the MG491 C-terminal peptide deletion mutant.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Escherichia coli XL1-Blue strain was used to amplify the plasmids, and E. coli BL21 (DE3) was used to recombinantly express MG200, MG491, and their variants. M. genitalium G37 WT and its isogenic Δp1 and Δp1TC mutant strains were grown in SP-4 broth (30) at 37 °C under 5% CO2 in tissue culture flasks until reaching the midlog phase of growth as described previously (31). Transformant colonies were isolated on SP-4 agar plates supplemented with 34 μg/ml chloramphenicol (Roche Applied Science) or 100 μg/ml gentamicin (Sigma-Aldrich).
Expression and Purification of Target Proteins
The MG491 coding sequence (GenBankTM accession number Q9ZB78; Fig. 1A) was cloned between NcoI and XhoI restriction sites of the pET21d expression vector (Novagen, Madison, WI). This plasmid was subsequently transformed in E. coli BL21 (DE3) and expressed overnight at 16 °C after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside when cells reached an A600 of 0.6. Cells were recovered by centrifugation at 4500 × g for 30 min, and the whole cell lysate was obtained by resuspending the pellet in lysis buffer (0.05 m Tris-HCl buffer (pH 8.0) containing 0.5 m sodium chloride, 0.01 m imidazole, and 0.3 mg/ml lysozyme) supplemented with a Complete Protease Inhibitor Mixture Tablet (Roche Applied Science) per liter of culture. The suspension was then sonicated and centrifuged twice at 25,000 × g for 30 min at 4 °C. MG491 was purified by immobilized metal ion affinity chromatography using the following buffer system: buffer A composed of 0.05 m Tris-HCl (pH 8.0) and 0.5 m sodium chloride and buffer B with the same composition as buffer A but also containing 0.5 m imidazole. An extra size exclusion chromatography (SEC) purification step followed, which was performed on a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in 0.05 m Tris-HCl (pH 8.0) containing 0.15 m sodium chloride. A truncated version of MG491, named MG491-Nt_1–308 (Fig. 1A), was cloned in a pOPINE vector (OPPF, Oxford, UK) and expressed and purified as the native protein. MG200 and its variants were prepared as described previously (28).
FIGURE 1.
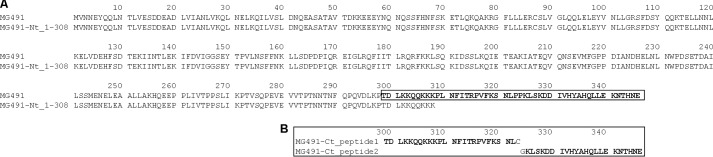
Primary amino acidic sequence of MG491 and variants. A, sequences of MG491 (residues Met1–Glu346) and MG491-Nt_1–308 (residues Met1–Lys308). The C-terminal end of MG491 (residues Thr299–Glu346), where the region responsible for the interaction with MG200 EAGR box is located, is highlighted in boldface type. B, sequences of two synthetic peptides from the C-terminal end of MG491: MG491-Ct_peptide 1 and MG491-Ct_peptide 2.
Surface Plasmon Resonance
The interaction between MG200 and MG491 as well as between some of their domains was analyzed by SPR in a BIAcore® 3000 (GE Healthcare). MG200, MG491, and MG491-Ct_peptide 1 (Fig. 1B) were immobilized as ligands at the carboxymethyldextran surface of a CM5 sensor chip (GE Healthcare) according to the manufacturer's instructions for amine coupling. MG200 and MG491 were diluted to 50 and 30 μg/ml, respectively, in 0.01 m sodium acetate buffer (pH 4.0) for immobilization, whereas MG491-Ct_peptide 1 was diluted to 160 μg/ml in 0.01 m HEPES buffer (pH 7.5). Protein immobilization was performed in three distinct flow cells for 8.5 min for MG200, 0.5 min for MG491, and 5.5 min for MG491-Ct_peptide 1 at a flow rate of 10 μl/min. The reference flow cell was prepared by amine coupling activation followed by deactivation. Measurements were performed at 25 °C in 0.02 m potassium phosphate buffer (pH 7.4) containing 0.1 m sodium chloride and 0.005% (v/v) surfactant P20. After each cycle, the flow cell surfaces were typically regenerated by injecting 10 μl of 0.5 m sodium chloride containing 0.005 m EDTA at 10 μl/min. Dissociation times were 3 min in all cases. Each response curve, shown as unit values, was obtained from the subtraction of the sensorgram of the reference flow cell and of a blank run without protein. For the kinetic measurement, six different concentrations of MG491 were injected for 3 min at a rate of 60 μl/min. Kinetic data analysis was performed using global curve fitting on the BIAevaluation software (version 4.1) assuming a Langmuir 1:1 binding model, shown to be supported by the obtained χ2 analysis. The study of the interaction of MG200 EAGR box with immobilized MG491 or MG491 variants by SPR was attempted, but the domain reacted nonspecifically, with the reference flow cell hindering the measurements.
NMR Spectroscopy
NMR experiments were carried out at 25 °C on a Bruker Avance spectrometer operating at proton frequencies of 700 MHz, equipped with cryoprobe. All NMR spectra were processed with the Topspin Bruker software (version 2.0) and analyzed with CARA.
Assignment of the MG200 EAGR Box
NMR experiments for resonance assignment were performed on 1 mm 13C,15N-MG200 EAGR box in 0.02 m potassium phosphate buffer (pH 6.5) containing 0.1 m sodium chloride and 10% D2O. Backbone resonance assignments of MG200 EAGR box, which were deposited at the BioMagResBank database with ID 25346, were carried out through conventional multidimensional NMR techniques based on triple resonance experiments. Amide protons and nitrogen chemical shifts were measured acquiring 1H,15N heteronuclear single quantum coherence (HSQC) spectra at different pH values (i.e. 6.5, 7.0, and 7.4). The pH-dependent changes in chemical shifts are small (<0.03 ppm) and limited to a few peaks (data not shown), allowing the assignment of the heteronuclear correlation maps at pH 7.0 and 7.4. The ϕ and ψ dihedral angle constraints were derived from the chemical shift analysis by using the TALOS+ program (32).
Relaxation Measurements
A full series of 15N relaxation experiments were carried out on 0.2 mm 15N-MG200 EAGR box prepared in 0.02 m potassium phosphate buffer (pH 7.4) containing 0.1 m sodium chloride to quantify backbone dynamics. Relaxation experiments on 15N-MG200 EAGR box were performed measuring 15N backbone longitudinal (R1) and transverse (R2) relaxation rates and the heteronuclear 1H,15N NOEs.
NMR Titration Experiments
Titrations of 15N-MG200 EAGR box samples with unlabeled MG491, MG491-Nt_1–308, MG491-Ct_peptide 1, or MG491-Ct_peptide 2 (Fig. 1B) were followed through 1H,15N HSQC until the respective ratios of 1:8, 1:4, 1:7, and 1:4 were reached. Experiments were performed in 0.02 m potassium phosphate buffer (pH 7.4) containing 0.1 m sodium chloride with 15N-MG200 EAGR box concentrations ranging from 25 to 50 μm. The CSP (in ppm) of the individual amide pairs was defined as the weighted average 1H and 15N CSPs according to Equation 1 (33, 34),
where ΔδHN and ΔδN are the CSPs between the free and the bound protein forms. KD values were calculated by plotting the weighted average 1H and 15N chemical shifts of the affected residues as a function of the concentration of the unlabeled partner. Curves were fitted considering a one-site binding model (35, 36).
Circular Dichroism (CD) Spectroscopy
CD spectra of MG491 and MG491-Nt_1–308 concentrated to 5 μm were acquired in a Jasco-J700 spectropolarimeter at 20 °C in a 0.1-cm path length cell. Each spectrum is an average of 20 scans recorded in 0.02 m potassium phosphate buffer (pH 7.5) containing 0.05 m sodium fluoride.
Construction of pΔp1cat Plasmid and Mutant Isolation
All of the following procedures were performed according to standard protocols (37). A two-step recombinant PCR was used to construct a fragment of DNA encompassing mg491 and mg219 and excluding the sequence from nucleotide 895 to 969, which encodes for the MG491-Ct_peptide 1. First, 5′ mg491 was amplified using primers mg491/5 and mg491Δp1N while a fragment including mg219 and the 3′-end of mg491 was amplified using primers mg491Δp1C and mg219/3. Second, the two previously obtained fragments were fused using the complementary ends included in primers mg491Δp1N and mg491Δp1C, and the fusion product was amplified using primers mg491/5 and mg219/3, which include XbaI and XhoI restriction sites, respectively. Finally, a second PCR fragment encompassing a 1-kb region downstream from mg219 was amplified using primers HRC/5 and HRC/3, which include XhoI and ApaI restriction sites, respectively. Both PCR products were cleaved to generate cohesive ends, ligated to a pBSK II plasmid digested with XbaI and ApaI, and transformed in E. coli XL1-Blue cells. The resulting pΔp1 plasmid was then cleaved with XhoI and ligated to a PCR fragment containing the cat (chloramphenicol acetyl transferase) gene under the control of the mg438 promoter (31). This PCR fragment was amplified from the pMTncat plasmid (28) using primers cat/5 and cat/3, both including an XhoI restriction site. The resulting pΔp1cat plasmid was used to transform M. genitalium G37 WT cells by electroporation as described previously (31), and cells were then plated in SP-4 medium containing chloramphenicol. Several transformant colonies were then selected for further analyses. The primers used are listed on Table 1.
TABLE 1.
Oligonucleotide list
| Primer name | Oligonucleotide sequencea |
|---|---|
| mg491/5 | GAAGATCTAGAATGGTTAATAATGAATATCAACAAC |
| mg491Δp1N | CTATGTCATCCTTACTTAGTTTCGGAGGTTTGAGATCAACTTGGGGTTGGA |
| mg491Δp1C | TCCAACCCCAAGTTGATCTCAAACCTCCGAAACTAAGTAAGGATGACATAG |
| mg219/3 | GAAGACTCGAGTTAAGATCTGGTTTTTTTATTGC |
| cat/5 | GAACTCTCGAGCAATTGTAGTATTTAGAATTAATAAAGTATG |
| cat/3 | GAAGACTCGAGTTACGCCCCGCCCTGCCACTC |
| HRC/5 | AATATCTCGAGTCTGGTTTTTTTAGTGTTAACAAC |
| HRC/3 | AATATGGGCCCACTAGCTAGATATTGTTCATATTG |
| mg491A | ACCTGAACTGACCTGATAGTG |
| p438mg491 | GGGGTCGACTAGTATTTAGAATTAATAAAGTATGGTTAATAATGAATATCAACAAC |
| mg491/3 | AAAGGCAATTGCTATTCATTATGGGTATTTTTTTC |
a Restriction sites are underlined.
PCR Analysis of the Transformant Colonies
To verify the deletion of the peptide 1-coding sequence, genomic DNAs from several pΔp1cat clones were amplified by PCR with primers mg491A and mg219/3. The intended deletion was identified by the size of the corresponding PCR products and confirmed by sequencing.
Construction of pMTnGm491 Plasmid and Complementation Assay
DNA coding for mg491 ORF was amplified using primers p438mg491 and mg491/3, incorporating the promoter region from mg438 (31) and SalI and MfeI restriction sites. The PCR fragment was cleaved with SalI and MfeI and inserted into pMTnGm (31) digested with SalI and EcoRI. The resulting pMTnGm491 plasmid was transformed in Δp1c1 cells as described previously, and transformant colonies were selected with chloramphenicol and gentamicin for further analyses.
SDS-PAGE and Western Immunoblotting
Total protein extracts from M. genitalium G37 WT and several Δp1 mutant cells were prepared for SDS-PAGE. Gels were stained with Coomassie Brilliant Blue or electrophoretically transferred to PVDF membranes. Membranes were probed as described previously (37) at a 1:1000 dilution with rabbit polyclonal anti-P41 (38) or with mouse polyclonal anti-MG219.7
Microcinematography
The gliding motility of G37 WT and mutant cells was analyzed by time lapse microcinematography as described previously (24) with some modifications. Briefly, several dilutions of the different mycoplasma cells were grown overnight in SP-4 medium on 8-well ibiTreat μ-slides (Ibidi). Then the medium was replaced by fresh SP-4 medium, and cell movement was examined at 37 °C using a Nikon Eclipse TE 2000-E inverted microscope. Frames were captured at 2-s intervals for a total observation time of 2 min with a Digital Sight DS-SMC Nikon camera controlled by NIS-Elements BR software. The frequency of motile cells was determined by examining 230 cells from each strain. The gliding speed of 50 cells and the diameter of the circular tracks from 100 cells of each strain were measured using the ImageJ software and the TrackJ plug-in.
Cell Morphology
M. genitalium G37 WT and mutant cells were grown over coverslips. Samples were fixed, dehydrated, and metalized as described previously (11). Scanning electron micrographs were acquired in a Merlin Zeiss microscope. At least 100 cells from each strain were analyzed using the ImageJ software. Cells smaller than 0.35 μm were classified as minute cells and enumerated for statistical analysis. Cells with a marked curvature in the cell body were also enumerated and classified as bipartite cells.
Statistical Analyses
Statistical analyses were performed using R software (R-project). Student's t tests were conducted to test the significance (p value <0.05) of differences in the mean velocity speed and the diameter of the circular tracks. χ2 tests were performed to examine the presence of significant differences between the frequency of motile cells from each strain and the number of minute or bipartite cells.
RESULTS
Identification of the Interaction between MG200 and MG491
The direct protein-protein interaction between the TO wheel complex proteins MG200 and MG491 was analyzed by SPR-injecting MG491 over MG200, which was covalently bound to a sensor chip by amine coupling to ∼6000 response units (also see “Experimental Procedures”). SPR response increased in proportion to increasing concentrations of MG491, and data were fit to a 1:1 binding model, resulting in a KD of 78.7 nm, which corresponds to association (ka) and dissociation (kd) rates of 2.48 × 104 m−1 s−1 and 1.95 × 10−3 s−1, respectively (Fig. 2). Given the modularity of MG200 and the hypothesis that its EAGR box could participate in protein-protein interactions (28), we also examined by SPR the interaction of MG491, immobilized on a sensor chip by amine coupling to ∼5300 response units, with MG200, with a construct of the MG200 protein lacking the EAGR box (MG200ΔEAGR box), and with the MG200 EAGR box. The injection of MG200ΔEAGR box over MG491 resulted in a flat curve, indicating that there is no interaction between the two protein constructs (data not shown), whereas the measurement of the injection of MG200 or MG200 EAGR box over MG491 was hindered by the nonspecific reactivity of the proteins with the reference flow cell. Solution NMR studies were performed to verify whether the interaction between MG491 and MG200 occurs through its EAGR box domain.
FIGURE 2.
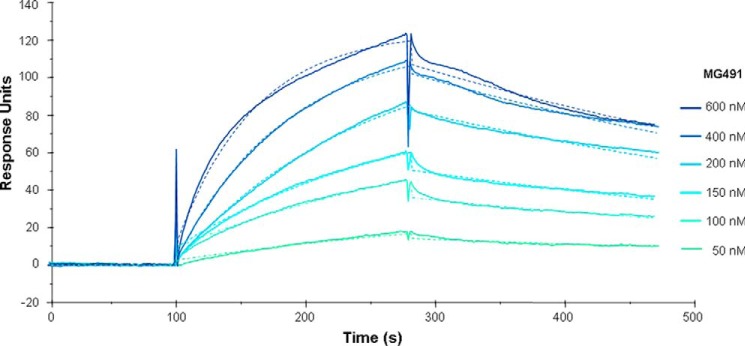
Interaction of immobilized MG200 with increasing concentrations of MG491 as measured by SPR. Colored lines, measured response units; colored dashed lines, calculated fit to a 1:1 Lagmuir binding model, producing calculated constants of ka = 2.48 × 104 m−1 s−1, kd = 1.95 × 10−3 s−1, KD = 78.7 nm, and χ2 = 1.62.
NMR Analysis of the MG200 EAGR Box
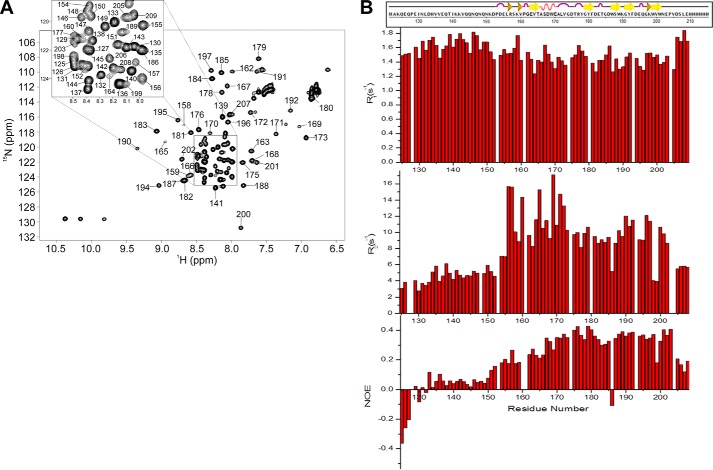
The NMR spectrum of a construct spanning MG200 residues from Lys124 to Ser207 plus a C-terminal His6 tag, the same used for the crystal structure determination of the MG200 EAGR box (28), was assigned. Combining the information derived from triple resonance experiments (39–41), 95% of the Cα, 85% of the CO, 94% of the backbone amides 1H and 15N, and 94% of the Cβ were assigned together with 88% of the side chain carbons and protons, excluding the C-terminal tag. Missing amide resonances correspond to the three first N-terminal residues, to His134, to Gly174, and to His210–His215. Secondary structure elements predicted by TALOS+ from the backbone torsion angle restraints, which were derived from the chemical shift index analysis, match the ones found in the MG200 EAGR box crystal structure (Fig. 3, A and B), suggesting that the domain maintains in solution the same secondary structural organization found in crystals.
FIGURE 3.
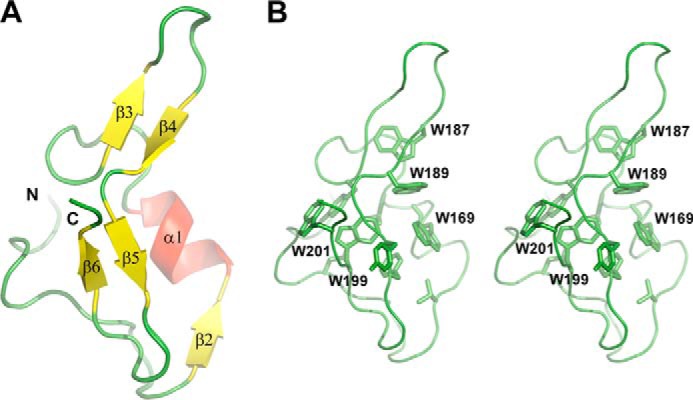
Structure of the MG200 EAGR box domain (Protein Data Bank code 4DCZ). A, schematic representation of the crystal structure of the MG200 EAGR box. The secondary structure element nomenclature is consistent with that previously reported (28). B, stereo view of the MG200 EAGR box with all of the tryptophan residues from the hydrophobic core depicted in stick representations.
In the HSQC spectrum of 15N-MG200 EAGR box (Fig. 4A) a large number of signals belonging to the N-terminal end are clustered in the central part of the spectrum (amide proton resonances between 8.0 and 8.5 ppm), all of them showing negative or low (<0.1) 1H,15N NOE values (Fig. 4B). Both properties indicate that the N-terminal end of the domain is unstructured and highly flexible, which is consistent with the crystallographic data. Detection and assignment of long range NOEs in unfolded polypeptide chains are extremely difficult (42), preventing the structural characterization of the N-terminal end of the MG200 EAGR box and the determination of its orientation with respect to the protein core. Analysis of the heteronuclear relaxation data (15N spin relaxation rates R1 and R2) and the heteronuclear 1H,15N NOE measurements (Fig. 4B) indicates an overall flexibility and fast mobility of the protein (NOEs < 0.5). The global correlation time for molecular tumbling was estimated from the R2/R1 ratio of the protein region showing NOE > 0.25 (residues Val164–Pro204) and is 6.90 ± 1.24 ns, consistent with a protein of the MG200 EAGR box size in monomeric state.
FIGURE 4.
1H,15N HSQC spectrum of 15N-MG200 EAGR box. The spectrum was acquired in 0.02 m potassium phosphate buffer (pH 6.5) containing 0.1 m sodium chloride. A, the proton peaks clustered in the spectral region between 8.0 and 8.5 ppm are enlarged at the top. Of the expected 90 1H,15N backbone resonances, 79 were assigned. The missing resonances are for the first three N-terminal residues, the C-terminal tag (His210–His215), and residues His134, Val136, and Gly174. B, 15N relaxation parameters R1 and R2 and heteronuclear 1H,15N NOEs for each residue of the MG200 EAGR box collected at 700 MHz.
Characterization of the Interaction between MG200 EAGR Box and MG491 by NMR
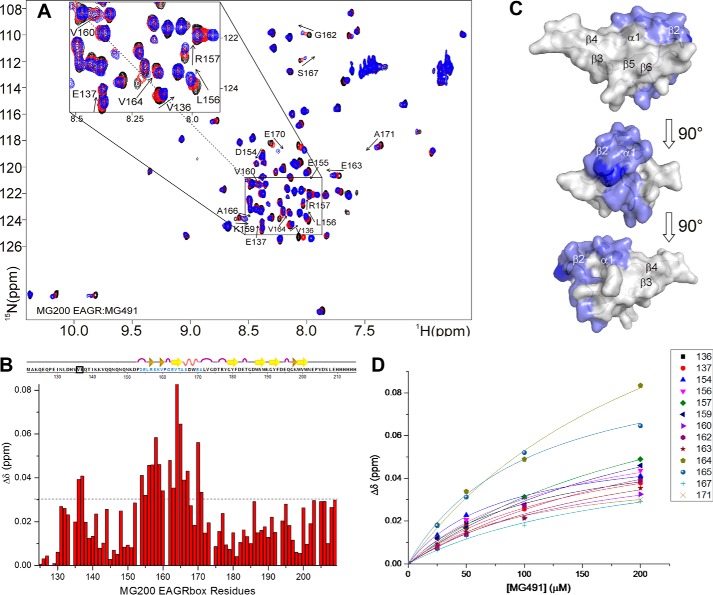
The interaction between MG200 EAGR box and MG491 was investigated at the molecular level by monitoring the complex formation by 1H,15N HSQC spectra. Backbone CSPs of 15N-MG200 EAGR box occurring upon the addition of increasing amounts of unlabeled MG491 (Fig. 5, A and B) reveal the formation of a specific complex in fast exchange between the free and the bound form on the NMR chemical shift time scale. The affected resonances are resolved in the 1H and 15N dimensions, allowing us to follow complex formation. During titration with MG491, the MG200 EAGR box residues experiencing large CSPs (Fig. 5B) cluster in the solvent-exposed face of helix α1 (Ala166, Ser167, Glu170, and Ala171), in strand β2 (Glu163–Thr165), and in the N-terminal residues Val136, Glu137, and Asp154–Gly162 (Fig. 5C). The linearity of the CSPs in the HSQC maps proves that a single binding event is occurring. A KD of 170 ± 40 μm has been estimated, fitting the CSP of selected residues as a function of the concentration of MG491 (Fig. 5D), using a one-site binding model.
FIGURE 5.
Analysis of the chemical shift perturbations of the 15N-MG200 EAGR box in complex with MG491. A, superimposition of 1H,15N HSQC spectra of MG200 EAGR box in the absence and in the presence of MG491. MG200 EAGR box/MG491 complex ratios of 1:0, 1:4, and 1:8 are represented in black, red, and blue traces, respectively. Assigned residues showing an average CSP higher than 0.03 ppm are labeled. Very low intensity peaks are below the displayed threshold in some of the spectra. B, plots of the weighted average CSPs of backbone amide resonances of 15N-MG200 EAGR box between the initial and the final point of the titration with MG491 as a function of residue number. The horizontal dashed line indicates the selected CSP threshold. Residues showing an average CSP higher than 0.03 ppm in the presence of MG491 are highlighted in pale blue in the graphical representation of the secondary structure elements. Pro161 is highlighted in dark blue. Residues above the threshold cluster in the region spanning from residue Asp154 to Ala171, except for residues Val136 and Glu137 (surrounded by a black square). C, CSPs mapping of 15N-MG200 EAGR box residues upon the addition of MG491 on the surface of the MG200 EAGR box structure (PDB accession code 4DCZ). Residues showing an average CSP higher than 0.03 ppm in the presence of MG491 are colored according to B. D, fitting of the weighted average CSPs of selected backbone amide resonances of 15N-MG200 EAGR box as function of the concentration of unlabeled MG491. The average KD for the adduct is 170 ± 40 μm.
To determine the regions of MG491 involved in the interaction with MG200 EAGR box, a soluble N-terminal construct of MG491 (MG491-Nt_1–308; Fig. 1A) was investigated. This construct was designed by using information from a limited proteolysis experiment performed on MG491, which produced a stable globular fragment when cleaved with trypsin in mild conditions, which was afterward identified by MALDI-TOF MS. Moreover, in order to ensure that this fragment preserves structural integrity despite lacking the last 48 residues of MG491, far-UV CD spectra of MG491 and MG491-Nt_1–308 were acquired (Fig. 7). The comparison of these spectra reveals that the MG491-Nt_1–308 variant maintains in solution the same secondary structure motifs found in the full-length protein.
FIGURE 7.
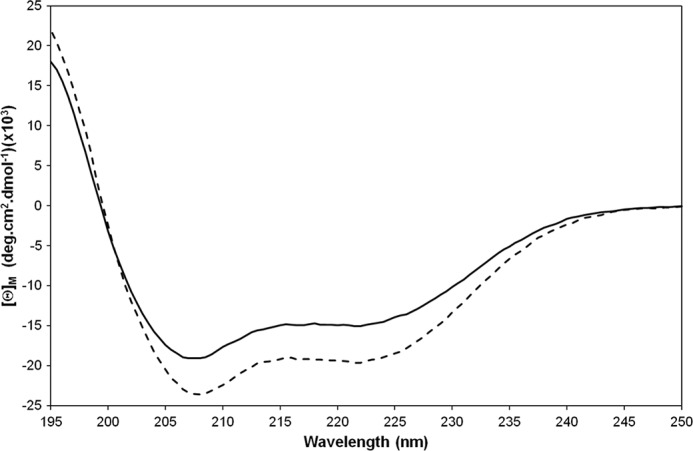
Superimposition of far-UV CD spectra of MG491 (solid line) and MG491-Nt_1–308 (dashed line). Spectra were acquired in 5 μm samples prepared in 0.02 m potassium phosphate buffer (pH 7.5) containing 0.05 m sodium fluoride at 20 °C.
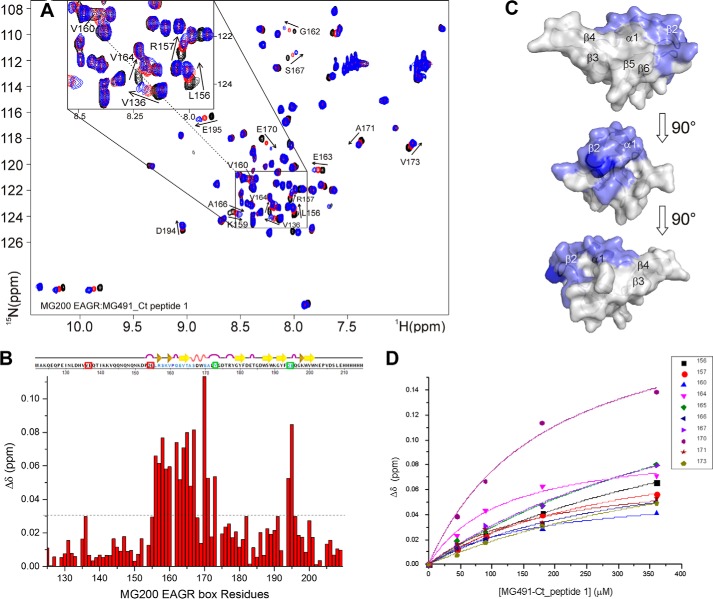
The recognition process was studied by monitoring the CSPs of 15N-MG200 EAGR box resonances in the HSQC spectra upon the addition of increasing concentrations of MG491-Nt_1–308. The absence of variations (data not shown) indicates that the C terminus of MG491 (residues Lys308–Glu346; Fig. 1A), missing in this construct, is critical for the interaction with MG200. To validate this result and given that all attempts to recombinantly express the C-terminal of MG491 failed, the interaction between MG200 EAGR box and two synthetic peptides from the C-terminal of MG491 was studied. The two peptides were custom-synthesized by GenScript (Piscataway, NJ) and correspond to residues Thr299–Leu322 (MG491-Ct_peptide 1) and residues Lys325–Glu346 (MG491-Ct_peptide 2) (Fig. 1B). The titration of the 15N-MG200 EAGR box with unlabeled MG491-Ct_peptide 2 does not show any significant shift (with the exception of the resonances belonging to the C-terminal tag; data not shown), indicating that no specific interaction is taking place. On the contrary, significant CSPs were observed on the titration with unlabeled MG491-Ct_peptide 1, confining the interaction area of MG491 to this region (Fig. 6, A and B). The shifts changes are large and progressive, which is indicative of a weak interaction occurring at a fast rate on the NMR chemical shift time scale, as confirmed by the calculated KD (370 ± 65 μm; Fig. 6D). The weighted average CSPs were plotted as a function of the 15N-MG200 EAGR box backbone resonances and mapped on the protein surface (Fig. 6C). Interacting residues are mainly located in the MG200 EAGR box region that binds to MG491 (i.e. in the solvent-exposed face of helix α1 (Ala166 and Ser167), in strand β2 (Glu163–Thr165), and in the N-terminal residues Leu156–Gly162). Three additional shifts were observed for Val173 and for residues Asp194 and Glu195 from the loop connecting the β-strands β5 and β6, whereas four others are missing (namely Val136, Glu137, Asp154, and Glu155 belonging to the N-terminal end). The MG200 EAGR box residues that differ in the two interactions are equivalent in nature but belong to different protein regions. Val173, Asp194, and Glu195, interacting with MG491-Ct_peptide 1, are adjacent to helix α1 and to strand β2 and are exposed on the same face of the MG200 EAGR box C-terminal end. In contrast, Asp154 and Glu155, involved in the interaction with MG491, are located on the opposite side of the MG200 EAGR box, where the N-terminal end is exposed, suggesting that the two partners adopt two preferential orientations in relation to the MG200 EAGR box surface.
FIGURE 6.
Analysis of the chemical shift perturbations of the 15N-MG200 EAGR box in complex with MG491-Ct_peptide 1. A, superimposition of 1H,15N HSQC spectra of MG200 EAGR box in the absence and in presence of MG491-Ct-peptide 1. MG200 EAGR box/MG491-Ct_peptide 1 complex ratios 1:0, 1:3.5, and 1:7 are represented in black, red, and blue traces, respectively. Assigned residues showing an average CSP higher than 0.03 ppm are labeled. Very low intensity peaks are below the displayed threshold in some of the spectra. B, plots of the weighted average CSPs of backbone amide resonances of 15N-MG200 EAGR box between the initial and the final point of the titration with MG491-Ct_peptide 1 as function of residue number. The horizontal dashed line indicates the selected CSP threshold. Residues showing an average CSP higher than 0.03 ppm in the presence of MG491-Ct_peptide 1 are highlighted in pale blue in the graphical representation of the secondary structure elements. Pro161 is highlighted in dark blue. Residues above the threshold affected by the presence of MG491 but not by peptide 1 are surrounded by red squares, whereas those showing CSPs in the presence of peptide 1 but not of MG491 are surrounded by green squares. C, CSP mapping of 15N-MG200 EAGR box residues upon the addition of MG491-Ct_peptide 1 on the surface of the MG200 EAGR box structure (PDB accession code 4DCZ). Residues showing an average CSP higher than 0.03 ppm in the presence of MG491-Ct_peptide 1 are colored according to B. D, fitting of the weighted average CSPs of selected backbone amide resonances of 15N-MG200 EAGR box as a function of the concentration of unlabeled MG491-Ct_peptide 1. The average KD for the adduct is 370 ± 65 μm.
Validation of the Interacting Regions in MG200 and MG491 by SPR
To validate the results obtained by NMR, the interaction of the MG200 EAGR box with MG491-Ct_peptide 1 was investigated by SPR. MG491 or MG491-Nt_1–308 (Fig. 7) was injected over immobilized MG200 (Fig. 8A), and MG200 or MG200ΔEAGR box was injected over immobilized MG491-Ct_peptide 1 (Fig. 8B). Injecting MG491 over MG200 gave a clear SPR response in contrast to the lack of signal resultant from the injection of MG491-Nt_1–308 over MG200. On the alternative experimental configuration, injecting MG200 over MG491-Ct_peptide 1 gave an SPR response that was nonexistent when MG200ΔEAGR box was injected on the same flow cell. Together, these results clearly demonstrate that MG200 binds MG491 through its EAGR box domain and that MG491 binds MG200 through a 25-residue-long peptide from its C-terminal region.
FIGURE 8.
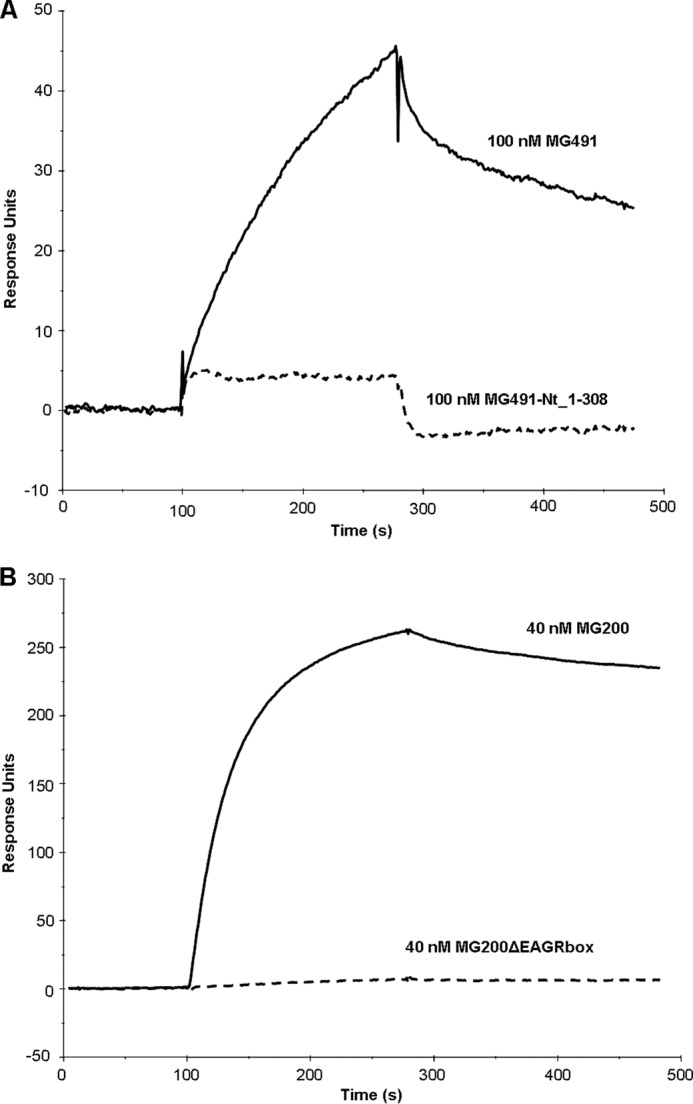
Comparative analysis of the interactions between variants of the MG200 and MG491 proteins by SPR. SPR sensorgrams of 100 nm MG491 and 100 nm MG491-Nt_1–308 injected over the MG200-immobilized flow cell (A) and of 40 nm MG200 and 40 nm MG200ΔEAGR box injected over the MG491-Ct_peptide 1-immobilized flow cell (B).
Isolation of an M. genitalium MG491-Ct_peptide 1-deficient Mutant
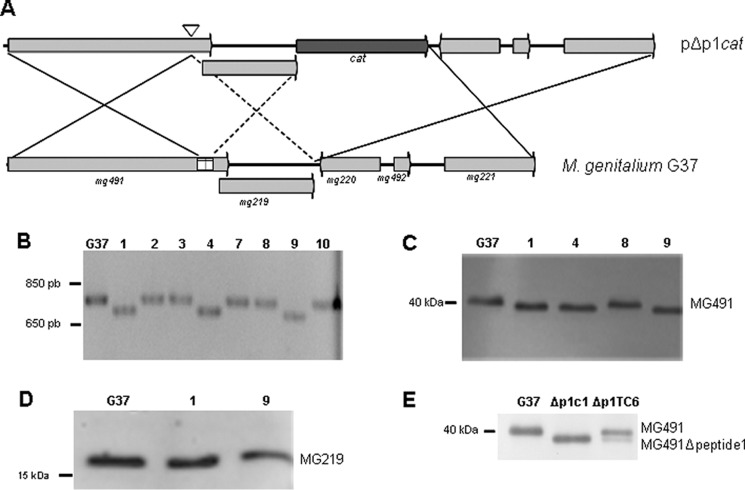
An M. genitalium G37 mutant strain lacking the MG491-Ct_peptide 1 was engineered to determine the in vivo function of peptide 1. Given the overlap of the sequence of mg491 with the 5′-end of the downstream mg219 gene (Fig. 9A), the cat marker, used to select the recombinant clones, was not placed at the 3′-end of mg491. Instead, the marker gene was included at the 3′-end of mg219 to avoid interferences with its transcription and/or translation. Because this strategy may result in a high frequency of non-recombinant clones (Fig. 9A), a PCR-based screening was designed to test the presence of the intended deletion among the transformant colonies. Approximately 30% of the recovered clones showed a PCR fragment consistent with the deletion of a 75-bp fragment at the 3′ region of mg491 (Fig. 9B), which was further confirmed by sequencing. The intended deletion was also demonstrated by Western blot analysis of the three recombinant clones using anti-P41 antibodies (38) and by a shift in the electrophoretic mobility of the MG491 protein band (Fig. 9C). Clones 1 and 9 were further investigated for the existence of polar effects affecting the expression of MG219, and no differences were detected by Western blot (Fig. 9D). Protein profiles from clone 1 cells were also examined by SDS-PAGE and Western blot, and no downstream events were detected affecting the levels of the main mycoplasma TO proteins HMW1, HMW2, P140, P110, MG386, and MG200 (data not shown). Clone 1 was named Δp1c1 and selected for further studies.
FIGURE 9.
Engineering of the Δp1cat mutant strain. A, possible crossover events between the WT G37 genome and the pΔp1 plasmid. Solid lines, crossover events resulting in the deletion of peptide 1 coding region and in the insertion of cat. Dashed lines indicate a crossover event resulting also in the insertion of cat but not in the deletion of peptide 1. B, screening of pΔp1cat transformant colonies by PCR using genomic DNA as template. The deletion of the region coding for peptide 1 was identified by the presence of a PCR band (∼700 bp) smaller than that of the WT (clones 1, 4, and 9). C, Western blot analysis of G37 WT and Δp1 clones 1, 4, 8, and 9 using a polyclonal antiserum against the M. pneumoniae P41 protein (ortholog of MG491). Clones 1, 4, and 9 show a shift in the MG491 band, consistent with the deletion of the 25 residues of the MG491-Ct_peptide 1. D, Western blot analysis of G37 WT and Δp1clones 1 and 9 using a polyclonal antiserum against MG219. Because no band shift is apparent, polar effects derived from the deletion of peptide 1 were discarded. E, Western blot analysis using anti-P41 antibodies of G37 WT, Δp1c1, and Δp1TC6 (Δp1 clone complemented with a copy of mg491) cells. Δp1TC6 cells present two bands corresponding to MG491 and MG491ΔCt_peptide 1 mutant protein.
Complementation Assay of the M. genitalium Strain Δp1c1
To discard the presence of other modifications in Δp1c1 cells, a WT copy of mg491 was inserted on these cells by transposon delivery. Western blot analysis of several of these colonies revealed the presence of both MG491 and MG491ΔCt_peptide 1 (Fig. 9E), and a complemented clone, named Δp1TC6, was selected for further studies.
Gliding Motility and Cell Morphology of the M. genitalium Mutant Strains
The motile properties of the Δp1c1 and Δp1TC6 cells were investigated by time lapse microcinematography, and the frequency of motile cells, the mean gliding velocity, and the diameter of the circular tracks were compared with those of G37 WT cells (Fig. 10, A–C, Table 2, and supplemental Movies S1–S3). The mean velocity and the ratio of motile cells of the Δp1c1 strain were similar to those found for WT cells. However, a significant reduction in the diameter of the circular tracks performed by Δp1c1 motile cells was found (Table 2), suggesting that cells would have less capacity to disseminate than WT cells that glide, drawing wider circular tracks, which confers on them a higher probability to move away from their original positions (Fig. 10, A and B) (11, 24). Upon the reinsertion of a WT copy of mg491 in Δp1c1 cells (Δp1TC6 strain), the diameter of the circular tracks recovered almost to WT levels (Table 2).
FIGURE 10.
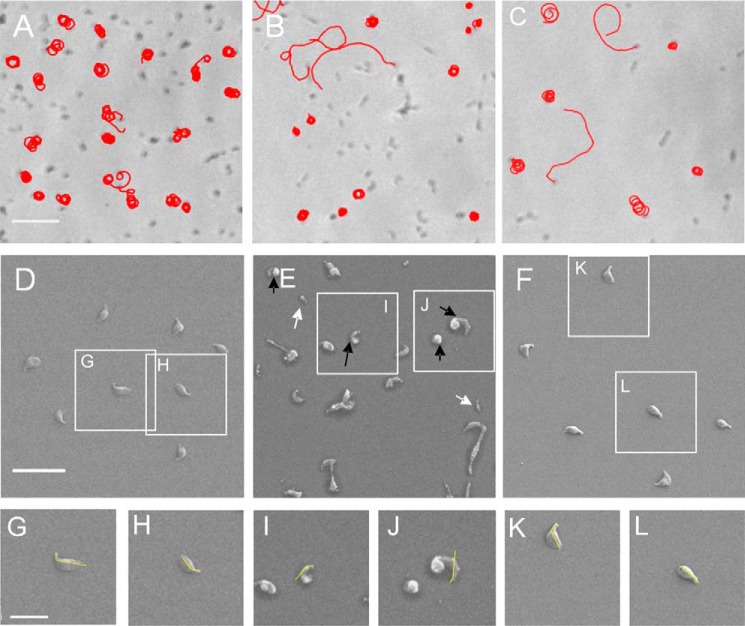
Gliding motility and cell morphology of Δp1c1 and Δp1TC6 cells. A–C, microcinematographs of G37 WT, Δp1c1, and Δp1TC6 cells, respectively. Bar, 5 μm. D–L, scanning electron micrographs of G37 WT, Δp1c1, and Δp1TC6 cells. D and insets G and H are from G37 WT cells; E and insets I and J are from Δp1c1 cells; and F and insets K and L are from Δp1TC6 cells. Yellow lines, TO-body axis angle. White and black arrows point to minute cells and bipartite cell bodies, respectively, detected in Δp1c1 cells. Bars, 2 μm in D and 1 μm in G.
TABLE 2.
Gliding properties and cell morphology of M. genitalium WT, Δp1, and Δp1C6 cells
| Cells | Diameter of circular tracksa | TO-body axis angle | Bipartite cells | Minute cells (0.35 μm) |
|---|---|---|---|---|
| μm | degrees | % | % | |
| G37 WT | 1.05 ± 0.02 | 136 ± 2 | 2 | 0 |
| Δp1 | 0.81 ± 0.03b | 142 ± 2 | 21b | 17.5b |
| Δp1C6 | 1.05 ± 0.04 | 142 ± 2 | 8 | 4 |
a Values shown include ± S.E.
b Statistical difference with G37 WT cells (p value < 0.05).
The presence of cells showing altered patterns of gliding motility prompted us to investigate cell morphology and TO architecture by scanning electron microscopy (Fig. 10, D–L). M. genitalium G37 WT cells have curved TOs, and this curvature correlates well with the diameter of the circular tracks found for Δp1TC6 cells (43). Although no significant differences were observed for the TO-body axis angle between Δp1c1 and G37 WT cells, the presence of cells showing bipartite cell bodies with a strong curvature between the two parts was noteworthy (Fig. 10E and Table 2). This provides an explanation for the narrow tracks observed for Δp1c1 cells. The bipartite cell bodies, which were never observed in G37 WT cells, could be in the origin of the large amount of minute cells detected when examining scanning electron micrographs of Δp1c1 cells (Fig. 10E). As expected, the frequency of bipartite and minute cells was strongly reduced in Δp1TC6 cells, but a significant number of these were still present (Table 2), indicating that reinserting a WT copy of MG491 does not fully complement the mutant phenotype, probably due to the presence of a small amount of MG491ΔCt_peptide 1 mutant protein in the Δp1TC6 cells (Fig. 9E). Minute cells are very uncommon in WT cells, suggesting that they are reminiscent of TOs detached from the cell body. Similar detachments were observed by Hasselbring and Krause in 2007 (22) when characterizing an M. pneumoniae mutant of P41 (ortholog of MG491) where P41 was determined to play an important role in the stabilization of the cytoskeleton by anchoring the TO to the cell body. In the present study, MG491 was observed to have a role similar to that assigned to P41. Moreover, the peptide 1 was found to be essential for the protein function, and consequently the interaction between MG491 and MG200 was determined to be critical for the stability of the mycoplasma TO cytoskeleton and for normal gliding motility.
DISCUSSION
The human pathogen M. genitalium and other motile mycoplasmas present a unique type of motility (gliding motility), which is related to a complex cytoskeleton known as the terminal organelle. M. genitalium, with the smallest genome among self-replicating organisms, appears as a suitable system to attempt a complete characterization of the structure and functioning of the TO, zooming in resolution from the atomic level to cell morphology. This work focused on two proteins of the TO wheel complex ultrastructure, MG200 and MG491.
The interaction between MG200 and MG491 was identified by SPR, with a KD in the 80 nm range. Because the MG200 EAGR box was previously described to be a protein-protein interaction domain (28), its interaction with MG491 was examined by NMR. The HSQC spectrum of 15N-MG200 EAGR box has been assigned, and the global correlation time for molecular tumbling derived from the analysis of the heteronuclear relaxation data and the heteronuclear 1H,15N NOE measurements indicated that the domain was mainly in a monomeric state, whereas dimers were found both in the crystal structure and in solution by SEC-multiangle light scattering (MALS). Detection of only the dimeric species by SEC-MALS might be due to the fact that light scattering has higher sensitivity to bigger particles. Moreover, a monomer/dimer equilibrium was also supported by nanoelectrospray ionization-MS analysis, which revealed the presence of both species in solution.
The CSPs occurring in the HSQC spectra of 15N-MG200 EAGR box when titrated with increasing amounts of MG491 allowed the determination of the residues from the MG200 EAGR box that participate in the complex formation (Fig. 5, A and B). The possible interaction of the MG200 EAGR box with a construct that lacked the last 48 residues from MG491 (MG491-Nt_1–308) was assayed, and the absence of CSPs indicated that the C-terminal end of MG491, missing in this construct, was critical for the interaction with MG200. This result was confirmed by titrating 15N-MG200 EAGR box with two peptides from the C-terminal end of MG491 (Fig. 1B). The significant CSPs observed for the titration with MG491-Ct_peptide 1 in contrast to the lack of variations for the titration with MG491-Ct_peptide 2 allowed the determination that that only the MG491-Ct_peptide 1 participates in the interaction with the MG200 EAGR box. Moreover, the shift trajectories of the MG200 EAGR box in complex with MG491-Ct_peptide 1 are similar to those observed for MG491 (Figs. 5 (A and B) and 6 (A and B)), suggesting that the chemical environment of the protein residues is equivalent in the two adducts. From the comparison of the changes observed for the titrations with MG491 or MG491-Ct_peptide 1, it is apparent that they are also significantly similar in magnitude and distribution on the protein surface (Figs. 5C and 6C). However, some additional shifts were observed when titrating with MG491-Ct_peptide 1 in comparison with MG491 for residues Asp194 and Glu195 from the loop connecting the β-strands β5 and β6, which can be due to new interactions in this region. Also, for MG491-Ct_peptide 1, there are a few missing shifts corresponding to residues from the N-terminal end (namely for Val136, Glu137, Asp154, and Glu155), very likely due to its smaller size with respect to MG491, which gives a smaller binding surface and a relatively higher KD (lower affinity). Altogether, these observations confirm that the EAGR box is a platform for specific protein-protein interactions between the proteins that form the TO ultrastructures, as suggested by the analysis of the MG200 EAGR box structure (28). However, and very surprisingly, the region of the domain that is prompt to interact with other proteins is not the solvent-exposed β3-β4 hairpin, as predicted from the analysis of the crystal structure, but a region at the N-terminal end of the domain, spanning from residue Asp154 to Ser167, that includes the solvent-exposed face of helix α1 (Ala166, Ser167, Glu170, and Ala171) and strand β2 (Glu163–Thr165), which are part of the dimerization interface (Fig. 11). Finally, the NMR data were completely corroborated by SPR, confirming that MG200 binds MG491 through its EAGR box domain and that MG491 binds MG200 through a 25-residue-long peptide from its C-terminal region. The biophysical and structural information, combined with the crystal structure of the MG200 EAGR box, results in a model of the MG200-MG491 interaction that affects cell motility and cell morphology in a very specific way, as determined by the in vivo study of the MG491-Ct_peptide 1 deletion mutant. Therefore, the interaction between the TO wheel complex proteins MG200 and MG491 was revealed to be critical to the stability of the TO, suggesting that it could affect mycoplasma infective capabilities.
FIGURE 11.
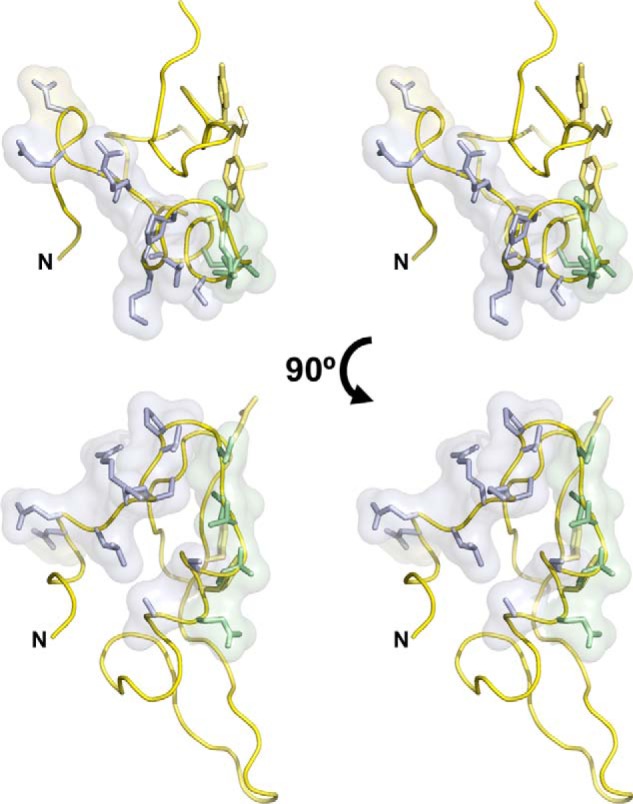
Binding surface of MG200 EAGR box with MG491. Stereo view representations of the MG200 EAGR box with the residues participating only in the interaction with MG491 depicted in blue sticks. Residues depicted in yellow sticks participate in the formation of the crystallographic dimer, whereas residues depicted in green participate both in the interaction with MG491 and in dimer formation.
Supplementary Material
Acknowledgments
We especially thank the Instruct Centre CERM (Italy) for access to the NMR Research Infrastructure. We also acknowledge Isabel Bally from the Institut de Biologie Structurale (CEA, CNRS, Université Joseph Fourier, Grenoble, France) for access to the SPR technical platform of the Grenoble Partnership for Structural Biology (PSB) and Nicole Thielens (CEA, CNRS, Université Joseph Fourier, Grenoble, France) for help in the acquisition and interpretation of SPR data. This work used the platforms of the Grenoble Instruct Centre (ISBG; UMS, 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble PSB. Many thanks are given to M. Pons (BioNMR Group, Facultat de Quimica, UB, Spain) and Daniele de Sanctis (Structural Biology Group, ESRF, France) for helpful scientific discussions.
This work was supported by Ministerio de Ciencia e Innovacion Grants BFU2012-36827 (to I. F.) and BFU2010-22209-C02-01 (to E. Q.), a grant from the Centre de Referencia de R+D de Biotecnologia (Generalitat de Catalunya, Spain) (to E. Q.), and by FEDER funds through the Operational Competitiveness Programme-COMPETE and by Portuguese national funds through FCT-Fundação para a Ciência e a Tecnologia under Project FCOMP-01-0124-FEDER-027581 (EXPL/BBB-BQB/0546/2012) (to B. C.). The NMR characterization was conducted through the FP7 Access to Research Infrastructures (Bio-NMR Contract 261863) and by Instruct, which is part of the European Strategy Forum on Research Infrastructures (ESFRI) and supported by national member subscriptions.

This article contains supplemental Movies S1–S3.
L. Martinelli, D. Lalli, L. García-Morales, M. Ratera, E. Querol, J. Piñol, I. Fita, and B. M. Calisto, manuscript in preparation.
- TO
- terminal organelle
- EAGR
- enriched in aromatic and glycine residues
- SPR
- surface plasmon resonance
- CSP
- chemical shift perturbation
- SEC
- size exclusion chromatography
- HSQC
- heteronuclear single quantum coherence
- MALS
- multiangle light scattering.
REFERENCES
- 1. Sirand-Pugnet P., Citti C., Barrec A., Blanchard A. (2007) Evolution of mollicutes: down a bumpy road with twists and turns. Res. Microbiol. 158, 754–766 [DOI] [PubMed] [Google Scholar]
- 2. Taylor-Robinson D., Bébéar C. (1997) Antibiotic susceptibilities of mycoplasmas and treatment of mycoplasmal infections. J. Antimicrob. Chemother. 40, 622–630 [DOI] [PubMed] [Google Scholar]
- 3. Johansson K. E., Pettersson B. (2002) in Molecular Biology and Pathogenicity of the Mycoplasmas, pp. 1–29, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 4. Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., Maruf M., Hutchison C. A., 3rd, Smith H. O., Venter J. C. (2006) Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 103, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanchard A., Bébéar C. (2011) The evolution of Mycoplasma genitalium. Ann. N.Y. Acad. Sci. 1230, E61–E64 [DOI] [PubMed] [Google Scholar]
- 6. McGowin C. L., Anderson-Smits C. (2011) Mycoplasma genitalium: an emerging cause of sexually transmitted disease in women. PLoS Pathog. 7, e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sweet R. L. (2012) Pelvic inflammatory disease: current concepts of diagnosis and management. Curr. Infect. Dis. Rep. 10.1007/s11908-012-0243-y [DOI] [PubMed] [Google Scholar]
- 8. Hatchel J. M., Balish M. F. (2008) Attachment organelle ultrastructure correlates with phylogeny, not gliding motility properties, in Mycoplasma pneumoniae relatives. Microbiology 154, 286–295 [DOI] [PubMed] [Google Scholar]
- 9. Kirchhoff H., Rosengarten R., Lotz W., Fischer M., Lopatta D. (1984) Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Isr. J. Med. Sci. 20, 848–853 [PubMed] [Google Scholar]
- 10. Balish M. F., Krause D. C. (2006) Mycoplasmas: a distinct cytoskeleton for wall-less bacteria. J. Mol. Microbiol. Biotechnol. 11, 244–255 [DOI] [PubMed] [Google Scholar]
- 11. Burgos R., Pich O. Q., Ferrer-Navarro M., Baseman J. B., Querol E., Piñol J. (2006) Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal-organelle development. J. Bacteriol. 188, 8627–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lluch-Senar M., Querol E., Piñol J. (2010) Cell division in a minimal bacterium in the absence of ftsZ. Mol. Microbiol. 78, 278–289 [DOI] [PubMed] [Google Scholar]
- 13. Kirchhoff H., Maass C., Runge M., Franz B., Schmidt R., Quentmeier H., Mühlradt P. F. (1992) Tetrazolium (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) reduction by mycoplasmas. Int. J. Syst. Bacteriol. 42, 506–508 [DOI] [PubMed] [Google Scholar]
- 14. Miyata M. (2008) Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16, 6–12 [DOI] [PubMed] [Google Scholar]
- 15. Hegermann J., Herrmann R., Mayer F. (2002) Cytoskeletal elements in the bacterium Mycoplasma pneumoniae. Naturwissenschaften 89, 453–458 [DOI] [PubMed] [Google Scholar]
- 16. Henderson G. P., Jensen G. J. (2006) Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60, 376–385 [DOI] [PubMed] [Google Scholar]
- 17. Seybert A., Herrmann R., Frangakis A. S. (2006) Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 156, 342–354 [DOI] [PubMed] [Google Scholar]
- 18. Catrein I., Herrmann R. (2011) The proteome of Mycoplasma pneumoniae, a supposedly “simple” cell. Proteomics 11, 3614–3632 [DOI] [PubMed] [Google Scholar]
- 19. Cloward J. M., Krause D. C. (2009) Mycoplasma pneumoniae J-domain protein required for terminal organelle function. Mol. Microbiol. 71, 1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hasselbring B. M., Krause D. C. (2007) Proteins P24 and P41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae. J. Bacteriol. 189, 7442–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenri T., Seto S., Horino A., Sasaki Y., Sasaki T., Miyata M. (2004) Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186, 6944–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasselbring B. M., Krause D. C. (2007) Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol. Microbiol. 63, 44–53 [DOI] [PubMed] [Google Scholar]
- 23. Jordan J. L., Chang H. Y., Balish M. F., Holt L. S., Bose S. R., Hasselbring B. M., Waldo R. H., 3rd, Krunkosky T. M., Krause D. C. (2007) Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect. Immun. 75, 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pich O. Q., Burgos R., Ferrer-Navarro M., Querol E., Piñol J. (2006) Mycoplasma genitalium mg200 and mg386 genes are involved in gliding motility but not in cytadherence. Mol. Microbiol. 60, 1509–1519 [DOI] [PubMed] [Google Scholar]
- 25. Cloward J. M., Krause D. C. (2010) Functional domain analysis of the Mycoplasma pneumoniae co-chaperone TopJ. Mol. Microbiol. 77, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Proft T., Hilbert H., Plagens H., Herrmann R. (1996) The P200 protein of Mycoplasma pneumoniae shows common features with the cytadherence-associated proteins HMW1 and HMW3. Gene 171, 79–82 [DOI] [PubMed] [Google Scholar]
- 27. Balish M. F., Hahn T. W., Popham P. L., Krause D. C. (2001) Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the Triton shell. J. Bacteriol. 183, 3680–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calisto B. M., Broto A., Martinelli L., Querol E., Piñol J., Fita I. (2012) The EAGR box structure: a motif involved in mycoplasma motility. Mol. Microbiol. 86, 382–393 [DOI] [PubMed] [Google Scholar]
- 29. Citti C., Blanchard A. (2013) Mycoplasmas and their host: emerging and re-emerging minimal pathogens. Trends Microbiol. 21, 196–203 [DOI] [PubMed] [Google Scholar]
- 30. Tully J. G., Rose D. L., Whitcomb R. F., Wenzel R. P. (1979) Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J. Infect. Dis. 139, 478–482 [DOI] [PubMed] [Google Scholar]
- 31. Pich O. Q., Burgos R., Planell R., Querol E., Piñol J. (2006) Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152, 519–527 [DOI] [PubMed] [Google Scholar]
- 32. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garrett D. S., Seok Y. J., Peterkofsky A., Clore G. M., Gronenborn A. M. (1997) Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry 36, 4393–4398 [DOI] [PubMed] [Google Scholar]
- 34. Grzesiek S., Bax A., Clore G. M., Gronenborn A. M., Hu J. S., Kaufman J., Palmer I., Stahl S. J., Wingfield P. T. (1996) The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 3, 340–345 [DOI] [PubMed] [Google Scholar]
- 35. Fielding L. (2007) NMR methods for the determination of protein-ligand dissociation constants. Prog. Nucl. Magn. Reson. Spectrosc. 10.1016/j.pnmrs.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 36. Meyer B., Peters T. (2003) NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 42, 864–890 [DOI] [PubMed] [Google Scholar]
- 37. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Krause D. C., Proft T., Hedreyda C. T., Hilbert H., Plagens H., Herrmann R. (1997) Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 179, 2668–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kay L. E., Ikura M., Tschudin R., Bax A. (2011) Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. 1990. J. Magn. Reson. 213, 423–441 [DOI] [PubMed] [Google Scholar]
- 40. Wider G., Wüthrich K. (1999) NMR spectroscopy of large molecules and multimolecular assemblies in solution. Curr. Opin. Struct. Biol. 9, 594–601 [DOI] [PubMed] [Google Scholar]
- 41. Wüthrich K. (1990) Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 265, 22059–22062 [PubMed] [Google Scholar]
- 42. Dyson H. J., Wright P. E. (2004) Unfolded proteins and protein folding studied by NMR. Chem. Rev. 104, 3607–3622 [DOI] [PubMed] [Google Scholar]
- 43. Burgos R., Pich O. Q., Querol E., Piñol J. (2008) Deletion of the Mycoplasma genitalium MG_217 gene modifies cell gliding behaviour by altering terminal organelle curvature. Mol. Microbiol. 69, 1029–1040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.