Background: Recruitment of peripheral proteins to the inner membrane complex (IMC) of the malaria parasite can be mediated by N-terminal acylation.
Results: Characterization of substrate determinants and identification of an IMC-localized palmitoyl acyltransferase PfDHHC1.
Conclusion: Residues close to palmitoylation sites interfere with specific IMC recruitment. PfDHHC1 represents an apicomplexan-specific PAT.
Significance: Dissection of palmitoylation for protein recruitment to the inner membrane complex in P. falciparum.
Keywords: Malaria, Membrane Trafficking, Phylogenetics, Plasmodium, Protein Acylation, Inner Membrane Complex, Palmitoyl Acyltransferase
Abstract
To survive and persist within its human host, the malaria parasite Plasmodium falciparum utilizes a battery of lineage-specific innovations to invade and multiply in human erythrocytes. With central roles in invasion and cytokinesis, the inner membrane complex, a Golgi-derived double membrane structure underlying the plasma membrane of the parasite, represents a unique and unifying structure characteristic to all organisms belonging to a large phylogenetic group called Alveolata. More than 30 structurally and phylogenetically distinct proteins are embedded in the IMC, where a portion of these proteins displays N-terminal acylation motifs. Although N-terminal myristoylation is catalyzed co-translationally within the cytoplasm of the parasite, palmitoylation takes place at membranes and is mediated by palmitoyl acyltransferases (PATs). Here, we identify a PAT (PfDHHC1) that is exclusively localized to the IMC. Systematic phylogenetic analysis of the alveolate PAT family reveals PfDHHC1 to be a member of a highly conserved, apicomplexan-specific clade of PATs. We show that during schizogony this enzyme has an identical distribution like two dual-acylated, IMC-localized proteins (PfISP1 and PfISP3). We used these proteins to probe into specific sequence requirements for IMC-specific membrane recruitment and their interaction with differentially localized PATs of the parasite.
Introduction
Malaria tropica, the most severe form of human malaria, is caused by the apicomplexan parasite Plasmodium falciparum and represents one of the major human health problems in endemic countries. Half of the world's population is at risk, and approximately one million people die annually, the majority being children under the age of 5 years in sub-Saharan Africa (1). From an anthropocentric point of view, the most important part of the parasite's complex life cycle is the asexual and intra-erythrocytic developmental cycle, which causes all symptoms associated with malaria. Invasion of human red blood cells by merozoites initiates this intracellular replication cycle. This process is coordinated and powered by the parasite and relies on a sophisticated protein network with various levels of hierarchy and control points (2). Of critical importance to host cell invasion are rhoptries and micronemes, the secretory organelles at the apical pole of the parasite, as well as the inner membrane complex (IMC),4 a membranous structure consisting of two layers located on the cell's sub-surface, underneath the plasma membrane (3). Although the secretory organelles are characteristic of all organisms belonging to the apicomplexan phylum, the IMC is a morphological trait of a large phylogenetic group called Alveolata (4). The Alveolata comprise the Dinoflagellata (typically marine flagellates), Ciliata (like Paramecium spp.), and the parasitic Apicomplexa, including the genera Theileria, Eimeria, Toxoplasma, and Plasmodium. For dinoflagellates and ciliates, largely aquatic groups, the IMC appears to have a predominantly structural role in cell architecture. However, for apicomplexans, an additional function is its role in cell motility and cytokinesis (5).
To date, over 25 IMC resident proteins have been identified (3, 5). Phylogenetic analysis has demonstrated that in addition to a common core set of conserved proteins, the IMC includes many lineage-specific proteins, reflecting additional adaptations to specialized requirements of the compartment within the respective phylogenetic group (6). Functional specialization of IMC proteins is exemplified by a group of well characterized proteins that form the so-called glideosome, the motor complex that drives the locomotion of all motile parasite stages (7–9). In addition to IMC resident proteins, studies have recently revealed an additional IMC-associated protein family termed IMC sub-compartment proteins (ISPs) (10). Originally discovered in P. falciparum (11), ISPs have also been the focus of detailed investigations in Toxoplasma gondii (11–13). Although the precise functions of ISPs are still unknown, knock-out studies in T. gondii and Plasmodium berghei suggest that they play a role in cell polarity and daughter cell formation and are not connected to cell motility (11, 12, 14).
PfISPs are relatively small (144 and 148 amino acids for PfISP1/PF10_0107 and PfISP3/PF14_0578, respectively) and display N-terminal myristoylation and palmitoylation motifs. It was previously shown that, in contrast to the glideosome-associated protein 45 (GAP45), the N-terminal domain of the ISPs is sufficient for IMC membrane recruitment (10, 15). Although myristoylation of proteins is catalyzed by a cytosolic N-myristoyltransferase (PF14_0127; see Ref. 16), palmitoylation is catalyzed by palmitoyl acyltransferases (PATs) and is associated with distinct membrane systems (17–19). The function of palmitoylation is not restricted to membrane attachment, but it also regulates protein-protein interaction and therefore multiple cellular processes (20). To date, over 400 P. falciparum proteins have been shown to be palmitoylated (21), including proteins essential for asexual development and erythrocyte invasion. PATs are polytopic membrane proteins, possessing four or more transmembrane domains (19, 22). Organism complements of PATs vary from over 20 members in metazoan species to only seven in the budding yeast Saccharomyces cerevisiae (23). Tetrahymena, Toxoplasma, and Plasmodium possess 38, 18, and 12 predicted PATs, respectively. PATs reside in different tissues and sub-cellular localizations and are thought to direct the recruitment of proteins to specific membranes: for example, Vac8 to vacuole membrane in yeast (24), RAS2 in mammalian cell lines to the plasma membrane (25), and TgARO in T. gondii to the rhoptry membrane (26, 27). A localization map of PATs in T. gondii and P. berghei was recently established, with two PATs found to localize to the IMC (26, 28). Here, we expand these studies to P. falciparum. We deliver a detailed phylogenetic analysis and dissect the specific sequence requirements that are essential for dual-acylated proteins to be localized to the IMC.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection of P. falciparum
P. falciparum (3D7) was cultured in human O+ erythrocytes according to standard procedures using complete Roswell Park Memorial Institute medium (29). Gametocytes were produced using a modified version of the established protocol (6, 30). For transfection, ring-stage parasites (10%) were electroporated with 100 μg of plasmid DNA resuspended in cytomix as described previously (31). Transfectants were selected using 10 nm WR99210 (for pARL-based vectors) or 30 nm blasticidin (for pBcam-based vectors), respectively. For single crossover integration, the following transfection parasites were alternately grown with and without WR99210 pressure (∼4 weeks for each interval off-drug) to promote integration of the transfection vectors in the endogenous loci. Double transfectant parasites expressing PfISP3mCherry and PfDHHC1-GFP were generated by transfecting 100 μg of pB-PfISP3-mCherry into the stable transgenic cell line expressing 3D7-PfDHHC1-GFP. Double transfectant parasites expressing PfISP3-GFP and GAP45-mCherry were generated by transfecting 100 μg of pB-GAP45mCherry into the transgenic cell line expressing PfISP3-GFP. Selection was carried out using 30 nm blasticidin.
Nucleic Acids and Constructs
3D7 parasites expressing full-length PfDHHC1 and PFDHHC2 as a GFP fusion protein were generated by PCR amplification of the respective genes. PCR amplifications were performed using Phusion DNA polymerase (New England Biolabs). PCR amplicons were digested with KpnI and AvrII, cloned into a derivative of pARL-1a-GFP. Expression of the GFP fusion construct is controlled by the late stage promoter of ama1 (32). To circumvent the internal KpnI site in the genes coding PfDHHC3 and PfDHHC9, both open reading frames were cloned into the BamHI/SalI of a derivative of pBcamR (33). This vector mediates the overexpression of mCherry fusion proteins under the control of the ama1 promoter (6). The GFP replacement construct for tagging endogenous PfDHHC1 was designed using pARL-1a-GFP vector. 1 kb of the PfDHHC1 gene was cloned into the NotI/AvrII site of the pARL-1a-GFP releasing the crt promoter that controls the expression of the fusion protein in the original vector (34). All constructs were sequenced for accuracy. Oligonucleotides used for these constructs are summarized in Table 1.
TABLE 1.
Oligonucleotides used in this study
Restriction sites are underlined.

Full-length and truncated versions of PfISP3, PfISP1, and GAP45 were previously cloned and localized in P. falciparum as overexpressing GFP- or mCherry fusion proteins (6, 10, 15). To dissect sequence requirements for membrane attachment nucleic acid substitutions introduced into these genes were generated by oligonucleotide directed mutagenesis. DNA fragments were digested with KpnI and AvrII, cloned into a derivative of pARL-1a-GFP (under the control of the ama1 promoter; 32) and sequenced for accuracy. The respective oligonucleotides summarized in Table 1.
Western Blot Analysis
Synchronized P. falciparum cultures were saponin-lysed using 0.03% saponin (Sigma). Parasite pellets were resuspended in adequate amount of PBS and 5× SDS loading dye. Proteins were separated on 10% SDS-PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell). Monoclonal mouse anti-GFP (Roche Applied Science) or monoclonal rat anti-mCherry (Chromotek) was diluted 1:1000 in 10% (w/v) skim milk. The secondary antibody was horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) (1:3000, Jackson ImmunoResearch) and HRP-conjugated goat anti-mouse IgG (1:3000, Jackson ImmunoResearch). The immunoblots were developed by chemiluminescence using Immobilon Western Chemiluminescent HRP substrate (Millipore) and visualized using the ChemiDoc XRS+ System (Bio-Rad).
Live Microscopy and Immunofluorescence Assays
Images of unfixed GFP-expressing parasites were captured using a Zeiss Axio Skop 2plus microscope with a Hamamatsu Digital camera (model C4742-95, Zeiss AxioVision) with 1 μg/ml DAPI (Roche Applied Science) for nuclear stain. Immunofluorescence microscopy was performed on parasites fixed with a mixture of 4% formaldehyde and 0.0075% glutaraldehyde. Parasites were permeabilized with 0.1% Triton X-100 and blocked with 3% bovine serum albumin (BSA). First, parasites were incubated for 1 h with anti-GAPM2 (1:500, see Ref. 6) or RALP (1:500, see Ref. 35). Subsequently, cells were washed three times with PBS and incubated for an hour with Alexa-Fluor 594 goat anti-rabbit IgG antibodies (1:2000, Molecular Probes) and 1 μg/ml DAPI (Roche Applied Science). After removal of unbound antibodies by washing three times with PBS, coverslips were mounted with Fluoromount G (Sigma) and kept at 4 °C until evaluation.
S-Acyl Biotin Switch Assay
Saponin-lysed parasite pellets derived from 100 ml of synchronized late stage parasites were subjected to biotin switch protocol as described previously (15, 21).
Solubility Assays
P. falciparum proteins were extracted using 0.03% saponin (Sigma) from a synchronized late stage parasite culture (10 ml) as described previously (15). In short, the extraction was done sequentially; the lysate was resuspended in 100 μl of double distilled H2O, freeze-thawed three times, and centrifuged at 21,000 × g for 5 min. After this hypotonic lysis, the pellet was washed once with PBS and resuspended in 100 μl of freshly prepared 0.1 m Na2CO3 and kept on ice for 30 min to extract peripheral membrane proteins. The pellet was washed once with PBS and extracted for 30 min with 100 μl of 1% Triton X-100 and centrifuged at 21,000 × g for 5 min to obtain the integral membrane protein fraction in the supernatant. The final pellet was washed once with PBS and resuspended on 100 μl of PBS containing the insoluble fraction. Equal amounts of all supernatants were analyzed by immunoblotting with adequate amounts of 5× SDS loading dye. Proteins were detected using anti-GFP antibodies, and the cytosolic protein GAPDH was used as a control.
Phylogenetic Analysis
To get a comprehensive understanding of the phylogenetic relation and evolutionary background of apicomplexan PATs, 16 apicomplexan proteomes were searched and the retrieved sequences analyzed.
Data Collection of Putative PATs
The catalog of putative Asp-His-His-Cys (DHHC) was identified by searching for proteins matching the zf-DHHC HMM profile (dCt region, ∼170-amino acid profile corresponding to region encompassing DPG-CRD-TTXE motif), with HMMSEARCH of HMMER 3.1 package (36) using default parameters. The apicomplexan organisms used for this analysis are as follows: Cryptosporidium hominis TU502; Cryptosporidium muris RN66; Cryptosporidium parvum IowaII; Babesia bovis T2Bo; Theileria annulata strain Ankara; Theileria parva strain Muguga; P. berghei ANKA; Plasmodium chabaudi chabaudi, Plasmodium cynomolgi strain B; P. falciparum 3D7; Plasmodium knowlesi strain H; Plasmodium vivax Sal-1; Plasmodium yoelii yoelii 17XNL; Neospora caninum Liverpool; T. gondii ME49; Eimeria tenella strain Houghton, and their proteomes were downloaded from EupathDB (37). The hits were then manually checked for the presence of cysteine-rich domain (CRD), an ∼50-amino acid region housing the catalytically active DHH(Y)C residues (38). Hits lacking this domain were discarded. The remaining hits were segregated into “canonical” and “pseudo” PATs based on whether the CRD was intact or incomplete. The latter group includes hits that were incomplete/degraded at the N or C terminus of the CRD region, as well as those where the “DH(H/Y)C” motif was substituted. Many of the pseudo-PATs contain the DPG motif and TTXE motifs (13/28) thus arguing that these sequences are either degenerate sequences or represent artifacts of sequence assembly. The catalog of canonical and pseudo-PATs for all the species are listed in Fig. 4. Using the same methodology, putative PATs were identified in the proteomes of closely related free-living protists Paramecium tetraurelia and Tetrahymena thermophila SB210 and categorized, to enable a comparison of the evolution of the parasitic apicomplexan PATs with their free-living counterparts. The proteomes of P. tetraurelia and T. thermophila were downloaded from UNIPROT (39). Furthermore, the complement of characterized DHHC PATs from Saccharomyces cerevisiae S288c and Homo sapiens were extracted from SGD (40) and Swiss-Prot, respectively, to serve as the reference set of PATs.
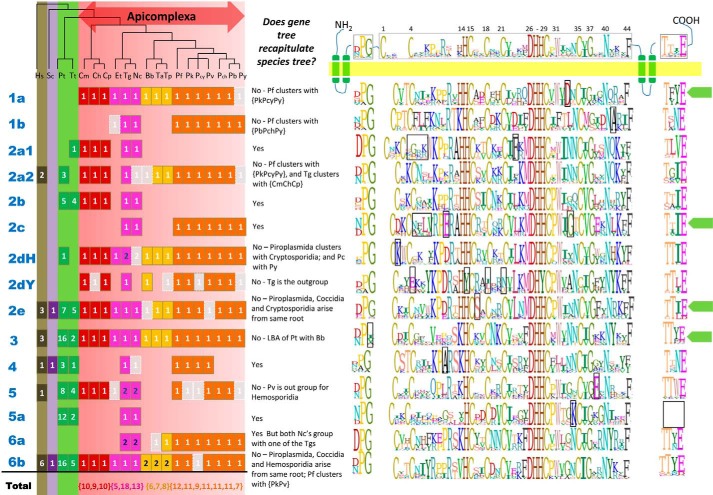
FIGURE 4.
Species distribution and sequence logos for the CRD region and DPG and TTXE motif for PAT clusters. The species tree for the apicomplexan organisms used in this analysis is shown (61, 62). The number of canonical PATs identified for each of these organisms in indicated in solid boxes, colored in brown, magenta, yellow, and orange to represent the clades of Cryptosporidia, Coccidia, Piroplasmida, and Hemosporidia, respectively. The pseudo-PATs are indicated in faded gray boxes. Information regarding whether the gene tree recapitulates the species tree is also provided. The sequence logos corresponding to the CRD region and the DPG and TTXE motif are provided alongside for each of the clusters, with residues unique to each class being boxed.
Identification of Evolutionarily Related Clusters of DHHC PATs
The evolutionary relationships between the set of PATs was resolved based on rigorous phylogenetic analyses. Because the PATs in free-living protists are likely to be very similar due to the duplication events, a representative set of PATs was generated by clustering at 40% sequence identity using CD-HIT (41) for use in further analysis. Multiple sequence alignments were generated using Muscle (42) and Probcons (43) for the dCt region (corresponding to the Pfam HMM profile). The CRD region was not used for phylogenetic analyses because it contained poorer signals than the other two sets, presumably due to its smaller size (∼50 amino acids in length), according to the likelihood mapping algorithm Tree-puzzle 5.2 (44). The alignment was manually edited using Geneious (created using Biomatters) and trimmed with TrimAl (45) using two cutoffs as follows: automated (which has been shown to be optimized for phylogenetic analyses using maximum likelihood method; Ref. 45) and gap threshold of 0.1 (which removes columns that contain gaps in >90% of sequences). The optimal model of substitution for each of the datasets was determined using ModelGenerator (46). Using the optimal model of substitution and allowing for rate heterogeneity at different sites modeled using γ-distribution, phylogenetic trees were then constructed using two methods, MrBayes and PhyML (47, 48). Bayesian phylogenies were generated using MrBayes for 3 million generations with a burn-in of 80,000 for two runs of four chains each. Maximum likelihood phylogenetic reconstruction was performed using PhyML with 1024 bootstrap replicates. This analysis resolved the PATs into six major clades and 14 subclades. Each of these clusters, including dCt region and full sequence, was then individually analyzed using the same series of steps outlined above to study whether the gene tree recaps the species tree or not.
Analysis and Annotation of PAT Clusters
The sequences in every cluster were analyzed for conservation of the following sequence motifs: consensus of CRD region; DPG motif, TTXE motif; and presence/absence of inserts between DPG motif and CRD region. Every cluster was also analyzed in terms of conservation of additional domains (wherever was present) and transmembrane helix distribution. Domain information was obtained from Interpro (49), whereas Tied Mixture Hidden Markov Model (TMHMM) (50) was used to predict transmembrane helices for the sequences.
RESULTS
Identification of an IMC-localized Palmitoyl Acyltransferase
Three known IMC-associated proteins in P. falciparum (PfGAP45, PfISP1, and PfISP3) displayed a conserved myristoylation and palmitoylation site within the first 20 amino acids that might be linked to IMC membrane association and may therefore be recognized by an IMC-residing PAT. Recent work in T. gondii and P. berghei revealed two IMC-localized PATs in each species as follows: TgDHHC2 (TGME49_278850, homolog of PfDHHC2) and TgDHHC14 (TGME49_293730, homolog of PfDHHC1) in T. gondii (26, 28) and PbDHHC3 (PBANKA_092730, homolog of PfDHHC3) and PbDHHC9 (PBANKA_093210, homolog of PfDHHC9) in P. berghei (28).
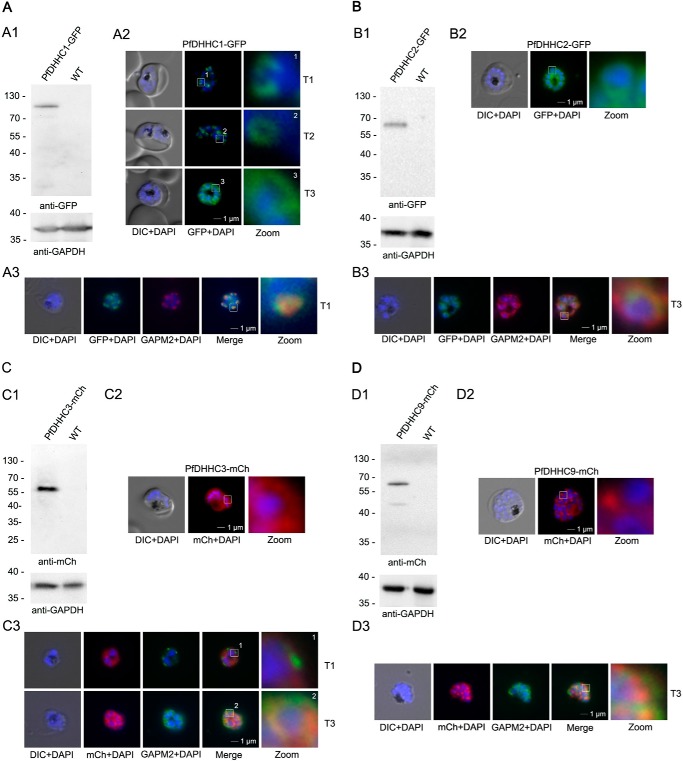
We aimed to localize all four of the homologous genes (PFC0160w/PfDHHC1, PFF0485c/PfDHHC2, PF11_0217/DHHC3, and PF11_0167/PfDHHC9) to validate their putative IMC localization in P. falciparum. We overexpressed the enzymes either as GFP or mCherry fusions, circumventing internal restrictions sites in the genes of interest. We used the ama1 promoter to mimic the late transcription of most IMC proteins (51). In agreement with this, the transcription of all four PATs appears to be up-regulated to some level in schizonts (51). Localization of the fusion proteins in unfixed parasites revealed that PfDHHC1-GFP has a localization pattern, resembling the IMC, and shows the IMC-typical dynamics during schizogony (6, 10, 52, 53); it commences as cramp-like structures, transforming to small ring-shaped formations that toward the end of schizogony expand and are then equally distributed underneath the plasma membrane (Fig. 1A). This IMC localization was supported by the co-localization with the IMC marker GAPM2 (PFD1110w, glideosome-associated protein with multiple membrane spans (6, 54)). The localization patterns of the other overexpressed PATs (PfDHHC2, -3, and -9) are distinct and do not co-localize with the IMC marker GAPM2 (Fig. 1, B–D).
FIGURE 1.
Overexpression and localization of PfDHHC1 (PFC0160w), PfDHHC2 (PFF0485c), PfDHHC3 (PF11_0217), and PFDHHC9 (PF11_0167) in late stage parasites. A, expression of PfDHHC1-GFP. Panel A1, Western blot analysis using anti-GFP antibodies. A single protein band of about 100 kDa (expected mass of 100 kDa) was detected in the transgenic but not in the parental parasite line. Panel A2, localization of PfDHHC1-GFP in unfixed parasites showing characteristic IMC dynamics during schizogony as follows: commencing as cramp-like structures (T1) and transforming to small ring-shaped formations (T2) that toward the end of schizogony expand and are then equally distributed underneath the plasma membrane (T3). Nuclei are stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as zoom. Scale bar, 1 μm. Panel A3, co-localization with the IMC marker GAPM2 (anti-GAPM2, red) in fixed cells confirmed PfDHHC1-GFP IMC localization shown here in an early stage of IMC biogenesis (T1). B, expression of PfDHHC2-GFP. Panel B1, Western blot revealed a 65-kDa (expected mass of 59 kDa) protein band detected by anti-GFP antibodies in the transgenic cell line but not in the control. Panel B2, live microscopy of PfDHHC2-GFP revealed a circular structure around the nucleus of the nascent merozoites reminiscent of the ER. Scale bar, 1 μm. Panel B3, co-localization with the IMC marker GAPM2 (anti-GAPM2, red) in fixed cells shows differential localization of PfDHHC2-GFP with the IMC shown here as nascent merozoites (T3). C, expression of PfDHHC3-mCherry. Panel C1, Western blot analysis with anti-mCherry antibody detected a 60-kDa (expected mass of 61 kDa). Panel C2, microscopic analysis located this GFP fusion protein in the periphery of the nascent merozoites. Panel C3, co-localization with the IMC marker GAPM2 (anti-GAPM2, green) in fixed cells shows differential localization of PfDHHC2-mCherry with the IMC localization in early stages (T1) and co-localization in nascent merozoites (T3) consistent with plasma membrane association. Scale bar, 1 μm. D, expression of PfDHHC9-mCherry. Panel D1, expression leads to 60 kDa and some degraded protein at about 45 kDa protein (expected mass of 61 kDa) detected in Western blot analysis using mCherry antibodies. Panel D2, microscopy localized this fusion protein mainly in apical structures in the parasite. Panel D3, co-localization with the IMC marker GAPM2 (anti-GAPM2, green) in fixed cells shows differential localization of PfDHHC9 with the IMC (T3). DIC, differential interference contrast. Scale bar, 1 μm.
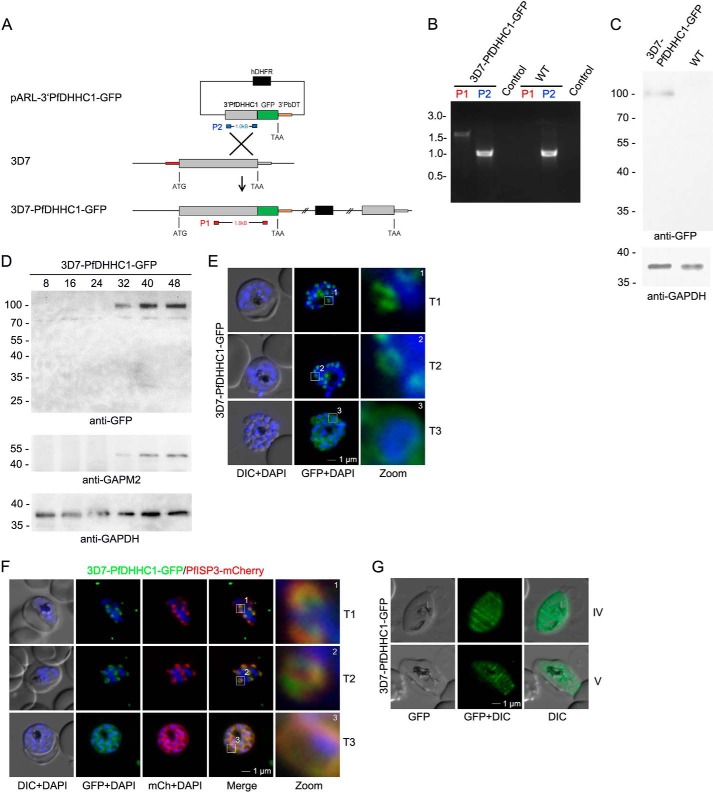
To confirm the IMC localization of PfDHHC1, the endogenous gene was tagged with GFP resulting in the parasite line 3D7-PfDHHC1-GFP (Fig. 2). Appropriate plasmid integration into the PfDHHC1 locus was shown by PCR (Fig. 2B), and expression of the fusion protein was verified by Western blotting (Fig. 2C). The stage-specific expression pattern of PfDHHC1 was analyzed using synchronized 3D7-PfDHHC1-GFP parasite material harvested 8, 16, 24, 32, 40, and 48 h post-invasion and probed with either anti-GFP or anti-GAPM2 antibodies as a stage-specific control in a Western blot (Fig. 2D). Anti-GAPDH antibodies were used as loading control. Microscopy of endogenously tagged PfDHHC1 confirmed IMC localization with its distinct biogenesis during schizogony (Fig. 2E). Co-localization with the dual-lipidated PfISP3 (PF14_0578) revealed identical dynamics of these two proteins (Fig. 2F), which would be congruent with a potentially specific interaction of these proteins. In pre-sexual gametocyte stages (Fig. 2G), PfDHHC1-GFP is not equally distributed within the nascent IMC but accumulates in transversal structures resembling the symmetric meshwork enclosing the gametocyte described previously as the localization of the Plasmodium-specific IMC protein MAL13P1.228 (6) that might reflect the sutures of individual IMC plates (55). Given the presence of multiple IMC-localized PATs in T. gondii and P. berghei and their differing distribution in P. falciparum, we performed a systematic phylogenetic analysis of PATs within the alveolate lineage to gain insights into their evolutionary relationships.
FIGURE 2.
PfDHHC1 is an IMC localized palmitoyl acyltransferase. A, schematic representation of the GFP replacement of the endogenous 3′ end of PfDHHC1 creating a 3D7-PfDHHC1-GFP cell line. The vector encompasses the selection cassette (black), and 1 kb of the PfDHHC1 gene (gray) was fused to GFP (green) accompanied by the 3′UTR of PbDT (orange) without any promoter. Integration of the vector takes place by homologous recombination (cross) into the PfDHHC1 locus creating a full-length PfDHHC1-GFP fusion under the control of the endogenous promoter. B, integration was confirmed by PCR using two different primer combinations on gDNA. One primer set (red) hybridizes in a PfDHHC1 region upstream of the integration and in the coding region of GFP. This primer combination can only amplify a 1.7-kb DNA fragment after recombination took place. The other set (blue) amplifies 1.1 kb of PfDHHC1 in the parental as well as in the transgenic parasite line. Control indicates PCRs with the red primer set in the absence of parasite DNA. C, expression of the transgene from the endogenous locus was shown by Western blot analysis using anti-GFP antibodies (upper panel) resulting in a fusion protein of ∼100 kDa (calculated mass of 100 kDa) using late stage parasite material. Anti-GAPDH was used as a loading control. D, stage-specific expression pattern of PfDHHC1-GFP using synchronized 3D7-PfDHHC1-GFP parasite material (harvested after 8, 16, 24, 32, 40, and 48 h) and anti-GFP antibodies. Antibodies directed against the IMC marker protein GAPM2 were used as a stage-specific control. Anti-GAPDH antibodies were used as loading control. E, PfDHHC1-GFP was localized in unfixed late stage parasites showing characteristic IMC dynamics during schizogony (T1 to T3). Nuclei stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as zoom. Scale bar, 1 μm. F, co-localization of PfDHHC1-GFP and PfISP3mCherry. PfISP3mCherry was episomally co-expressed and shows identical spatial distribution compared with PfDHHC1-GFP. G, PfDHHC1-GFP distribution within the nascent IMC during gametocytogenesis (stage IV and V). These symmetric sutures of the IMC are expanding with ongoing growth of the IMC vesicles and maturation of the gametocytes (stage IV and V). DIC, differential interference contrast. Scale bars, 1 μm.
Phylogenetics Reveals Six Major Families of PATs in the Alveolata
A recent study used the conserved, yet relatively short DHHC-CRD to construct a phylogeny of PAT proteins derived from several species of Apicomplexa (28). However, due to the shortness of the domain (∼77 amino acids), phylogenetic insights were largely confined to comparisons of five families across four species. Here, we applied the hidden Markov model for zf-DHHC (PF01529; a 170-amino acid profile encompassing the DPG, CRD, and TTXE motifs) to retrieve 297 canonical and 28 pseudo (lacking an intact CRD and/or lacking the DH(HY)C motif) PATs from 18 alveolates. Compared with S. cerevisiae (seven PATs), apicomplexans tend to possess a larger repertoire of PATs (Fig. 3), with almost double the number in Hemosporidia (7–12) and even more in Coccidia (e.g. 18 in T. gondii). Furthermore, the free-living ciliates P. tetraurelia and T. thermophila show a much larger expansion in their DHHC PAT complement (100 and 38, respectively), likely related to documented duplication events (56, 57). These expansions highlight the potential importance of PATs to alveolate and apicomplexan innovations.
FIGURE 3.
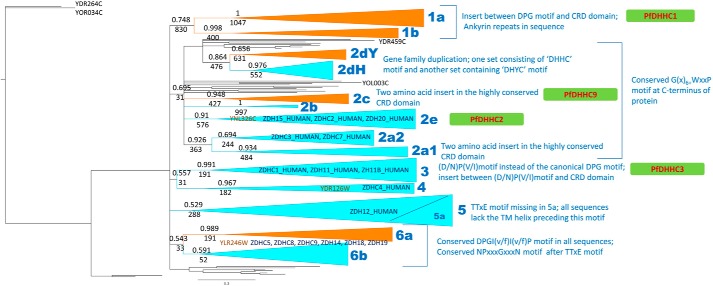
Phylogenetic analyses of PATs from Apicomplexa, free-living protists, yeast, and human. The phylogenetic tree reconstructed using MrBayes based on dCt region of all canonical PATs from apicomplexa, yeast, and human, and nonredundant representative set from free-living protists. The clusters with posterior probabilities greater than ∼0.6 are shown as clades, with posterior probabilities and bootstrap replicates indicated at the branching point. Clusters consisting only of apicomplexan members are shown in orange, and clusters consisting of sequences from both apicomplexan organisms and the free-living protists are shown in cyan. The yeast PATs that are orthologs of specific clusters are labeled in brown, and human orthologs are shown in blue. Features characteristic of a cluster are indicated alongside the cluster.
For information on evolutionary relationships between PATs, we performed maximum likelihood and Bayesian phylogenetic analyses on the region encompassed by the zf-DHHC domain (Fig. 4). This analysis allowed us to define six main clades of PATs, with clade 2 further divided into seven further sub-clades. Each clade and subclade is associated with distinct sequence features, including the presence of unique repeats, motifs, and clade-defining residues (Figs. 3 and 4). Three yeast PATs are found to associate with three of these clades (2e, 4 and 6), whereas 16 human PATs are found to cluster with the clades 2e, 2a2, and 3–6, suggesting the involvement of clade members in conserved cellular processes.
PfDHHC1 Is a Member of a Highly Conserved Apicomplexan-specific Clade of PATs
Across the 14 sub-clades are five that appear apicomplexan-specific (designated clusters 1a, 1b, 2c, 2dY, and 6a, Fig. 3). Cluster 1a, which includes PfDHHC1 (PFC0160w), is highly conserved, yet specific to apicomplexans, with a single ortholog identified in all species with the exception of P. yoelii, in which the ortholog is a pseudo-PAT. However, comparison with its closest ortholog in P. berghei indicates that this is likely due to a poorly predicted gene model. Inspection of the sequences indicated conservation over the entire sequence length (data not shown). Furthermore, all members of cluster 1a (as well as 1b) are defined by the presence of ankyrin repeats at the N terminus, in addition to a distinct conserved insert between the DPG motif and the CRD region. Interestingly, the Toxoplasma ortholog TgDHHC14 (TGME49_293730) has also been associated with the IMC (26, 28). Cluster 2dY is similarly unique with the distinguishing feature that all members possess, i.e. “DHYC” motifs instead of “DHHC” motifs. Although DHYC-PATs have been identified in yeast (YDR264C/AKR1_YEAST and YOR034C/AKR2_YEAST), the apicomplexan DHYC-PATs represent a distinct clade, suggesting an instance of convergent evolution. From sequence logos constructed for each clade (Fig. 4), we observe that Cys-1 of the CRD region and Asn-40 are invariant, along with the active site residues DH(H/Y)C. In addition, Trp-31, Gly-37, and Phe-44 are also largely conserved, exhibiting only conservative substitutions. In the case of DHHC PATs, Cys-4, His-14, Cys-15, Cys-18, Cys-21, and Cys-35 are also invariant; however, for DHYC PATs, Cys-4, Cys-18, and Cys-21 are not conserved, whereas His-14 is substituted by Tyr-14, suggesting an alternative route for catalysis. Finally, it is worth noting that PfDHHC9 (PF11_0167), a member of cluster 2c, is restricted to the coccidians and hemosporidians. Toxoplasma and P. berghei orthologs, TGME49_217870 and PBANKA_093210, have previously been shown to localize to the ER and the IMC, respectively, compared with the apical membrane/food vacuole for PfDHHC9 (28). Given the disparity in localization of these orthologs, we speculate that after their emergence the members of this clade have undergone adaptive radiation resulting in the evolution of lineage-specific functions.
Outside the apicomplexan-specific clusters, we note several instances of alveolate-specific clusters (2a1, 2b, 2dH, and 5a, a subclade of clade 5 lacking the TTXE motif, see Fig. 3). Intriguingly, with the exception of 2dH, members from these clusters appear to have been lost in the hemosporidians. Cluster 2dH is noteworthy as orthologs are present for all alveolates except N. caninum, and therefore likely represent a conserved alveolate innovation.
For the most part, lineage-specific duplications within subclades are rare with the exception of duplicates in T. gondii in clades 2dH, 5, and 6a, although piroplasmids contain two copies of PATs in cluster 6b. Finally, clusters 2a2, 2e, 3–5, and 6b appear conserved outside the alveolates with yeast and human orthologs, offering the opportunity to gain insights into functions of cluster members. This is of particular relevance with two of the clades, 2e and 3, that contain the non-IMC membrane-localized PATs PF11_0217/PfDHHC3 and PFF0485c/PfDHHC2, respectively.
Palmitoylation of the IMC Localized PfISPs
The two members of the ISP family in P. falciparum, PfISP1 and PfISP3, homologs of TgISP1 and TgISP3 (10, 11), encode N-terminal myristoylation and palmitoylation motifs. The N-terminal 20 amino acids of both proteins are sufficient for IMC membrane association (15), suggesting that they are recognized and modified by the IMC-residing PATs.
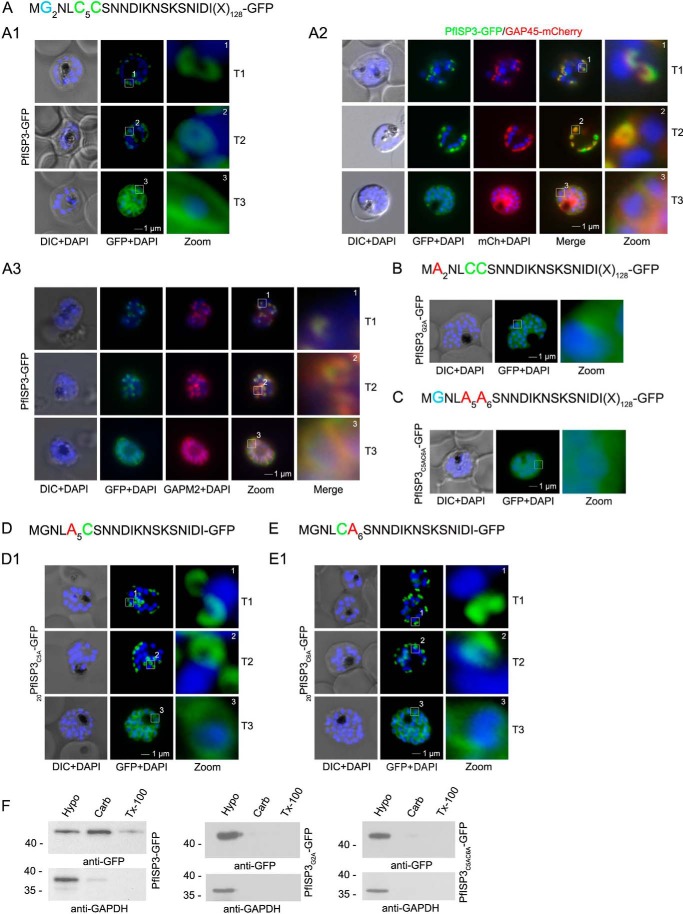
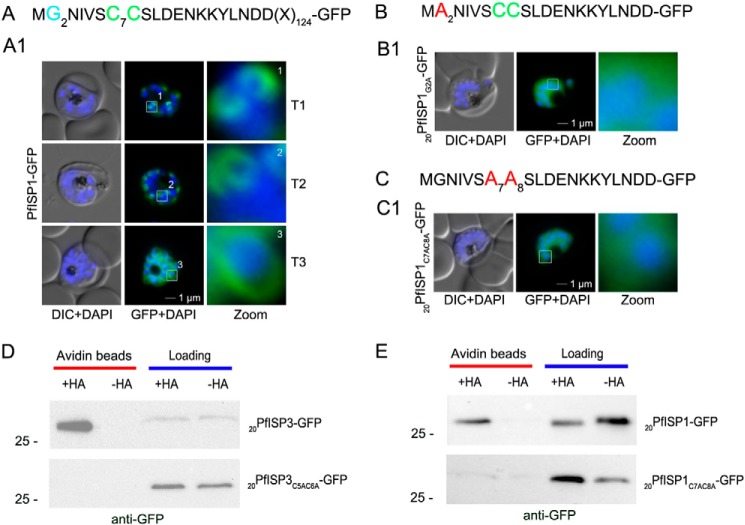
First, we verified the role of N-terminal myristoylation and palmitoylation motifs for IMC membrane recruitment by expressing and analyzing the subcellular distribution of mutants by microscopy (Figs. 5 and 6). Expression of full-length PfISP3-GFP shows a typical IMC dynamic during schizogony (Fig. 5A) and co-localizes with the IMC markers GAP45 and GAPM2 (Fig. 5, panels A2 and A3). Alanine substitutions of the myristoylation or palmitoylation motifs led to the expected cytosolic distribution of the mutant proteins, underlining the synergistic effect of myristoylation and palmitoylation for membrane association (Fig. 5, B and C). Noteworthy, single point mutations of either C5 or C6 in PfISP3 do not interfere with IMC membrane localization, which might point toward a redundancy of the palmitoylation sites (Fig. 5, D and E). Cytosolic distribution of PfISP3G2A-GFP and PfISP3C5AC6A-GFP was additionally verified by solubility assays. Although overexpressed PfISP3-GFP is mainly in the carbonate fraction, confirming its membrane association, both mutants are exclusively found in the hypotonic fraction, resembling the cytosolic GAPDH control. (Fig. 5F). The pertinent alanine substitutions (20PfISP1G2A-GFP and 20PfISP1C7AC8A-GFP) were also introduced in PfISP1 and resulted in similar phenotypes (Fig. 6, A–C).
FIGURE 5.
Role of N-terminal acylation for IMC membrane association of PfISP3. A, overexpression of PfISP3-GFP showing characteristic IMC dynamics during schizogony (T1–T3) in unfixed parasites. Nuclei are stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as zoom. Scale bar, 1 μm (panel A1). Panel A2, co-localization of PfISP3-GFP (green) and GAP45mCherry (red) in unfixed parasites revealed their identical dynamic during schizogony. Panel A3, co-localization with the IMC marker GAPM2 (α-GAPM2, red) in fixed cells. Putative myristoylation and palmitoylation sites are highlighted in light blue (G2) or green (C5C6). B and C, IMC membrane association depends on the presence of N-terminal myristoylation and palmitoylation motifs. N-terminal myristoylation (B, PfISP3G2A-GFP) and palmitoylation motif mutants (C, PfISP3C5AC6A-GFP) were expressed in P. falciparum and localized in unfixed parasites. Mutation of either the myristoylation or the palmitoylation motifs resulted in a cytosolic distribution of the GFP fusion protein (panel B1 and C1). D and E, mutation of the individual palmitoylation sites C5 (PfISP3C5A-GFP) or C6 (PfISP3C6A-GFP) does not interfere with IMC recruitment of the fusion protein. F, solubility assays confirmed the essential role of both myristoylation and palmitoylation modifications for membrane association of PfISP3-GFP. In contrast to wild type proteins, the mutant proteins are, like GAPDH (lower panels), exclusively detectable in the hypotonic fraction (soluble, H2O/SN; carbonate, Carb/supernatant; and membrane Triton X-100, Tx100/SN). DIC, differential interference contrast.
FIGURE 6.
Role of N-terminal acylation for IMC membrane association of PfISP1 and acyl biotin exchange assays. A, overexpression of PfISP1-GFP showing the same characteristic IMC dynamics as PfISP3-GFP. Putative myristoylation and palmitoylation sites are highlighted in light blue (G2) or green (C7C8). B and C, mutation of either one of the acylation motifs (20PfISP1G2A-GFP and 20PfISP1C7AC8A-GFP) resulted in a cytosolic variant shown by microscopy. Nuclei are stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as zoom. Scale bar, 1 μm. D, 20PfISP3-GFP (upper panel) and 20PfISP3C5AC6A-GFP (lower panel) fusion proteins are present in the 2 aliquots that were incubated with biotinylation reagent with or without hydroxylamine after N-ethylmaleimide treatment (Loading). After elution from the NeutrAvidin beads (Avidin beads), only 20PfISP3-GFP but not the cysteine mutant 20PfISP3C5AC6A-GFP is enriched in the hydroxylamine (+HA)-treated sample when compared with the untreated sample (−HA). E, 20PfISP1-GFP, 20PfISP1-GFP fusion proteins are present in the 2 aliquots that were incubated with biotinylation reagent with or without hydroxylamine after N-ethylmaleimide treatment (Loading). After elution from the NeutrAvidin beads (Avidin beads), only 20PfISP1-GFP but not the cysteine mutant 20PfISP1C5AC6A-GFP is enriched in the hydroxylamine (+HA)-treated sample when compared with the untreated sample (−HA). DIC, differential interference contrast.
Second, to confirm the predicted palmitoylation in vivo, parasites expressing either the wild type sequence (20PfISP3-GFP and 20PfISP1-GFP) or the corresponding cysteine/alanine substitutions (20PfISP3C5AC6A-GFP and 20PfISP1C7AC8A-GFP) were used in acyl biotin exchange assays (21, 58). This assay substitutes thioester-linked acyl groups with biotin that can subsequently be used for affinity purification. These experiments showed that both PfISPs (20PfISP3-GFP and 20PfISP1-GFP) are biotinylated and are enriched in the fraction that is expected to contain all S-acylated proteins assayed by this method. The corresponding mutants without palmitoylation motifs were not enriched (Fig. 6, D and E). Taken together, these results show that IMC membrane recruitment of PfISP3 and PfISP1 depends on N-terminal lipid modifications.
Minimal Sequence Requirements for IMC Localization of PfISP3
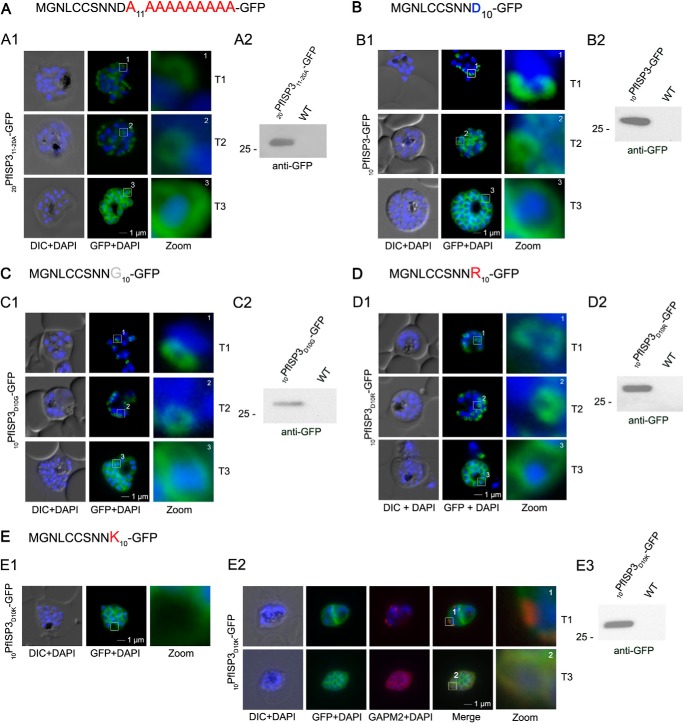
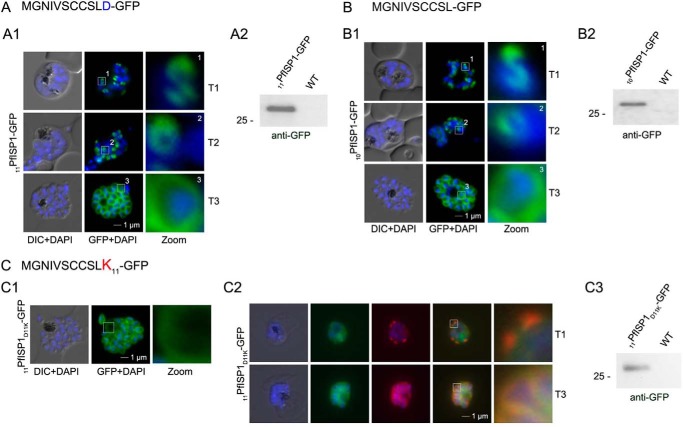
Although fatty acylation is essential for IMC localization of the ISPs, it is unknown how the membrane specificity is achieved to exclusively trap these proteins at the IMC. To identify potential sequence features that may contribute to such specificity, we generated a construct, where the last 10 amino acids were exchanged to alanines (20PfISP311–20A-GFP; Fig. 7A). This mutant is still truthfully located at the IMC upon its expression in the parasite (Fig. 7A) and associates with the specific intermediates (T1 and T2) of nascent IMC during schizogony. Hence, the N-terminal alanine substitutions in 20PfISP311–20A-GFP do not appear to interfere with IMC membrane recruitment and protein acylation itself.
FIGURE 7.
Mutational analysis of the minimal sequence requirements for IMC association of PfISP3. A and B, expression and localization of the 20-amino acid N terminus with the last 10 (20PfISP311–20A-GFP) amino acids substituted with alanines. Panel A1, 20PfISP311–20A-GFP was localized in unfixed parasites and showed an identical dynamic as the wild type protein. Panel A2, Western blot analysis of the expression using anti-GFP antibodies. B, first 10 amino acids of PfISP3 (10PfISP3-GFP) are sufficient for IMC localization (panel B1). Panel B2, Western blot analysis of the expression 10PfISP3-GFP. C and D, mutation of the sole charged residue D10 into a neutral glycine (10PfISP3D10G-GFP, panel C1) or a positively charged arginine (10PfISP3D10R-GFP, panel D1) does not interfere with IMC targeting. Western blot analysis (panels C2 and D2) of GFP fusion protein expression. E, substitution of D10 with a positively charged lysine re-directs the GFP fusion protein to the periphery of the nascent merozoites. The zoom highlights the additional association with the food vacuole membrane characteristic for plasma membrane proteins (panel E1). Co-localization was with the IMC marker GAPM2. Nascent IMC (anti-GAPM2, red) is clearly distinguishable from the peripherally associated 10PfISP3D10K-GFP in early stages (T1) and congruent in late stages (T3). Western blot analysis (panel E3) of 10PfISP3D10K-GFP. DIC, differential interference contrast.
Lysine but Not Arginine within Close Proximity of the Palmitoylation Sites of PfISP3 Prevents IMC Membrane Targeting
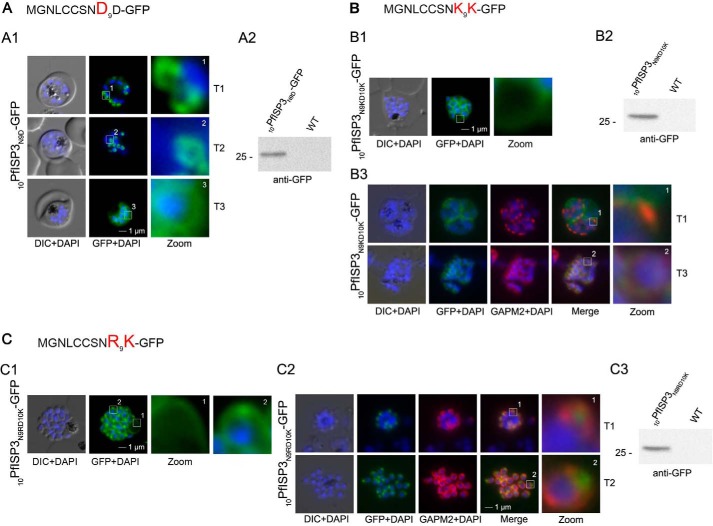
The distribution of 20PfISP311–20A-GFP points toward a crucial role of the first 10 amino acids for IMC targeting (Fig. 7A). This was validated by the expression and localization of the first 10 amino acids of PfISP3 fused with GFP (10PfISP3-GFP). Consistent with the previous findings, 10PfISP3-GFP is localized to the IMC (Fig. 7B). Next, the only charged residue (Asp-10) in the 10 amino acids was exchanged either to a neutral glycine (Fig. 7C) or to a positively charged arginine (Fig. 7D) or lysine (Fig. 7E). Although neither the exchange with glycine (10PfISP3D10G-GFP) nor arginine (10PfISP3D10R-GFP) interfered with correct targeting, the substitution with a lysine (10PfISP3D10K-GFP) led to a re-direction of the GFP fusion to the periphery of the nascent merozoites, resembling plasma membrane localization. The aberrant localization of 10PfISP3D10K-GFP was confirmed by co-localization using the IMC marker protein GAPM2 (Fig. 7E). Interestingly, previous work showed an essential role for an arginine and a lysine in close proximity downstream of the palmitoylated cysteine for rhoptry membrane targeting of PfARO (15). To probe into a putative discriminative role of an arginine in combination with a lysine, we attempted to re-direct 10PfISP3-GFP. As expected, the exchange of Asn-9 with aspartate (10PfISP3N9D-GFP) did not change IMC localization (Fig. 8A), and the exchange of Asn-9 and Asp-10 with lysines (10PfISP3N9KD10K-GFP) re-directed the protein to the periphery of the merozoites (Fig. 8B). Nevertheless, the substitution of Asn-9 and Asp-10 with an arginine and lysine, respectively (10PfISP3N9RD10K-GFP), led to a partial re-direction of the GFP fusion to a structure that co-localizes with the rhoptry marker RALP (Fig. 8C).
FIGURE 8.
Re-direction of the IMC 10PfISP3-GFP fusion protein to the rhoptry membrane. A, substitution of Asn-9 with Asp (10PfISP3N9D-GFP, A) does not interfere with correct IMC recruitment of the GFP fusion protein (panel A1). Nuclei were stained with DAPI (blue). Panel A2, Western blot analysis using anti-GFP antibodies. B and C, substitution of N9 and D10 with lysine and its expression in the parasite (10PfISP3N9KD10K-GFP) leads to a peripheral localization (panel B1). The zoom highlights the additional association of this mutant protein with the food vacuole membrane. Panel B2, Western blot analysis using anti-GFP antibodies. Panel B3, co-localization with the IMC marker GAPM2. Nascent IMC (anti-GAPM2, red) is clearly distinguishable from the peripherally associated 11PfISP1D11K-GFP in early stages (T1) and congruent in late stages (T3). C, in contrast, the substitution of N9 with arginine (10PfISP3N9RD10K-GFP) re-directs the fusion protein to cell periphery (zoom 1) and the apical pole (zoom 2, panel C1). Panel C2, co-localization with the IMC marker GAPM2. Nascent IMC (anti-GAPM2, red) is clearly distinguishable from the apical 10PfISP3N9RD10K-GFP in early stages (T1) and late stages (T3). Panel C3, Western blot analysis using anti-GFP antibodies. DIC, differential interference contrast.
PfISP1 Uses the Same N-terminal Signature for IMC Targeting as PfISP3
Given the similar N-terminal sequence signature of both PfISP proteins, we probed into the minimal sequence requirements for IMC-specific membrane recruitment of PfISP1 (Fig. 9). First, we expressed minimal targeting sequences by using only the first 11 (including the negative charged residue) or the 10 amino acids of PfISP1. As expected, based on the previous results from PfISP3, both constructs, 11PfISP1-GFP and 10PfISP1-GFP, directed GFP to the IMC (Fig. 9, A and B). Again, the introduction of a lysine in this short targeting sequence (11PfISP1D11K-GFP) aborted IMC trafficking and led to an aberrant membrane association (Fig. 9C).
FIGURE 9.
Mutational analysis of the minimal sequence requirements for IMC association of PfISP1. A and B, expressions of the first 11 (11PfSP1-GFP, A) or 10 (10PfISP1-GFP, B) amino acids are sufficient to direct GFP to the IMC (panels A1–B1). C, substitution of D11 with lysine (11PfISP1D11K-GFP, C) abolishes IMC targeting (panel C1) and leads to peripheral localization. The zoom highlights its additional association with the food vacuole membrane consistent with plasma membrane association. Panel C2, co-localization with the IMC marker GAPM2. Nascent IMC (anti-GAPM2, red) is clearly distinguishable from the peripherally associated 11PfISP1D11K-GFP in early stages (T1) and congruent in late stages (T3). Panel C3, Western blot analysis using anti-GFP antibodies. DIC, differential interference contrast.
DISCUSSION
The specific recruitment of proteins to endomembrane systems such as the IMC is a prerequisite for their physiological role. The IMC has three main functions as follows: (i) it plays a major role in motility and invasion; (ii) it confers stability and shape to the cell; and (iii) it provides a scaffolding framework during cytokinesis. Therefore, it is not surprising that it harbors a structurally, phylogenetically, and functionally diverse proteome. To date, about 30 IMC proteins have been identified so far in Apicomplexa, with the majority being associated with the cytosolic face of the IMC (3). Most of these peripheral membrane proteins display predicted palmitoylation motifs either in combination with an N-terminal myristoylation motif (e.g. PfISP1, PfISP3, and GAP45) or without (e.g. alveolins).
Palmitoyl acyltransferases embedded in distinct membranes are thought to play an important role for membrane-specific recruitments of these proteins. Here, we show that PfDHHC1 is, like its homolog TgDHHC14 in T. gondii (26), IMC-localized. Future work has to address the localization of PfDHHC2, PfDHHC3, and PfDHHC9. Although previous analyses reveal PfDHHC1 to be a member of a conserved clade (28), our comparative analyses involving ciliates establishes that this PAT is an apicomplexan-specific innovation.
By augmenting the PAT repertoire in apicomplexans with those from alveolates, yeast and human, our study enables us to differentiate those PATs representing alveolate and apicomplexan-specific innovations from those mediating core conserved eukaryotic functions. Furthermore, our phylogenetic analysis defines six distinct clades of PATs, supported by the presence of distinguishing molecular signatures. In general, PATs exhibit few lineage-specific innovations and are largely conserved across many species. This conservation, when considered in combination with the presence of clade-specific residues in the catalytic domain, argues that alveolates and apicomplexans have recruited this ubiquitous family of enzymes to perform a central role in their biology.
Although considerable efforts were undertaken to identify specific sequence requirements for substrate specificity of PATs, a clear consensus sequence for individual PATs has not yet been identified. On the contrary, experimental data suggest overlapping substrate specificity between enzymes (23, 35, 59, 60), leaving us with the question of how peripheral membrane proteins are recruited to specific membrane systems such as the IMC in the malaria parasite. In a previous study, we tracked 17 proteins predicted to be myristoylated and palmitoylated in the first 20 N-terminal amino acids in the malaria parasite (15). The use of this short sequence excluded putative interference with other determinants for membrane specificity like protein-protein interaction or structural features and focused on primary sequence motifs. Interestingly, although most of the corresponding GFP fusion proteins were trafficked to the parasite plasma membrane, three were sorted to the apical organelles and two, PfISP1 and PfISP3, were trafficked to the IMC. Both groups use a different molecular signature. Although rhoptry membrane attachment depends on positively charged residues in addition to an arginine two amino acids apart from the palmitoylated cysteines, a positively charged lysine appears to be the only exclusion criterion for an IMC localization (Figs. 8 and 9). In agreement with this, all experimentally localized rhoptry fusion proteins (PF08_0062, PFL1110c), including PfARO (PFD0720w), display an arginine residue two amino acids apart from the palmitoylated cysteines (15). It will be highly interesting to dissect the discriminative parameters between IMC and plasma membrane-localized proteins.
To help define substrate specificity, we generated a structural model of the CRD domain (Fig. 10). The structure features two lobes, with one lobe containing a set of conserved Cys and His residues that are spatially close and may therefore form the zinc-binding pocket. The zinc finger domain is a short independent domain present in many proteins, consisting of a conserved mosaic of cysteine and histidine residues, a characteristic feature of the CRD domain of DHHC PATs, which requires zinc binding to stabilize its structure (63). Recently, zinc binding has been experimentally shown to be necessary for stabilizing the structure of the CRD domain in a yeast DHHC PAT, the Swf1 protein (64). Within the other lobe, the invariant Asn-40 appears to form a hydrogen bond with the active site His-27, whereas the conserved Gly-37, located close to His-27 at a constrained turn, is likely important for packing. The conserved Trp-31 and Phe-44 may interact through their aromatic side chains, helping to maintain orientation of the nearby active site residues. Outside these conserved residues, we identified a number of cluster-defining residues that may serve to define substrate specificity. We note that different clades of PATs are associated with distinct sets of clade-specific residues and motifs (Fig. 10). Interestingly, both PfDHHC1 and PfDHHC9, members of apicomplexan specific groups, feature a clade-defining residue close to the active site of the CRD (Asp-33 and Gly-33, respectively). We speculate that these residues mediate discriminatory roles for the DHHC catalytic site of these two proteins. In addition, we also show that PfDHHC9 and PfDHHC2 (PFF0485c) feature clade-defining residues close to the likely zinc-binding pocket (Glu-11 and Ser-17, respectively), which may have implications for the function of this site. For example, Glu-11 may be involved in interactions with positively charged side chain residues associated with substrates targeted to the plasma membrane (e.g. via the formation of stabilizing salt bridges). Finally, although PfDHHC3 (PF11_0127) does not feature any conserved clade-defining residues in the CRD region, we do note that cluster 3 members differ from all other PATs as the only group without the well conserved Gly of the DPG motif, present next to the second transmembrane helix. This might have potential consequences for its embedment into the membrane.
FIGURE 10.
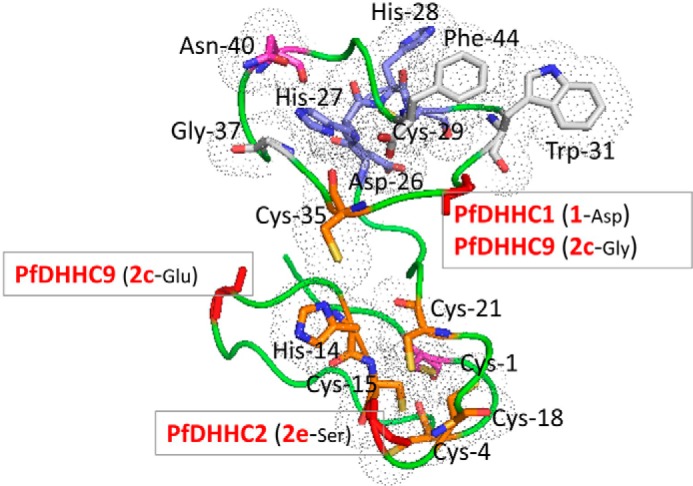
Structural model for the CRD region. Based on PfDHHC7 (PF3D7_0528400, 2dH), a model was generated using I-TASSER (65), the best performing web server for protein structure prediction according to the community-wide Critical Assessment of Protein Structure Prediction (CASP) experiment (66). The fit of the sequence to the modeled structure was verified using PROSA (67), which returned a Z-score of −4.54, comparable with those observed for experimental structures of this size. The conserved residues of the CRD region are shown as sticks. The different residues are colored as follows: DHHC motif in blue, invariant Cys-1 and Asn-40 in magenta, highly conserved Cys and His residues in orange, and the largely conserved Gly-37, Trp-31, and Phe-44 in white. The residues are marked according to the number shown in the consensus sequence logo for the CRD region in Fig. 3. The backbone of cluster defining residues for the PATs of interest are shown as red sticks.
Acknowledgments
We are grateful for WR99210 provided by Jacobus Pharmaceuticals and the support of the Hamilton Health Sciences McMaster Hospital Transfusion Medicine. We thank Drs C. Daubenberger and P. Sharma for providing antibodies against GAPDH and GAP45.
This work was supported by Canadian Institutes of Health Research Grants MOP111196 (to T. W. G.) and MOP84556 (to J. P.), the Canadian Foundation for Innovation (to T. W. G.), the Natural Sciences and Engineering Research Council (to T. W. G.), University of Osnabrück (GK UOsBio), Boehringer Ingelheim Fonds (to A. T. J. P.), Deutsche Forschungsgemeinschaft Grant SFB 944, P11 (to C. U.), the Hans-Mühlenhoff Foundation (to C. U.), and the M. G. DeGroote Institute for Infectious Disease Research Summer Student Fellowship awards (to O. R.).
- IMC
- inner membrane complex
- PAT
- palmitoyl acyltransferase
- CRD
- cysteine rich domain
- ER
- endoplasmic reticulum
- ISP
- IMC sub-compartment protein.
REFERENCES
- 1. Shetty P. (2012) The numbers game. Nature 10.1038/484S14a [DOI] [PubMed] [Google Scholar]
- 2. Cowman A. F., Crabb B. S. (2006) Invasion of red blood cells by malaria parasites. Cell 124, 755–766 [DOI] [PubMed] [Google Scholar]
- 3. Kono M., Prusty D., Gilberger T. W. (2013) A membranous system in the spotlight: the apicomplexan inner membrane complex and its adaptation to an endoparasitic life style. Front. Biosci. 18, 982–992 [DOI] [PubMed] [Google Scholar]
- 4. Cavalier-Smith T. (1993) Kingdom protozoa and its 18 phyla. Microbiol. Rev. 57, 953–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harding C. R., Meissner M. (2014) The inner membrane complex through development of Toxoplasma gondii and Plasmodium. Cell. Microbiol. 16, 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kono M., Herrmann S., Loughran N. B., Cabrera A., Engelberg K., Lehmann C., Sinha D., Prinz B., Ruch U., Heussler V., Spielmann T., Parkinson J., Gilberger T. W. (2012) Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol. Biol. Evol. 29, 1–20 [DOI] [PubMed] [Google Scholar]
- 7. Keeley A., Soldati D. (2004) The glideosome: a molecular machine powering motility and host-cell invasion by Apicomplexa. Trends Cell Biol. 14, 528–532 [DOI] [PubMed] [Google Scholar]
- 8. Baum J., Gilberger T.-W., Frischknecht F., Meissner M. (2008) Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol. 24, 557–563 [DOI] [PubMed] [Google Scholar]
- 9. Frénal K., Polonais V., Marq J. B., Stratmann R., Limenitakis J., Soldati-Favre D. (2010) Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe 8, 343–357 [DOI] [PubMed] [Google Scholar]
- 10. Beck J. R., Rodriguez-Fernandez I. A., Cruz de Leon J., Huynh M.-H., Carruthers V. B., Morrissette N. S., Bradley P. J. (2010) A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 10.1371/journal.ppat.1001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu G., Cabrera A., Kono M., Mok S., Chaal B. K., Haase S., Engelberg K., Cheemadan S., Spielmann T., Preiser P. R., Gilberger T. W., Bozdech Z. (2010) Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat. Biotechnol. 28, 91–98 [DOI] [PubMed] [Google Scholar]
- 12. Fung C., Beck J. R., Robertson S. D., Gubbels M.-J., Bradley P. J. (2012) Toxoplasma ISP4 is a central IMC sub-compartment protein whose localization depends on palmitoylation but not myristoylation. Mol. Biochem. Parasitol. 184, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tonkin M. L., Brown S., Beck J. R., Bradley P. J., Boulanger M. J. (2012) Purification, crystallization and preliminary x-ray diffraction analysis of inner membrane complex (IMC) subcompartment protein 1 (ISP1) from Toxoplasma gondii. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 68, 832–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poulin B., Patzewitz E.-M., Brady D., Silvie O., Wright M. H., Ferguson D. J., Wall R. J., Whipple S., Guttery D. S., Tate E. W., Wickstead B., Holder A. A., Tewari R. (2013) Unique apicomplexan IMC sub-compartment proteins are early markers for apical polarity in the malaria parasite. Biol. Open 2, 1160–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cabrera A., Herrmann S., Warszta D., Santos J. M., John Peter A. T., Kono M., Debrouver S., Jacobs T., Spielmann T., Ungermann C., Soldati-Favre D., Gilberger T. W. (2012) Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite. Traffic 13, 1335–1350 [DOI] [PubMed] [Google Scholar]
- 16. Gunaratne R. S., Sajid M., Ling I. T., Tripathi R., Pachebat J. A., Holder A. A. (2000) Characterization of N-myristoyltransferase from Plasmodium falciparum. Biochem. J. 348, 459–463 [PMC free article] [PubMed] [Google Scholar]
- 17. Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 18. Roth A. F., Feng Y., Chen L., Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohno Y., Kihara A., Sano T., Igarashi Y. (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761, 474–483 [DOI] [PubMed] [Google Scholar]
- 20. Linder M. E., Jennings B. C. (2013) Mechanism and function of DHHC S-acyltransferases. Biochem. Soc. Trans. 41, 29–34 [DOI] [PubMed] [Google Scholar]
- 21. Jones M. L., Collins M. O., Goulding D., Choudhary J. S., Rayner J. C. (2012) Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Planey S. L., Zacharias D. (2009) Palmitoyl acyltransferases, their substrates, and novel assays to connect them (Review). Mol. Membr. Biol. 26, 14–31 [DOI] [PubMed] [Google Scholar]
- 23. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Global analysis of protein palmitoylation in yeast. Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dietrich L. E., Ungermann C. (2004) On the mechanism of protein palmitoylation. EMBO Rep. 5, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rocks O., Gerauer M., Vartak N., Koch S., Huang Z.-P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P. I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 [DOI] [PubMed] [Google Scholar]
- 26. Beck J. R., Fung C., Straub K. W., Coppens I., Vashisht A. A., Wohlschlegel J. A., Bradley P. J. (2013) A Toxoplasma palmitoyl acyltransferase and the palmitoylated armadillo repeat protein TgARO govern apical rhoptry tethering and reveal a critical role for the rhoptries in host cell invasion but not egress. PLoS Pathog. 9, e1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mueller C., Klages N., Jacot D., Santos J. M., Cabrera A., Gilberger T. W., Dubremetz J.-F., Soldati-Favre D. (2013) The Toxoplasma protein ARO mediates the apical positioning of rhoptry organelles, a prerequisite for host cell invasion. Cell Host Microbe 13, 289–301 [DOI] [PubMed] [Google Scholar]
- 28. Frénal K., Tay C. L., Mueller C., Bushell E. S., Jia Y., Graindorge A., Billker O., Rayner J. C., Soldati-Favre D. (2013) Global analysis of apicomplexan protein S-acyltransferases reveals an enzyme essential for invasion. Traffic 14, 895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trager W., Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 30. Fivelman Q. L., McRobert L., Sharp S., Taylor C. J., Saeed M., Swales C. A., Sutherland C. J., Baker D. A. (2007) Improved synchronous production of Plasmodium falciparum gametocytes in vitro. Mol. Biochem. Parasitol. 154, 119–123 [DOI] [PubMed] [Google Scholar]
- 31. Fidock D. A., Wellems T. E. (1997) Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. 94, 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Treeck M., Struck N. S., Haase S., Langer C., Herrmann S., Healer J., Cowman A. F., Gilberger T. W. (2006) A conserved region in the EBL proteins is implicated in microneme targeting of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 281, 31995–32003 [DOI] [PubMed] [Google Scholar]
- 33. Flueck C., Bartfai R., Niederwieser I., Witmer K., Alako B. T., Moes S., Bozdech Z., Jenoe P., Stunnenberg H. G., Voss T. S. (2010) A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 6, e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Struck N. S., de Souza Dias S., Langer C., Marti M., Pearce J. A., Cowman A. F., Gilberger T. W. (2005) Re-defining the Golgi complex in Plasmodium falciparum using the novel Golgi marker PfGRASP. J. Cell Science 118, 5603–5613 [DOI] [PubMed] [Google Scholar]
- 35. Haase S., Cabrera A., Langer C., Treeck M., Struck N., Herrmann S., Jansen P. W., Bruchhaus I., Bachmann A., Dias S., Cowman A. F., Stunnenberg H. G., Spielmann T., Gilberger T. W. (2008) Characterization of a conserved rhoptry-associated leucine zipper-like protein in the malaria parasite Plasmodium falciparum. Infect. Immun. 76, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finn R. D., Clements J., Eddy S. R. (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aurrecoechea C., Barreto A., Brestelli J., Brunk B. P., Cade S., Doherty R., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Hu S., Iodice J., Kissinger J. C., Kraemer E. T., Li W., Pinney D. F., Pitts B., Roos D. S., Srinivasamoorthy G., Stoeckert C. J., Jr., Wang H., Warrenfeltz S. (2013) EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res. 41, 684–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. (2006) Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 39. Magrane M., Consortium U. (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011 database/bar009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cherry J. M., Adler C., Ball C., Chervitz S. A., Dwight S. S., Hester E. T., Jia Y., Juvik G., Roe T., Schroeder M., Weng S., Botstein D. (1998) Saccharomyces Genome Database. Nucleic Acids Res. 26, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W., Godzik A. (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 [DOI] [PubMed] [Google Scholar]
- 42. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Do C. B., Mahabhashyam M. S., Brudno M., Batzoglou S. (2005) ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 15, 330–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 [DOI] [PubMed] [Google Scholar]
- 45. Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keane T. M., Creevey C. J., Pentony M. M., Naughton T. J., Mclnerney J. O. (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., Huelsenbeck J. P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guindon S., Delsuc F., Dufayard J. F., Gascuel O. (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137 [DOI] [PubMed] [Google Scholar]
- 49. Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A. F., Selengut J. D., Sigrist C. J., Thimma M., Thomas P. D., Valentin F., Wilson D., Wu C. H., Yeats C. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 51. Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yeoman J. A., Hanssen E., Maier A. G., Klonis N., Maco B., Baum J., Turnbull L., Whitchurch C. B., Dixon M. W., Tilley L. (2011) Tracking glideosome-associated protein 50 reveals the development and organization of the inner membrane complex of Plasmodium falciparum. Eukaryot. Cell 10, 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ridzuan M. A., Moon R. W., Knuepfer E., Black S., Holder A. A., Green J. L. (2012) Subcellular location, phosphorylation and assembly into the motor complex of GAP45 during Plasmodium falciparum schizont development. PLoS One 7, e33845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bullen H. E., Tonkin C. J., O'Donnell R. A., Tham W. H., Papenfuss A. T., Gould S., Cowman A. F., Crabb B. S., Gilson P. R. (2009) A novel family of Apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. J. Biol. Chem. 284, 25353–25363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meszoely C. A., Erbe E. F., Steere R. L., Trosper J., Beaudoin R. L. (1987) Plasmodium falciparum: freeze-fracture of the gametocyte pellicular complex. Exp. Parasitol. 64, 300–309 [DOI] [PubMed] [Google Scholar]
- 56. Aury J. M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B. M., Ségurens B., Daubin V., Anthouard V., Aiach N., Arnaiz O., Billaut A., Beisson J., Blanc I., Bouhouche K., Câmara F., Duharcourt S., Guigo R., Gogendeau D., Katinka M., Keller A. M., Kissmehl R., Klotz C., Koll F., Le Mouël A., Lepère G., Malinsky S., Nowacki M., Nowak J. K., Plattner H., Poulain J., Ruiz F., Serrano V., Zagulski M., Dessen P., Bétermier M., Weissenbach J., Scarpelli C., Schächter V., Sperling L., Meyer E., Cohen J., Wincker P. (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444, 171–178 [DOI] [PubMed] [Google Scholar]
- 57. Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., Badger J. H., Ren Q., Amedeo P., Jones K. M., Tallon L. J., Delcher A. L., Salzberg S. L., Silva J. C., Haas B. J., Majoros W. H., Farzad M., Carlton J. M., Smith R. K., Jr., Garg J., Pearlman R. E., Karrer K. M., Sun L., Manning G., Elde N. C., Turkewitz A. P., Asai D. J., Wilkes D. E., Wang Y., Cai H., Collins K., Stewart B. A., Lee S. R., Wilamowska K., Weinberg Z., Ruzzo W. L., Wloga D., Gaertig J., Frankel J., Tsao C. C., Gorovsky M. A., Keeling P. J., Waller R. F., Patron N. J., Cherry J. M., Stover N. A., Krieger C. J., del Toro C., Ryder H. F., Williamson S. C., Barbeau R. A., Hamilton E. P., Orias E. (2006) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 59. Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M. R., El-Husseini A. (2009) Neuronal palmitoyl acyltransferases exhibit distinct substrate specificity. FASEB J. 8, 2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hou H., John Peter A. T., Meiringer C., Subramanian K., Ungermann C. (2009) Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic 10, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 61. Wasmuth J., Daub J., Peregrín-Alvarez J. M., Finney C. A., Parkinson J. (2009) The origins of apicomplexan sequence innovation. Genome Res. 19, 1202–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martinsen E. S., Perkins S. L., Schall J. J. (2008) A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol. Phylogenet. Evol. 47, 261–273 [DOI] [PubMed] [Google Scholar]
- 63. Laity J. H., Lee B. M., Wright P. E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 [DOI] [PubMed] [Google Scholar]
- 64. González Montoro A., Quiroga R., Valdez Taubas J. (2013) Zinc co-ordination by the DHHC cysteine-rich domain of the palmitoyltransferase Swf1. Biochem. J. 454, 427–435 [DOI] [PubMed] [Google Scholar]
- 65. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moult J., Fidelis K., Kryshtafovych A., Schwede T., Tramontano A. (2014) Critical assessment of methods of protein structure prediction (CASP)–round x. Proteins 82, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wiederstein M., Sippl M. J. (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]