Background: Helix-helix interactions commonly mediate a host of biological functions in membrane proteins.
Results: A stapled peptide sequentially complementary to a helix-helix interaction motif disrupted a bacterial multidrug resistance efflux pump.
Conclusion: Stapling membrane-penetrating peptides represents a viable way to target protein-protein interactions within membranes while retaining metabolic stability.
Significance: Stapled peptides are prospective therapeutics for inhibiting membrane protein interactions involved in disease states.
Keywords: Cell-penetrating Peptide (CPP), Membrane Bilayer, Membrane Protein, Multidrug Transporter, Peptide Chemical Synthesis, Peptide Conformation, Protein Drug Interaction
Abstract
Bacterial cell membranes contain several protein pumps that resist the toxic effects of drugs by efficiently extruding them. One family of these pumps, the small multidrug resistance proteins (SMRs), consists of proteins of about 110 residues that need to oligomerize to form a structural pathway for substrate extrusion. As such, SMR oligomerization sites should constitute viable targets for efflux inhibition, by disrupting protein-protein interactions between helical segments. To explore this proposition, we are using Hsmr, an SMR from Halobacter salinarum that dimerizes to extrude toxicants. Our previous work established that (i) Hsmr dimerization is mediated by a helix-helix interface in Hsmr transmembrane (TM) helix 4 (residues 90GLALIVAGV98); and (ii) a peptide comprised of the full TM4(85–105) sequence inhibits Hsmr-mediated ethidium bromide efflux from bacterial cells. Here we define the minimal linear sequence for inhibitor activity (determined as TM4(88–100), and then “staple” this sequence via Grubbs metathesis to produce peptides typified by acetyl-A-(Sar)3-88VVGLXLIZXGVVV100-KKK-NH2 (X = 2-(4′-pentenyl)alanine at positions 92 and 96; Z = Val, Gly, or Asn at position 95)). The Asn95 peptide displayed specific efflux inhibition and resensitization of Hsmr-expressing cells to ethidium bromide; and was non-hemolytic to human red blood cells. Stapling essentially prevented peptide degradation in blood plasma and liver homogenates versus an unstapled counterpart. The overall results confirm that the stapled analog of TM4(88–100) retains the structural complementarity required to disrupt the Hsmr TM4-TM4 locus in Hsmr, and portend the general validity of stapled peptides as therapeutics for the disruption of functional protein-protein interactions in membranes.
Introduction
Protein-protein interactions (PPIs)3 have gained appreciation as new targets for drug development. However, due to binding motifs that involve large continuous or discontinuous stretches at the interface of the interacting proteins, the quest for effective inhibitors of these interactions has proven challenging (1). For soluble proteins, a number of innovative ideas to target PPIs have emerged, such as fragment-based strategies, antibody-aided technologies, and scaffolding (2). In membranes, PPIs predominantly occur between two or more α-helices, because this is the preferred secondary structure element of membrane proteins. In most cases, the driving force for the formation of helix-helix interactions is to maximize van der Waals interactions by forming “knobs into holes”-like complexes (3); this situation is epitomized, as examples, by the right-handed GASright motif, that follows the sequence pattern G(A,S)XXXG(A,S); the small residue heptad motif (GXXXXXXG); and the leucine zipper. Other factors that contribute to membrane PPIs are hydrogen bonds, π-π interactions (4, 5), and cation-π interactions (6). Although targeting transmembrane (TM) domains remains challenging, approaches with linear peptides have already led to significant successes in laboratory settings. For example, peptides based on TM helix IV of the Class II G-protein coupled secretin receptor inhibited oligomerization and thereby secretin binding and biological function of the receptor (7). Peptides derived from an efflux pump from the ATP-binding cassette family led to successful blocking of tertiary interactions between its 12 membrane helices and efflux inhibition (8).
More recently, stapling of peptides derived from helical protein domains are receiving increased attention for the manipulation of helix-helix interactions (9). In this approach, peptide fragments that may otherwise be unstructured are constrained into a helical shape, often complementary to the target pharmacophore, by introduction of an all-hydrocarbon backbone tether. This decreases the entropic cost of peptide folding and facilitates recognition of the helix-helix binding site. Additionally, by masking the peptide backbone in the inner helix, the peptide is protected from proteolytic cleavage, generally increasing peptide metabolic stability. Moreover, some peptides carrying this helix-staple have been found to traverse membranes by pinocytosis and to be able to bind to intracellular targets and inhibit PPIs that potentially lead to diseases (10, 11). The fact that the amphiphilic helices formed by stapling can target the membrane has encouraged application of this approach for antimicrobial peptides (12, 13). However, to date, helix staples that inhibit helix-helix interactions in membrane proteins have not been described.
The aim of the present study was to apply the approach of all-hydrocarbon helix stapling that has been successfully used for manipulation of PPIs in soluble proteins (14) to inhibit the membrane-embedded bacterial multidrug efflux pumps of the small multidrug resistance (SMR) family. Efflux pumps constitute a major mechanism by which bacteria become multidrug resistant, an ever increasing peril to public health. To date, five families of multidrug efflux pumps have been categorized, including the major facilitator superfamily, the adenosine triphosphate (ATP)-binding cassette (ABC) family, the multidrug and toxic compound extrusion family, the resistance-nodulation-division superfamily, and the above mentioned SMR family (15). The best characterized SMR protein is the Escherichia coli efflux pump EmrE that predominantly extrudes lipophilic cationic compounds such as ethidium bromide and benzalkonium, a commonly used clinical disinfectant, as well as the uncharged antibiotic tobramycin (16). EmrE consists of 110 amino acids that form a four-helix bundle in the membrane environment.
Due to their relatively small size, SMRs generally need to oligomerize to form a structural pathway through which to extrude relatively large substrates. The smallest functional unit of EmrE is a dimer (17), for which data increasingly point to a dual topology, antiparallel dimer (18–21). Although TM helices 1 to 3 play a major role in the binding and transport process with Glu14 being conserved throughout the whole SMR family as the major docking station for substrates and protons (22), transmembrane helix 4 (TM4) has been found to be the key mediator of SMR oligomerization (23). Work in our lab on the EmrE homologue Hsmr from Halobacter salinarum established that the minimal sequence necessary for efficient dimerization is a TM4-TM4 heptad interface, namely TM4 residues 90Gly-Leu-X-Leu-Ile-X-X-Gly97-Val98 (24). This finding led to the hypothesis that residues Gly90 and Gly97 in combination form a “hole” into which Val98 docks in as a “knob” (24).
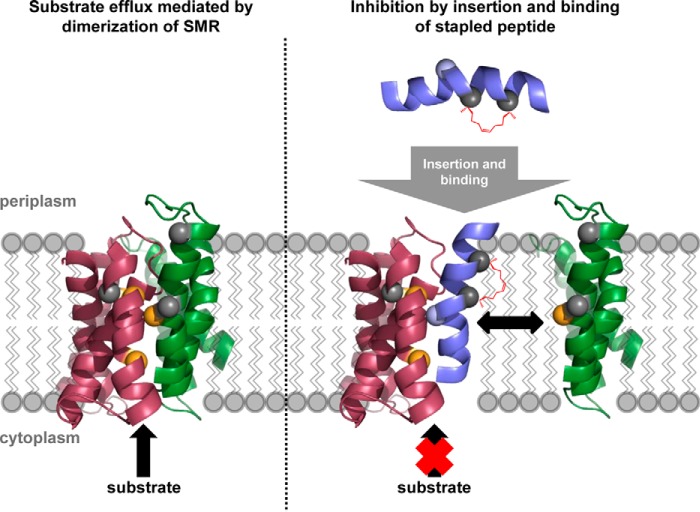
One can envision two broad approaches through which to inhibit the function of SMR efflux pumps: either directly attack the binding site, or disrupt protein dimerization. The SMR binding site, however, accommodates a huge variety of substrates that differ in chemical composition, size, spatial orientation and charge. Thus, the binding site is a “moving target” that is not conducive to a universal approach. Specific disruption of Hsmr dimerization, in contrast, would render the whole pump inactive and should therefore be the more powerful approach (Fig. 1). Focusing on the TM4-TM4 heptad dimerization sequence, we report here the development of stapled peptides that are significantly shorter than a 28-residue linear full-length TM4 peptide (25), and display specific efflux inhibition and resensitization of Hsmr-expressing E. coli cells to ethidium bromide.
FIGURE 1.
Inhibition of membrane-based protein-protein interactions by a stapled peptide. Wild type EmrE, an SMR homologue of Hsmr, relies on dimerization for function (depicted is EmrE antiparallel dimer from Protein Data Bank entry 3B5D) (36). Dimerization depends on interaction between TM4 of both monomers. A peptide resembling the interaction sequence of TM4 can compete out this helix-helix interaction, by inserting into the membrane and interacting at the target locus with TM4 of a given monomer, thereby preventing dimerization and consequently efflux of substrate. Introducing a staple renders the peptide into a preformed helical shape that enables optimal interaction of the hydrophobic staple moiety with both the target TM4-TM4 interaction motif and the lipid environment.
MATERIALS AND METHODS
Peptide Synthesis and Purification
Automated peptide synthesis was carried out using a PS3 peptide synthesizer (Protein Technologies, Tucson, AZ) applying the fluorenylmethoxycarbonyl (Fmoc)/tert-butyl (tBu) protection group strategy. Procedures were carried out as described previously (25) with some slight modifications. Generally, per reaction cycle 400 μmol (8 eq) of Fmoc-protected amino acid, 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxidhexafluorophosphate (HATU; 8 eq, 152 mg), and N,N′-diisopropylethylamine were used. Coupling time was 45 min. For Val and Ile as well as amino acids following them, coupling was repeated. At desired stapling positions, either (S)-N-Fmoc-2-(4′-pentenyl)alanine (Fmoc-S5-OH) or (R)-N-Fmoc-2-(7′-octenyl)alanine (Fmoc-R8-OH) (OKeanos Tech. Co. Ltd., Beijing, China) was incorporated by dissolving 150 μmol (3 eq) of amino acid and 6-chloro-benzotriazole-1-yl-oxy-tris-pyrrolidinophosphonium hexafluorophosphate (Pyclock) (3 eq, 83.2 mg) in 2 ml of N,N′-dimethylformamide/dichloromethane (3:7 (v/v)) and coupling at room temperature for 2 h. The following amino acid (400 μmol, 8 eq) was then coupled twice with 400 μmol (8 eq, 222 mg) Pyclock for 2 h at room temperature. Stapling of the peptides was carried out as described in the literature (26). For the metathesis reaction, the resin was swollen in dichloroethane. 10 μmol (20%, 8.2 mg) of Grubbs catalyst 1st generation (Sigma) was dissolved in 2 ml of dichloroethane and added to the resin. The suspension was agitated for 2 h at room temperature by stirring with a magnetic stir bar and nitrogen bubbling. Dichloroethane was added during this time period as it evaporated. The whole procedure was repeated once. Acetylation was carried out as described (25). N-terminal fluorescence labeling was achieved by adding a solution of 200 μmol (4 eq, 86 mg) of 5(6)-carboxy-tetramethylrhodamine (TAMRA) (EMD Millipore, Billerica, Massachusetts, USA), HATU (4 eq, 76 mg), and N,N′-diisopropylethylamine (4 eq, 34.2 μl) in 2 ml of N,N′-dimethylformamide to the resin and shaking for 2 h at room temperature. Cleavage was carried out as described previously. Purification was carried out by reversed-phase high performance liquid chromatography (RP-HPLC) using a Jupiter C4 column (250 × 21.2 mm, particle size 10 μm, pore size 300 Å, Phenomenex, Torrance, CA) and a gradient of solvent A (94.95% water, 4.95% acetonitrile, 0.1% TFA) and solvent B (94.95% acetonitrile, 4.95% water, 0.1% TFA). Identity of the peptides was confirmed by MALDI-TOF mass spectrometry. Purity (generally >95%) was confirmed by RP-HPLC using a Jupiter C4 column (250 × 4.6 mm, particle size 10 μm, pore size 300 Å, Phenomenex, Torrance, CA).
Circular Dichroism (CD) Spectroscopy
Secondary structure determination was carried out on a Jasco J-815 circular dichroism spectrometer (Jasco Applied Sciences Ltd., Halifax, Nova Scotia, Canada) utilizing a 1-mm path length quartz cuvette following published protocols (25). Helicity was determined by mean residue molar ellipticity at 222 nm for at least two independent experiments.
Ethidium Bromide Efflux Assay
Ethidium bromide efflux assays were performed as described previously with slight modifications (25). Briefly, after overexpression of Hsmr in E. coli BL21(DE3) cells harboring the Hsmr_pT7–7 expression vector, cells were harvested, resuspended, and diluted to an A600 nm = 0.1 in Minimal Medium A. Cells were then treated with 40 μm carbonyl cyanide m-chlorophenyl hydrazine (CCCP) for 5 min at room temperature, before adding 1 μg/ml of ethidium bromide and 1 μm peptide solution in water. Cells were incubated at 37 °C and 220 rpm for 30 min, centrifuged for 2 min, resuspended in Minimal Medium A supplemented with 1 μg/ml of ethidium bromide and quickly measured for fluorescence on a Photon Technology International C-60 spectrofluorimeter (Photon Technology International, Birmingham, NJ) over a time period of 1250 s while stirring (λex = 530 nm, slit width 2 nm, λem = 600 nm, slit width 4 nm, 1-s intervals). Blank fluorescence as well as plateau values for the Hsmr control curve were subtracted from fluorescence values. Curves were then linearized using Equation 1.
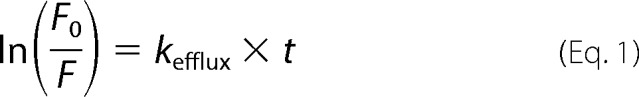 |
The efflux rate constant, kefflux, was determined by linear regression using GraphPad Prism 6, version 6.01. Experiments were carried out at least three times. All values are given as mean ± S.E. Statistical analysis was performed using one-way analysis of variance (GraphPad Prism 6, version 6.01). Significance values refer to comparison of all peptides to TM4(88–100)scr.
Ethidium Resensitization (ER) Assay and Antimicrobial Assay (AA)
Assays were performed as previously described with slight modifications (25). Briefly, 2-fold serial dilutions of peptides ranging from 0 to 40 μm (AA) or 0 to 20 μm (ER) were incubated for 20 h at 37 °C with 50,000 cfu/well of an overnight culture of E. coli BL21(DE3) (Invitrogen) harboring the Hsmr_pT7-7 expression vector in Mueller-Hinton broth with (ER) or without (AA) 30 μg/ml of ethidium bromide. In the case of ER, 100 μm isopropyl β-d-1-thiogalactopyranoside was added to the medium. Cell suspension in the presence or absence of ethidium bromide served as positive controls. Negative controls were Mueller-Hinton broth in the presence or absence of ethidium bromide. Optical density was measured at 600 nm with a SpectraMax Plus384 absorbance microplate reader (Molecular Devices, Downingtown, PA). All values were baseline subtracted. Results are given as minimal inhibitory concentrations (MIC). Assays were carried out at least three times in duplicates.
Hemolysis Assay
Hemolysis assays (RBC) were performed as described previously (27). The highest final peptide concentration was 200 μm. All values were baseline subtracted and normalized to a positive control. Results are given as minimum hemolytic concentration (MHC), which is the lowest peptide concentration at which hemoglobin concentrations reached at least 10% of the positive control. Assays were carried out at least twice in duplicate.
Peptide Stability in Blood Plasma and Liver Homogenates
Stability assays in blood plasma and liver homogenates were performed as previously described with some slight variations (28, 29). TAMRA-labeled peptide was dissolved to 10 μm in either blood plasma or beef liver homogenate. Samples were incubated at 37 °C with shaking. At the indicated time points, 300 μl were withdrawn, mixed with 600 μl of solvent C (acetonitrile/ethanol = 1:1 (v/v)), and frozen at −20 °C overnight. Samples were centrifuged at 16,000 × g for 30 s, supernatants were incubated for further 20 min at −20 °C and centrifuged again. Supernatants were analyzed by RP-HPLC on a Jupiter C4 column (250 × 4.6 mm, particle size 10 μm, pore size 300 Å, Phenomenex, Torrance, CA) at a wavelength of 565 nm using a gradient of 20 to 70% solvent B in A over 40 min at a flow rate of 1 ml/min. Peak identity was proven by MALDI-TOF mass spectrometry. Peaks were integrated using Adobe® Photoshop® CS4 Extended 11.0. Data points are presented as % intact peptide relative to the control at 0 h incubation time. Curves were linearized by Equation 2 and kdecay was determined by linear regression using GraphPad Prism 6, version 6.01.
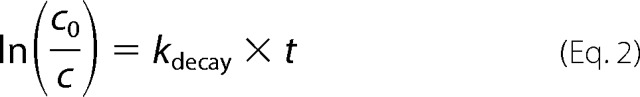 |
Half-lives were calculated by applying Equation 3 and are given as mean ± S.E. of two independent experiments.
 |
RESULTS
Synthesis of Hsmr TM4-based Peptides
The sequences for the peptides synthesized in this study were derived from TM4 of the efflux pump Hsmr found in the archaea H. salinarum (30). Given that Poulsen and Deber (25) had created a linear 28-residue peptide based on the full-length of the Hsmr TM4 that indeed inhibited EtBr efflux by more than 60%, we first sought to find shorter linear sequences derived from TM4 that displayed similar inhibiting activity.
To increase peptide aqueous solubility, while not compromising subsequent entry into the bacterial membrane, tags were applied to both termini of our designed peptides (31). Although the C terminus consisted of three positively charged Lys residues that specifically target the peptides to the negatively charged bacterial membrane, the uncharged N-terminal tag, consisting of three Sar (N-methyl-Gly) residues, one Ala, and an N-terminal acetyl group, is anticipated to aid in peptide partitioning by “burrowing” into the lipid bilayer. The peptides synthesized in this work are given in Fig. 2). TM4(90–98) consists of the minimal dimerization motif previously identified (24). Subsequent elongation of this motif based on the TM4 sequence to identify a minimal sequence that is able to inhibit Hsmr-mediated ethidium efflux comparable with the initial TM4 peptide, led to synthesis of TM4(89–99) and TM4(88–100).
FIGURE 2.
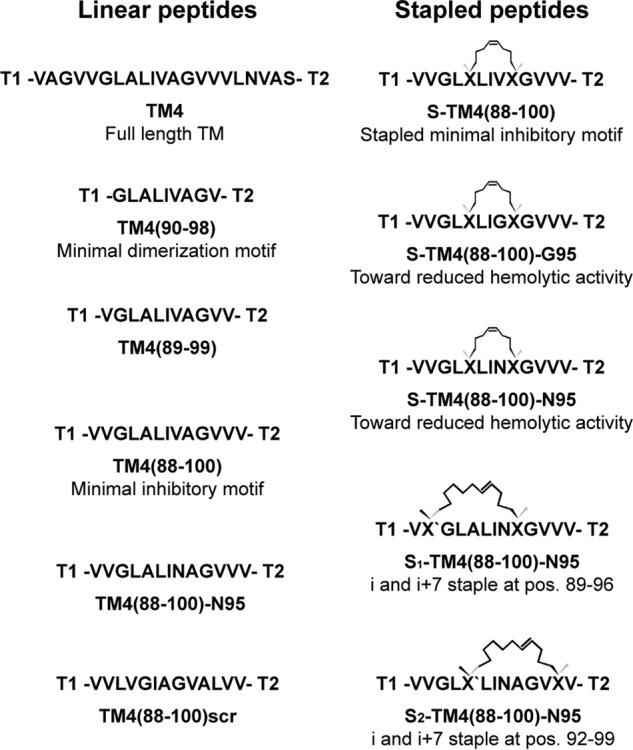
Overview of synthesized peptides. Solubility tags: T1, CH3CO-Ala-Sar3-; T2, -Lys3-NH2; X, (S)-N-Fmoc-2-(4′-pentenyl)alanine; X′, (R)-N-Fmoc-2-(7′-octenyl)alanine.
TM4(88–100) was identified as the minimal inhibitory motif (see below) and was therefore selected to undergo helix stapling by introduction of alkenylalanyl residues at the i and i+4 positions, followed by subsequent Grubbs metathesis (26), leading to a set of peptides stapled at positions 92 and 96 (S-TM4(88–100)). To reduce overall peptide hydrophobicity in the stapled peptides, Val at position 95 was mutated to either Gly (S-TM4(88–100)-G95) or Asn (S-TM4(88–100)-N95). For the latter, an unstapled control peptide was synthesized (TM4(88–100)-N95). Furthermore, i and i+7 staples were tested at positions 89 and 96 (S1-TM4(88–100)-N95) and at 92 and 99 (S2-TM4(88–100)-N95). To determine specificity of the observed biological effects, a control peptide that ostensibly will not bind to Hsmr TM4 was created, consisting of a “scrambled” variant of the minimal inhibitory peptide (TM4(88–100)scr). All peptides were synthesized by solid-phase peptide synthesis applying the Fmoc/tBu-protection group strategy. Staples were introduced according to published protocols (26).
Conformations of Linear and Stapled Hsmr TM4 Peptides in Aqueous and Membrane-mimetic Environments
TM4 was largely unstructured in aqueous solution (25), but displayed almost full α-helicity in circular dichroism spectra recorded in the micellar environment of SDS (MRE = about −30,000° at 222 nm) (Fig. 3). However, helicity of the “pure” dimerization motif peptide (TM4(90–98)) dropped significantly in SDS as compared with TM4. Subsequent stepwise elongation of this sequence based on wild type residues in the Hsmr TM4 sequence gave peptide TM4(88–100), which displayed an incremental increase in helicity, reverting back to that of full-length TM4. Extension of the stapling approach to yield the peptide S-TM4(88–100) similarly resulted in helicity in the presence of detergent that was comparable with TM4.
FIGURE 3.
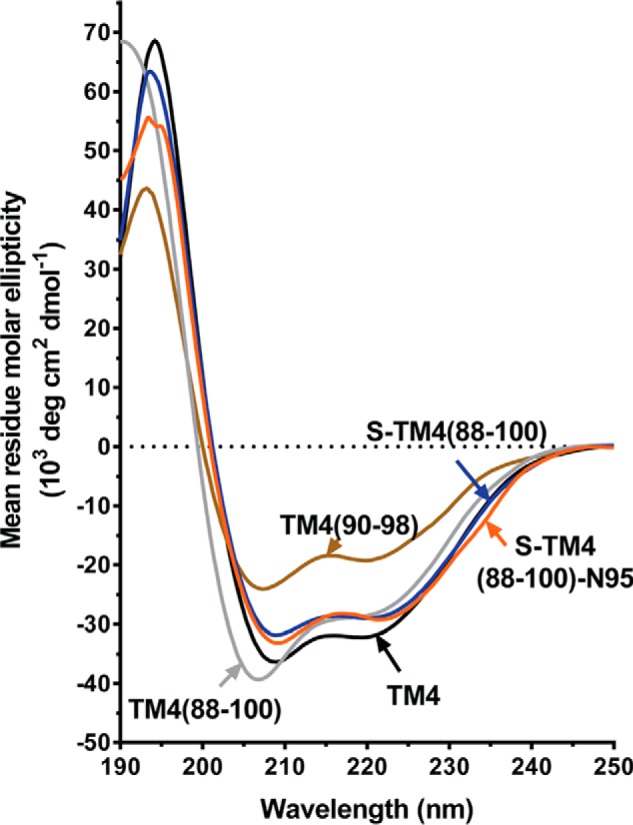
Circular dichroism spectra of selected linear and stapled Hsmr TM4 peptides. Spectra were obtained with 20 μm peptide in 20 mm SDS. See text for further discussion.
Ellipticity values for the full peptide library are given in Table 1. Here it may be noted that the scrambled peptide displays about 25% less helicity than the other “88–100”-length peptides, likely because the WT TM4 sequence is expected to dimerize: and this process aids in stabilizing the helix along the full peptide sequence. This situation has been previously noted in CD spectra of WT versus scrambled full-length TM4 peptide (25).
TABLE 1.
Structural and functional characterization of linear and stapled peptides
| Peptide | Ellipticity, CD spectra, (MRE222 nm)a | Ethidium efflux, rel. rateb | Ethidium resensitization, MICERc | Antimicrobial activity, MICAAc | Hemolytic activity, MHCd |
|---|---|---|---|---|---|
| μm | |||||
| TM4 | −30,300° | 0.37 | 1.25 | > 40 | 12.5 |
| TM4(90–98) | −18,300° | 0.91 | >20 | > 40 | > 200 |
| TM4(89–99) | −18,600° | 0.87 | >20 | >40 | >200 |
| TM4(88–100) | −32,100° | 0.51 | 5–10 | 20 | >200 |
| TM4(88–100)-N95 | −26,200° | 0.90 | >20 | >40 | >200 |
| TM4(88–100)scr | −20,000° | 1.07 | 20 | 40 | NDe |
| S-TM4(88–100) | −32,100° | 0.24 | 0.63–1.25 | 5 | 12.5 |
| S-TM4(88–100)-G95 | −26,200° | 0.56 | 1.25–2.5 | 10 | 25 |
| S-TM4(88–100)-N95 | −28,700° | 0.67 | 1.25–2.5 | 20 | >200 |
| S1-TM4(88–100)-N95 | −25,700° | 0.52 | 0.31 | 5 | 100 |
| S2-TM4(88–100)-N95 | −30,400° | 0.60 | 0.31–0.63 | 5 | 100 |
a MRE = mean residue molar ellipiticity (deg cm2 dmol−1) in circular dichroism spectra at 222 nm, recorded on samples of 20 μm peptide in 20 mm SDS. See also Fig. 3.
b For each experiment, relative efflux rate constants were determined using first-order exponential decay and normalized to ethidium efflux in Hsmr-expressing E. coli BL21(DE3) cells without addition of peptide (kaverage = 2.4 ± 0.4 s−1 with n = 11). Cells not expressing Hsmr have a relative efflux rate of 0.15 ± 0.02 (25).
c Minimal inhibitory concentration (MIC) represents the lowest concentration at which bacterial growth of Hsmr-expressing E. coli BL21(DE3) was completely inhibited with (MICER) or without (MICAA) the presence of ethidium bromide. See “Materials and Methods” for further details.
d MHC is the lowest concentration at which hemoglobin concentrations reached at least 10% of the positive control.
e ND, not determined.
Inhibition of Hsmr-mediated Ethidium Efflux by TM4 Peptides
As illustrated in Fig. 4 for selected peptides, and listed in Table 1 for the full peptide library, TM4 was able to inhibit ethidium efflux by 63%, whereas TM4(90–98) and TM4(89–99) did not show any activity. However, stapling of TM4(88–100) to give S-TM4(88–100), resulted in significant inhibition of ethidium efflux to 24%. This effect was reduced in both less hydrophobic variants S-TM4(88–100)-G95 (56%) and S-TM4(88–100)-N95 (67%); however, inhibition of efflux remained significant. In contrast, the scrambled variant TM4(88–100)scr did not show significant efflux inhibition (not shown). Importantly, the peptide concentration used in the efflux inhibition assays (=1 μm) was not toxic to the cells, and hence not a source of any observed antimicrobial activity.
FIGURE 4.
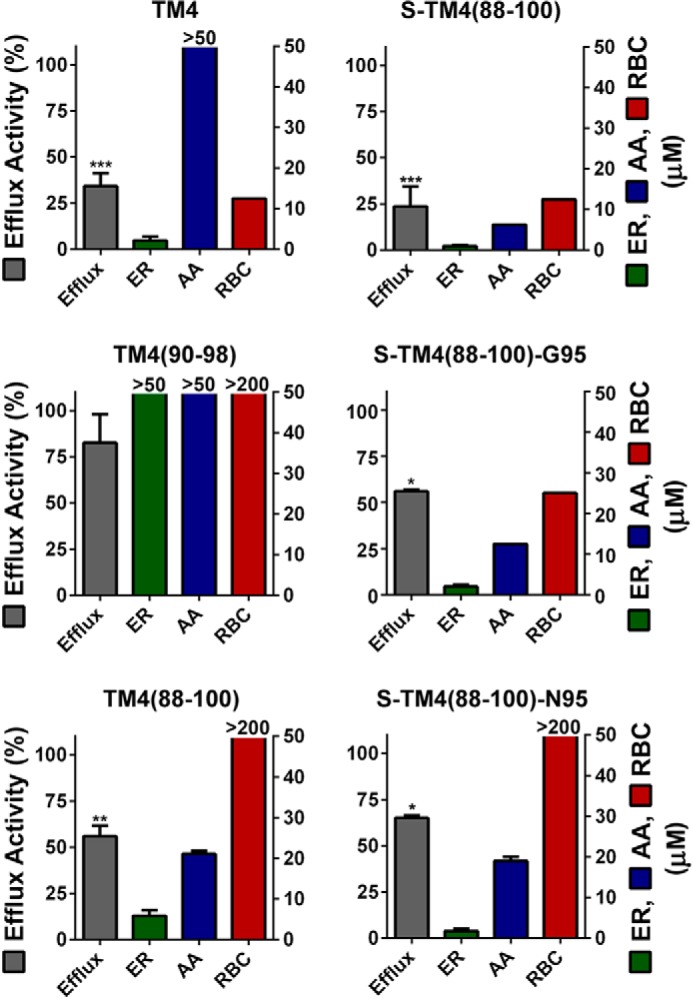
Profiles of peptide biological activity. Four assays, ethidium efflux activity (Efflux), ER, AA, and hemolytic activity in human RBC are depicted for linear peptides TM4, TM4(90–98), and TM4(88–100) (left panels); and stapled peptides S-TM4(88–100), S-TM4(88–100)-G95, and S-TM4(88–100)-N95 (right panels). Efflux assays were run using samples of 1 μm peptide and 1 μg/ml of ethidium. Efflux rate constants were determined using first-order exponential decay and normalized to ethidium efflux in bacteria without addition of peptide (kaverage = 2.4 ± 0.4 s−1 with n = 11). Cells not expressing Hsmr have a relative efflux rate of 0.15 ± 0.02 (25). Values are given as mean ± S.E. of at least two independent experiments and significance values refer to comparison of all peptides to TM4(88–100)scr (****, p < 0.001; ***, p < 0.005; *, p < 0.05). Values for ER, AA, and RBCs are given as the minimal inhibitory or hemolytic concentration, which is the lowest concentration at which complete growth inhibition occurred or hemoglobin concentrations reached at least 10% of the positive control. Values for all assays were determined in at least two independent experiments.
Ethidium Resensitization and Antimicrobial Activity of Hsmr TM4-based Peptides
Resensitization, the ability of a given peptide to kill the bacteria upon overnight incubation with ethidium, and intrinsic antimicrobial activity, the ability of a given peptide to kill bacteria in the absence of ethidium, were determined by incubation of Hsmr-expressing E. coli BL21(DE3) and 2-fold dilutions of peptide in the presence or absence, respectively, of ethidium bromide. EtBr concentration was chosen to be low enough to enable growth of Hsmr-expressing bacteria but high enough to completely inhibit growth of bacteria that expressed a non-functional Hsmr mutant. As presented for selected peptides in Fig. 4, full-length TM4 displayed a resensitization MICER of 1.25 μm and did not show any antimicrobial activity (MICAA > 50). TM4(88–100) was able to inhibit growth of bacteria in the presence of ethidium in the range of 5–10 μm. However, it also showed some intrinsic antibacterial activity with an MICAA of 20 μm.
Stapling of the peptides generally led to an enhancement of resensitization in the presence of ethidium bromide. Thus, stapling of TM4(88–100) in the same positions yielded S-TM4(88–100) with an MICER of 0.63–1.25 μm, but with a gain in intrinsic toxicity against bacterial cells. Decreasing the overall hydrophobicity of the peptide by substituting Val95 for Asn resulted in peptide S-TM4(88–100)-N95 that showed only a slight decrease in resensitization but a considerable decrease in intrinsic antibacterial activity compared with S-TM4(88–100). Resensitization and antibacterial data for the full peptide library are presented in Table 1.
Evaluation of TM4 Peptide Hemolytic Activity in Human Red Blood Cells
Suitable therapeutics must be highly selective for bacteria versus human host cells. Thus, for each peptide, hemolytic activity was assessed by incubation of 2-fold dilutions of peptide with a 4% suspension of freshly prepared human red blood cells in PBS. Results are presented for selected peptides in Fig. 4, and for the full library in Table 1. Interestingly, TM4 itself showed high hemolytic activity with an MHC of 12.5 μm. Although all other linear peptides were not hemolytic in the concentration range tested, stapling of TM4(88–100) resulted in an MHC comparable with TM4. However, when the hydrophobicity of this peptide was reduced to TM(88–100)-G95, and ultimately to S-TM4(88–100)-N95, there was no detectable hemolytic activity (Fig. 4, lower right panel).
Stapled Peptides Display Proteolytic Stability in Blood Plasma and Liver Homogenates
To quantitate the effect of a helix staple on peptide stability against proteolytic degradation, peptides TM4(88–100) and S-TM4(88–100)-N95 were N terminally labeled, instead of acetylated, with the fluorescent dye TAMRA. Peptides were incubated with pure human blood plasma or beef liver homogenate. As depicted in Fig. 5, TAMRA-labeled TM4(88–100) showed relatively rapid degradation profiles with half-lives around 52 h in human blood plasma and 50 min in liver homogenate. In contrast, the TAMRA-labeled S-TM4(88–100)-N95 showed a 4-fold prolonged blood plasma half-life and virtually no degradation in liver homogenates over the observed time periods.
FIGURE 5.
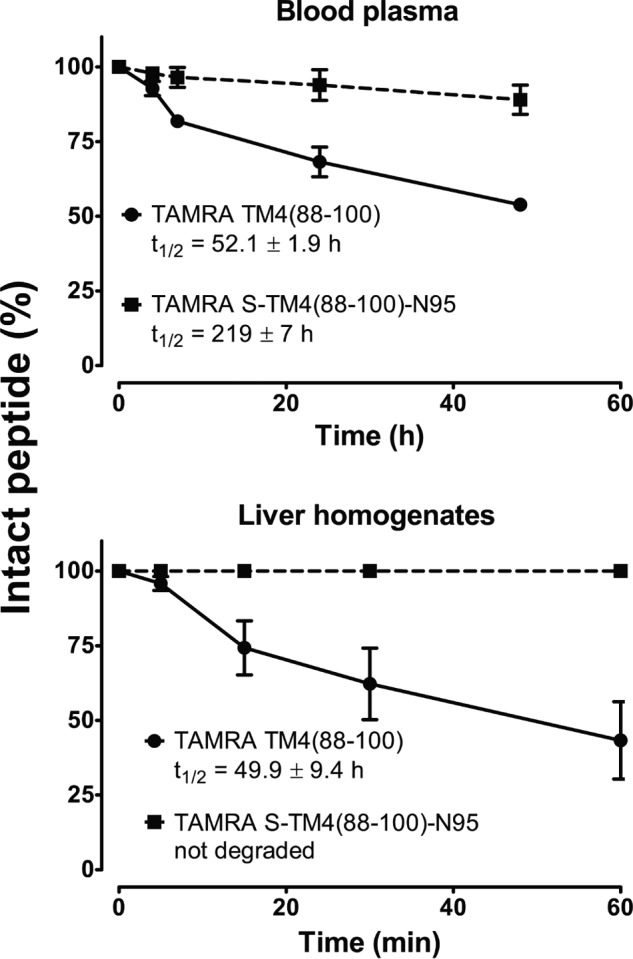
Metabolic stability of unstapled versus stapled peptide in human blood plasma and in bovine liver homogenates. 10 μm peptides (TAMRA-labeled TM4(88–100) and TAMRA-labeled S-TM4(88–100)-N95) were incubated in the respective medium for the indicated time periods. Cleavage products were separated by reversed phase HPLC at 565 nm and identified by MALDI-TOF mass spectrometry. Decay of the intact peptide (%) was determined by peak integration. See ”Materials and Methods“ for further details.
DISCUSSION
We describe the first application, to our knowledge, of the use of peptide stapling to inhibit and disrupt specific protein-protein interactions, and concomitantly protein function, in a membrane-embedded target. These are important observations because (i) protein-protein interactions in membrane proteins generally involve interactions of helical segments; and (ii) the “staple” itself is a membrane-compatible purely hydrocarbon bridge. As such, membrane-embedded oligomerization sites should constitute viable targets for stapled peptides.
The overall aim of this study was to synthesize peptides that enter the membrane and bind to Hsmr monomers or displace monomeric subunits within the dimer. Previous work with the full-length Hsmr TM4 peptide had shown that both the all-D version of TM4, and a scrambled version of TM4, were both inactive in ethidium efflux assays, strongly implying that the activity observed is due to specific binding of (all-L) TM4 to the intrinsic TM4-TM4 dimer motif (25). Thus, starting from our knowledge of the ethidium efflux inhibition by a solubility tagged peptide derived from full-length TM4 of Hsmr, we systematically reduced its sequence, initially limiting peptide length to essentially the heptad dimerization motif (TM4(90–98)), but ultimately elongating the peptide by two wild type Val residues on both N and C termini (TM4(88–100). This strategy yielded a peptide increased in helicity back to the level of TM4, with a concomitant gain of activity in EtBr efflux inhibition and resensitization. More than likely, the TM4-TM4 binding locus in the intact Hsmr dimer is in a “mid-membrane” location, where it would be accessible by our most active peptides (TM4 residues 88–100; 13 residues + possibly 3 Sar residues). We may further note that shorter peptides (such as those with TM4 residues 90–98) were much less active (and less helical), supporting the contention that the inhibitor peptide must substantially penetrate the membrane to align with the TM4 dimerization motif.
The observation that increased efflux values and lower MICER values do trend with lower MICAA values (with TM4 as an outlier) (Table 1), likely indicates the facile membrane-partitioning power of the stapled peptides versus the linear peptides. However, one may discriminate between resensitization and antimicrobial activity by considering the properties of the control peptide (TM4(88–100)scr), which is not able to inhibit efflux (Table 1), but displays both antimicrobial and resensitization activity, with a ratio of MICAA/MICER = 2. This result suggests that a ratio MICAA/MICER greater than 2 indicates, in principle, a contribution of specific Hsmr dimerization interruption rather than solely disruption of the bacterial membrane as the source of resensitization; indeed this ratio is about 16 for the stapled N95 peptide (Table 1). The prolonged metabolic stability of the stapled compounds likely also contributes to this increased ratio.
Although the novel stapled peptides were largely able to inhibit efflux to the extent of TM4 and resensitize bacteria to ethidium at a concentration of about 1 μm, one further needs to address the impact of the overall hydrophobic character of the peptide, which is modulated by the contribution of the helix staple itself. Hemolysis is expected to become significant when the hydrophobicity threshold for partitioning spontaneously into the zwitterionic membranes of erythrocytes has been exceeded (32). To decrease the hemolytic character of S-TM4(88–100), we opted to lower the overall peptide hydrophobicity by introducing less hydrophobic amino acids at position 95, as this was a position in the dimerization motif sequence that was identified as non-conserved throughout the SMR family (24). We chose Gly and Asn as a stepwise approach to introduce modest polarity into this TM sequence. This strategy led to peptide S-TM4(88–100)-N95, which was still able to inhibit efflux and resensitize bacteria in the low micromolar range, but now displayed lower antimicrobial, and importantly, no hemolytic, activity (Fig. 4). Visual comparison with the four-assay profiles obtained experimentally for peptides TM(88–100) and S-TM(88–100)-N95 (bottom two panels, Fig. 4) indicates that these two peptides each display the sought after properties. However, the excellent metabolic stability of S-TM4(88–100)-N95 (Fig. 5) renders this peptide the de facto “lead” structure. These findings correlate well with published data on the stability of stapled BID BH3 peptide in mouse serum (11) and tryptic digests of antimicrobial peptides (12). The gap between resensitization (ER) and intrinsic antimicrobial (AA) activity values for both TM4(88–100) and S-TM4(88–100)-N95 further indicates that these peptides specifically interact with the PPI TM-TM4 of the Hsmr efflux pump and do not exert their function solely by disrupting the bacterial membrane. This key result confirms that the geometric complementarity required to interact with, and disrupt, the TM4-TM4 locus in Hsmr is retained by the stapled analog of the TM4(88–100) peptide.
CONCLUSIONS
The present work has led to the design of a stapled peptide derived from the oligomerization TM4-TM4 site in the bacterial efflux pump Hsmr that displays specific efflux inhibition and resensitization of Hsmr-expressing E. coli cells to ethidium bromide, whereas being non-toxic to mammalian blood cells. Although the small residue heptad motif of TM4 in Hsmr was the basis for the peptides designed in this work, this motif is largely conserved throughout the SMR family (23); accordingly, this general approach should be transferable to homologues of Hsmr, such as PAsmr from Pseudomonas aeruginosa or Tbsmr from Mycobacterium tuberculosis (33).
The overall significance of our study lies not only in these findings, but also represents an initial application of the helix stapling technique to disrupt helix-helix interactions in helical membrane proteins. Such functionality may be of considerable utility in situations beyond the present instance of efflux pump expressing multidrug-resistant bacteria, to multidrug-resistant cancer cells that use efflux pumps of the ABC binding cassette for transport of chemotherapeutics (34), or to oncogenic G-protein coupled receptors that constitutively signal upon dimerization (e.g. epidermal growth factor receptor) (35). Our work therefore represents not only an important step in the development of peptide therapeutics against diseases caused by tertiary or quaternary helix-helix interactions in membrane proteins, but portends the validity of similar approaches to the disruption of protein-protein interactions in membranes.
Acknowledgment
We thank Dr. Li Zhang for recording of MALDI-TOF mass spectra.
This work was supported, in part, by Canadian Institutes of Health Research Grant CIHR FRN-5810 (to C. M. D.).
- PPI
- protein-protein interactions
- Fmoc
- fluorenylmethoxycarbonyl
- Fmoc-R8-OH
- (R)-N-Fmoc-2-(7′-octenyl)alanine
- Fmoc Fmoc-S5-OH
- (S)-N-Fmoc-2-(4′-pentenyl)alanine, HATU, 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxidhexafluorophosphate
- MHC
- minimal hemolytic concentration
- MIC
- minimal inhibitory concentration
- Pyclock
- 6-chloro-benzotriazole-1-yloxy-tris-pyrrolidinophosphonium hexafluorophosphate
- Sar
- sarcosine
- SMR
- small multidrug resistance family
- TAMRA
- 5(6)-carboxy-tetramethylrhodamine
- TIS
- triisopropylsilane
- TM4
- transmembrane helix 4
- ER
- ethidium resensitization
- AA
- antimicrobial assay.
REFERENCES
- 1. Wilson A. J. (2009) Inhibition of protein-protein interactions using designed molecules. Chem. Soc. Rev. 38, 3289–3300 [DOI] [PubMed] [Google Scholar]
- 2. Higueruelo A. P., Jubb H., Blundell T. L. (2013) Protein-protein interactions as druggable targets: recent technological advances. Curr. Opin. Pharmacol. 13, 791–796 [DOI] [PubMed] [Google Scholar]
- 3. Melnyk R. A., Partridge A. W., Deber C. M. (2002) Transmembrane domain mediated self-assembly of major coat protein subunits from Ff bacteriophage. J. Mol. Biol. 315, 63–72 [DOI] [PubMed] [Google Scholar]
- 4. Fink A., Sal-Man N., Gerber D., Shai Y. (2012) Transmembrane domains interactions within the membrane milieu: principles, advances and challenges. Biochim. Biophys. Acta 1818, 974–983 [DOI] [PubMed] [Google Scholar]
- 5. Ng D. P., Poulsen B. E., Deber C. M. (2012) Membrane protein misassembly in disease. Biochim. Biophys. Acta 1818, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 6. Johnson R. M., Hecht K., Deber C. M. (2007) Aromatic and cation-π interactions enhance helix-helix association in a membrane environment. Biochemistry 46, 9208–9214 [DOI] [PubMed] [Google Scholar]
- 7. Harikumar K. G., Pinon D. I., Miller L. J. (2007) Transmembrane segment IV contributes a functionally important interface for oligomerization of the Class II G protein-coupled secretin receptor. J. Biol. Chem. 282, 30363–30372 [DOI] [PubMed] [Google Scholar]
- 8. Maurya I. K., Thota C. K., Verma S. D., Sharma J., Rawal M. K., Ravikumar B., Sen S., Chauhan N., Lynn A. M., Chauhan V. S., Prasad R. (2013) Rationally designed transmembrane peptide mimics of the multidrug transporter protein Cdr1 act as antagonists to selectively block drug efflux and chemosensitize azole-resistant clinical isolates of Candida albicans. J. Biol. Chem. 288, 16775–16787 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Verdine G. L., Hilinski G. J. (2012) Stapled peptides for intracellular drug targets. Methods Enzymol. 503, 3–33 [DOI] [PubMed] [Google Scholar]
- 10. Chang Y. S., Graves B., Guerlavais V., Tovar C., Packman K., To K. H., Olson K. A., Kesavan K., Gangurde P., Mukherjee A., Baker T., Darlak K., Elkin C., Filipovic Z., Qureshi F. Z., Cai H., Berry P., Feyfant E., Shi X. E., Horstick J., Annis D. A., Manning A. M., Fotouhi N., Nash H., Vassilev L. T., Sawyer T. K. (2013) Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 110, E3445–E3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walensky L. D., Kung A. L., Escher I., Malia T. J., Barbuto S., Wright R. D., Wagner G., Verdine G. L., Korsmeyer S. J. (2004) Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chapuis H., Slaninová J., Bednárová L., Monincová L., Budĕs̆ínský M., C̆er̆ovský V. (2012) Effect of hydrocarbon stapling on the properties of α-helical antimicrobial peptides isolated from the venom of hymenoptera. Amino Acids 43, 2047–2058 [DOI] [PubMed] [Google Scholar]
- 13. Pham T. K., Kim D. H., Lee B. J., Kim Y. W. (2013) Truncated and constrained helical analogs of antimicrobial esculentin-2EM. Bioorg. Med. Chem. Lett. 23, 6717–6720 [DOI] [PubMed] [Google Scholar]
- 14. Walensky L. D., Bird G. H. (2014) Hydrocarbon-stapled peptides: principles, practice, and progress. J. Med. Chem. 57, 6275–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piddock L. J. (2006) Multidrug-resistance efflux pumps: not just for resistance. Nat. Rev. Microbiol. 4, 629–636 [DOI] [PubMed] [Google Scholar]
- 16. Nasie I., Steiner-Mordoch S., Schuldiner S. (2012) New substrates on the block: clinically relevant resistances for EmrE and homologues. J. Bacteriol. 194, 6766–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elbaz Y., Steiner-Mordoch S., Danieli T., Schuldiner S. (2004) In vitro synthesis of fully functional EmrE, a multidrug transporter, and study of its oligomeric state. Proc. Natl. Acad. Sci. U.S.A. 101, 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kolbusz M. A., Slotboom D. J., Lolkema J. S. (2012) Role of individual positive charges in the membrane orientation and activity of transporters of the small multidrug resistance family. Biochemistry 51, 8867–8876 [DOI] [PubMed] [Google Scholar]
- 19. Morrison E. A., DeKoster G. T., Dutta S., Vafabakhsh R., Clarkson M. W., Bahl A., Kern D., Ha T., Henzler-Wildman K. A. (2012) Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rapp M., Seppälä S., Granseth E., von Heijne G. (2007) Emulating membrane protein evolution by rational design. Science 315, 1282–1284 [DOI] [PubMed] [Google Scholar]
- 21. Seppälä S., Slusky J. S., Lloris-Garcerá P., Rapp M., von Heijne G. (2010) Control of membrane protein topology by a single C-terminal residue. Science 328, 1698–1700 [DOI] [PubMed] [Google Scholar]
- 22. Muth T. R., Schuldiner S. (2000) A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 19, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elbaz Y., Salomon T., Schuldiner S. (2008) Identification of a glycine motif required for packing in EmrE, a multidrug transporter from Escherichia coli. J. Biol. Chem. 283, 12276–12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poulsen B. E., Rath A., Deber C. M. (2009) The assembly motif of a bacterial small multidrug resistance protein. J. Biol. Chem. 284, 9870–9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poulsen B. E., Deber C. M. (2012) Drug efflux by a small multidrug resistance protein is inhibited by a transmembrane peptide. Antimicrob. Agents Chemother. 56, 3911–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim Y. W., Grossmann T. N., Verdine G. L. (2011) Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat. Protoc. 6, 761–771 [DOI] [PubMed] [Google Scholar]
- 27. Yin L. M., Edwards M. A., Li J., Yip C. M., Deber C. M. (2012) Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 287, 7738–7745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bellmann-Sickert K., Beck-Sickinger A. G. (2011) Palmitoylated SDF1α shows increased resistance against proteolytic degradation in liver homogenates. ChemMedChem 6, 193–200 [DOI] [PubMed] [Google Scholar]
- 29. Bellmann-Sickert K., Elling C. E., Madsen A. N., Little P. B., Lundgren K., Gerlach L. O., Bergmann R., Holst B., Schwartz T. W., Beck-Sickinger A. G. (2011) Long-acting lipidated analogue of human pancreatic polypeptide is slowly released into circulation. J. Med. Chem. 54, 2658–2667 [DOI] [PubMed] [Google Scholar]
- 30. Ninio S., Schuldiner S. (2003) Characterization of an archaeal multidrug transporter with a unique amino acid composition. J. Biol. Chem. 278, 12000–12005 [DOI] [PubMed] [Google Scholar]
- 31. Melnyk R. A., Partridge A. W., Yip J., Wu Y., Goto N. K., Deber C. M. (2003) Polar residue tagging of transmembrane peptides. Biopolymers 71, 675–685 [DOI] [PubMed] [Google Scholar]
- 32. Glukhov E., Burrows L. L., Deber C. M. (2008) Membrane interactions of designed cationic antimicrobial peptides: the two thresholds. Biopolymers 89, 360–371 [DOI] [PubMed] [Google Scholar]
- 33. Bay D. C., Rommens K. L., Turner R. J. (2008) Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778, 1814–1838 [DOI] [PubMed] [Google Scholar]
- 34. Kunjachan S., Rychlik B., Storm G., Kiessling F., Lammers T. (2013) Multidrug resistance: physiological principles and nanomedical solutions. Adv. Drug Deliv. Rev. 65, 1852–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yewale C., Baradia D., Vhora I., Patil S., Misra A. (2013) Epidermal growth factor receptor targeting in cancer: a review of trends and strategies. Biomaterials 34, 8690–8707 [DOI] [PubMed] [Google Scholar]
- 36. Chen Y. J., Pornillos O., Lieu S., Ma C., Chen A. P., Chang G. (2007) X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. U.S.A. 104, 18999–19004 [DOI] [PMC free article] [PubMed] [Google Scholar]