Background: Dysregulation of microRNAs (miRNAs) is associated with acute myeloid leukemia (AML).
Results: miR-638 up-regulation inhibited proliferation and promoted myeloid differentiation in AML leukemic cells by targeting cyclin-dependent kinase 2.
Conclusion: miR-638 is a novel player in myeloid differentiation.
Significance: Our findings may provide new insights into the regulatory role of miRNAs in normal hematopoiesis and leukemogenesis.
Keywords: Cyclin-dependent Kinase (CDK), Differentiation, Hematopoiesis, Leukemia, MicroRNA (miRNA)
Abstract
MicroRNAs have been extensively studied as regulators of hematopoiesis and leukemogenesis. We identified miR-638 as a novel regulator in myeloid differentiation and proliferation of leukemic cells. We found that miR-638 was developmentally up-regulated in cells of myeloid but not lymphoid lineage. Furthermore, significant miR-638 down-regulation was observed in primary acute myeloid leukemia (AML) blasts, whereas miR-638 expression was dramatically up-regulated in primary AML blasts and leukemic cell lines undergoing forced myeloid differentiation. These observations suggest that miR-638 might play a role in myeloid differentiation, and its dysregulation may contribute to leukemogenesis. Indeed, ectopic expression of miR-638 promoted phorbol 12-myristate 13-acetate- or all-trans-retinoic acid-induced differentiation of leukemic cell lines and primary AML blasts, whereas miR-638 inhibition caused an opposite phenotype. Consistently, miR-638 overexpression induced G1 cell cycle arrest and reduced colony formation in soft agar. Cyclin-dependent kinase 2 (CDK2) was found to be a target gene of miR-638. CDK2 inhibition phenotypically mimicked the overexpression of miR-638. Moreover, forced expression of CDK2 restored the proliferation and the colony-forming ability inhibited by miR-638. Our data suggest that miR-638 regulates proliferation and myeloid differentiation by targeting CDK2 and may serve as a novel target for leukemia therapy or marker for AML diagnosis and prognosis.
Introduction
Acute myeloid leukemia (AML)3 is a group of malignant blood neoplasms characterized by uncontrolled proliferation of myeloid precursor cells that fail to undergo terminal differentiation to produce mature blood cells (1). To date, various genetic aberrations, including translocations, inversions, deletions, point mutations (2), and epigenetic alterations (3, 4), have been identified in AML. Mechanistic studies in mouse models reveal how these genetic or epigenetic abnormalities cause leukemia. Such studies significantly facilitate the development of anti-leukemia therapies and encourage further investigations.
As epigenetic regulators, microRNAs (miRNAs) are non-coding RNAs of 19–25 nucleotides in length that suppress gene expression by repressing translation or accelerating the degradation of target mRNAs (5). Numerous studies show that miRNAs play pivotal roles in multiple biological processes, including proliferation, differentiation, and apoptosis (6–8). In addition, accumulating data suggest that miRNAs play crucial roles in normal hematopoiesis (9–11). Dysregulation of miRNAs is associated with AML (12, 13). For example, miR-26a (14), miR-199b-5p (15), miR-29a, and miR-142–3p (7) are significantly down-regulated in patients with AML and may be therapeutic targets or markers for diagnosis and/or prognosis.
Recently, miR-638 was proposed as an important regulator of development, tumorigenesis, hematopoiesis, and leukemogenesis. Differential expression of miR-638 occurs during early human embryonic development (16). miR-638 is highly expressed in human vascular smooth muscle cells, promoting proliferation and migration through suppression of NOR1 (17). Furthermore, miR-638 is frequently down-regulated in various solid tumors, such as stomach adenocarcinoma (16). Indeed, miR-638 represses BaP-induced carcinogenesis by targeting BRCA1 (18). The presence of miR-638 in human plasma samples suggests that miR-638 may function in blood cells (19). miR-638 is also found in exosomes, which suggests that miR-638 may be selectively packaged within these microparticles to serve as an antiviral molecule (20) or vector for intercellular communication between hematological and non-hematological cancer cells (21). The potential function of miR-638 in hematopoiesis or leukemia is further supported by reports that miR-638 is significantly up-regulated in leukemic cells upon induction of terminal myeloid differentiation (7, 22, 23). These results indicate a potential role of miR-638 in normal or abnormal hematopoiesis.
In this study, we focused on the function of miR-638 in the differentiation and proliferation of leukemic cells. We found that miR-638 was differentially expressed in myeloid cells but not in lymphoid cells. Dysregulation of miR-638 was observed in AML patients, implying that down-regulation of miR-638 may contribute to leukemogenesis. Indeed, overexpression of miR-638 inhibited proliferation and promoted differentiation of leukemic cell lines and primary AML blasts. Conversely, inhibition of miR-638 achieved the opposite effect. Further studies identified CDK2 as a target of miR-638; miR-638 repressed colony formation of HL-60 cells, and CDK2 overexpression rescued this phenotype. Thus, we have identified miR-638 as a new player whose down-regulation may contribute to leukemogenesis. miR-638 may serve as a therapeutic target or diagnostic/prognostic marker for leukemia therapy.
EXPERIMENTAL PROCEDURES
Cell Cultures, Blood Samples, and Induced Myeloid Differentiation
Human leukemic cell lines HL-60, NB4, and THP-1 were obtained from the Chinese Center for Type Culture Collection (Wuhan, China) and maintained in RPMI 1640 media (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). Myeloid differentiation was induced by phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) or all-trans-retinoic acid (ATRA) (Sigma-Aldrich) as described previously (7, 24, 25). Myeloid differentiation was detected by staining cells with the FITC-labeled anti-CD14 or anti-CD11b antibody or the corresponding isotype control antibody and analyzed by flow cytometry. May-Grünwald-Giemsa staining was applied to determine the morphology changes of leukemic cell undergoing myeloid differentiation. For each sample, the total cells and mature myeloid cells were counted under microscopy in three fields.
HEK293T cells were cultured with Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% FBS. Peripheral blood or cord blood was obtained from informed healthy donors or newly diagnosed AML patients, respectively. Primary AML blasts or mononuclear cells (MNCs) from healthy donors were isolated from peripheral blood samples on Ficoll-Hypaque density gradients and cultured as described previously (26, 27). Blood cells with specific surface markers were further purified from MNCs of healthy donors by cell sorting with antibodies (APC-CD11b, PE-CD14, FITC-CD45R, and PE-CD3ϵ). CD34+ stem cells were purified from cord blood using a human cord blood CD34-positive selection kit (StemCell Technologies, Vancouver, Canada); the purification efficiency was further checked with an APC-CD34 antibody by flow cytometry. All antibodies used for flow cytometry were purchased from Biolegend (San Diego, CA), BD Pharmingen, and eBiosciences (San Diego, CA). Experiments involving human blood samples were approved by the Medical Ethics Committee of Zhongnan Hospital of Wuhan University, Tongji Hospital of Huazhong Technology University, and Jiangsu Province Hospital of TCM. An informed consent form was obtained from each patient.
Quantitative Real-time RT-PCR
Total RNA was obtained using RNAiso Plus (TaKaRa, Tokyo, Japan), and 1 μg of total RNA was subjected to reverse transcription using the ReverTra Ace qPCR RT kit (TaKaRa). Quantitative analyses of miR-638 were performed using Bulge-LoopTM miRNA quantitative RT-PCR primer (RiboBio, Guangzhou, China) and Bestar® SYBR Green quantitative PCR master mix (DBI Bioscience, Shanghai, China) on an ABI Prism 7500 real-time PCR system (Applied Biosystems). PCR amplifications were performed in triplicate at 95 °C for 10 s and subjected to 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s followed by a standard melting curve analysis procedure. U6 snRNA and GAPDH were used as internal controls for miRNA and mRNA quantification, respectively. The expression of miRNAs and mRNAs was determined by the 2−ΔΔCt method. All of the reactions were performed in a 20-μl reaction volume in triplicate. The primer set for ITGAM (CD11b) was listed in Table 1. Primers for CD14, CSF1R, CSF3R, MPO, and GAPDH were described previously (7).
TABLE 1.
Primers used in this study
Underlined boldface characters indicate regions of the miR-638 seed sequences for mutation in luciferase reporter assay.
| Name | Sequences (5′–3′) |
|---|---|
| Primers used in quantitative RT-PCR | |
| ITGAM-F | ACTGCTGCTCCTGGCCCTC |
| ITGAM-R | GAAGCCCAAGCCCGTCCT |
| Primers used in plasmid construction | |
| MDH1–638-F | AATATCTCGAGCCCGGGAGCAACGGCTACAGA |
| MDH1–638-R | GAGTTGAATTCAATTAATTTTTATGGACAGCCCGAAAGAG |
| 3′UTR -CDK2-F | ACTTACGCGTGGGCTATTTGGACTCAGG |
| 3′UTR -CDK2-R | TAACAAGCTTAGCAAGAGCACTCAAGGAC |
| 3′UTR-CDK2–1F | CCAGACCGGGAAGCCTCCTGCTGCC |
| 3′UTR-CDK2–1R | GCTTCCGGGTCTGGTCATCCCAAAC |
| 3′UTR-CDK2–2F | CCCTACCGGGATTTTCCTCTGACGT |
| 3′UTR-CDK2–2R | AAATCCGGGTAGGGATCCTTGGCAA |
| 638_sites-F | CGCGTAGGGATCGCGGGCGGGTGGCGGCCTAGGGATCGCGGGCGGGTGGCGGCCTA |
| 638_sites-R | AGCTTAGGCCGCCACCCGCCCGCGATCCCTAGGCCGCCACCCGCCCGCGATCCCTA |
Oligonucleotide Transfection
The miR-638 mimic and a scrambled miR control, which is not homologous to any human genes; the miR-638 inhibitor (2′-O-methylated antisense oligonucleotides designed to specifically target mature miR-638); a nonspecific miRNA inhibitor control; and small interfering RNAs (siRNAs) targeting human CDK2 (si-CDK2) were all designed and synthesized by RiboBio. Mimics, inhibitors, or siRNAs were transfected into the suspension leukemic cells at working concentrations of 100, 200, and 100 nm, respectively, using TheraSilenceTM lipopolyamine (Celsion Corp., Lawrenceville, NJ) following the manufacturer's manual. BLOCK-iTTM fluorescent oligonucleotide (Invitrogen) was co-transfected to determine the transfection efficiency.
Generation of Stably Transfected HL-60 Cells
Retrovirus stocks were prepared, and retrovirus infection was performed as described previously (28). For overexpression of miR-638, the miR-638 gene (610-bp genomic DNA harboring miR-638 mature sequence and flanking sequences) was cloned into the MDH1-PGK-GFP 2.0 retroviral vector. For overexpression of CDK2, the human CDK2 cDNA was cloned into pMSCV-puro retroviral vector. Stable cell lines were selected by cell sorting or puromycin treatment. Primers for vector construction are listed in Table 1.
Cell Cycle Profiling and Proliferation Assay
For cell cycle distribution analysis, cells transfected with miRNA mimics were plated in 6-well plates and incubated for 48 h. Cells were harvested, washed twice with PBS, and fixed in 75% ethanol at 4 °C overnight. After wash, cells were incubated with RNase A (20 μg/ml) at 37 °C for 30 min and stained with 500 μg/ml propidium iodide at 4 °C for 30 min. DNA content was detected with a Beckman Coulter flow cytometer. For the proliferation assay, the transiently transfected (24 h) or stably transfected HL-60 cells (10,000) were replated in triplicates and manually counted at different time points. For experiments using the CCK-8 kit, 1,000 (vehicle-treated) or 5,000 (inducer-treated) cells were seeded into 96-well plates. After cells were incubated with 10 μl of CCK-8 for 3 h at 37 °C, the optical density was read at 450 nm with a microplate reader (Biotek). Each group had six replicates.
Colony-forming Assay
For colony-forming assays, 500 cells were suspended in RPMI 1640 medium containing 0.35% low melting point agarose and 10% FBS and plated onto a bottom layer containing 0.6% agarose and 10% FBS in 6-well plates. After about 2 weeks of incubation, 1 ml of 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (1 mg/ml) was added, and the plates were photographed 4 h later. The number of colonies was counted by ImageJ software (National Institutes of Health).
Luciferase Reporter Assay
Fragment (815 bp) from the 3′-UTR of CDK2 mRNA (NM_001798.3) harboring putative miR-638 binding sites was amplified from the human genome and subcloned into the pMIR-REPORTTM luciferase vector (Applied Biosystems) to obtain the wild-type reporter plasmid (CDK2-WT). Constructs with mutations in either of two putative binding sites or both were generated by overlapping PCR and designated as CDK2-M1, CDK2, or CDK2-DM, respectively. A construct with a double mutation in two binding sites (CDK2-DM) was cloned by using the template CDK2-M1 and the primers (listed in Table 1). A synthetic binding site with two tandem repeats complementary to the mature sequences of miR-638 was also cloned into the pMIR-REPORTTM vector (638_sites) and used as a positive control. HEK293T (2 × 105) cells were seeded in a 24-well plate and transfected with 100 ng of reporter vectors, 10 ng of pRL-TK, and a final concentration of 100 nm miR-638 or negative control mimic using Lipofectamine 2000 reagent (Invitrogen). Forty-eight hours later, cells were collected for luciferase activity assays with the Dual-Luciferase assay system (Promega).
Immunoblot Analysis
Total cell extracts (10 μg) were fractionated by electrophoresis on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Millipore). Membranes were further probed with antibodies against human CDK2, cyclin D1, PCNA (1:1,000) (Cell Signaling Technology), HSC70, or β-actin (1:5,000 dilution) (Santa Cruz Biotechnology, Inc.) followed by incubation with a 1:5,000 dilution of HRP-conjugated secondary antibodies (Santa Cruz Biotechnology). After wash, membranes were incubated with Pierce ECL Western blotting substrate (Thermo Fisher Scientific), and the protein bands were visualized by autoradiography.
Statistical Analysis
Data are presented as means ± S.D. of triplicates. Student's t test (unpaired, two tails) was used to analyze differences in two groups, and a p < 0.05 value was considered to be significant. Statistical analysis was performed with GraphPad Prism version 5.0 (GraphPad Software). The correlation between miR-638 and CDK2 expressions in AML blasts was tested with a two-tailed Pearson's correlation test. A p value of <0.05 was considered to be significant.
RESULTS
Differential Expression of miR-638 in Myeloid Lineage and miR-638 Dysregulation May Contribute to AML
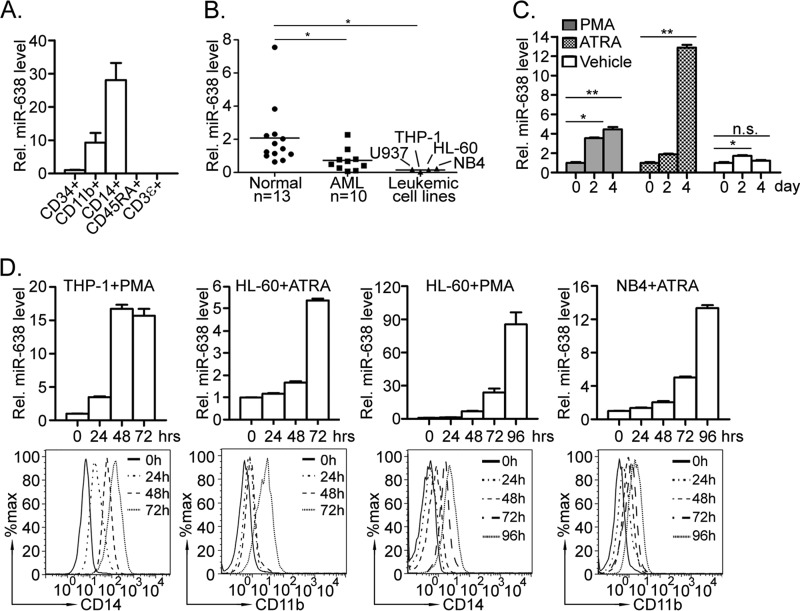
We first measured the expression of miR-638 in different blood cell lineages to assess its potential role in hematopoietic cell development. We found that miR-638 expression was developmentally up-regulated in myeloid lineages compared with that of CD34+ cells (∼10-fold increase in CD11b+ cells and ∼30-fold increase in CD14+ cells). In contrast, miR-638 was barely detectable in cells of the lymphoid lineage (CD45RA+ B-cells and CD3ϵ+ T-cells) (Fig. 1A). Interestingly, miR-638 was significantly down-regulated in leukemic cells in both primary AML samples (n = 10) and cell lines, including THP-1, HL-60, NB4, and U937, compared with that of MNCs isolated from the peripheral blood of healthy volunteers (n = 13) (Fig. 1B). These observations suggest that miR-638 may play a role in human myeloid differentiation, and its dysregulation may be involved in leukemogenesis.
FIGURE 1.
Lineage-specific expression of miR-638 and its dysregulation may contribute to myeloid leukemia. A, CD34+ cells and cells of different lineages (CD11b+, CD14+, CD45RA+, or CD3ϵ+) were purified by cell sorting. miR-638 expression was measured by quantitative RT-PCR. B, MNCs were isolated from peripheral blood of healthy volunteer donors (Normal; n = 13) and acute myeloid leukemia patients (AML; n = 10). Expression levels of miR-638 in MNCs, HL-60, THP-1, NB4, and U937 cells were analyzed by quantitative RT-PCR; *, p < 0.05 compared with control. C, MNCs were isolated from AML patients and treated with PMA, ATRA, or vehicle for days as indicated. Expression of miR-638 in treated cells was measured by quantitative RT-PCR. Representative results from one patient sample are presented. *, p < 0.05; **, p < 0.01; n.s., not significant. D, THP-1, HL-60 and NB4 cells were treated with PMA or ATRA for hours as indicated. CD14 or CD11b expression was measured by flow cytometry (bottom), and expression of miR-638 was quantitated by quantitative RT-PCR (top). Error bars, S.D.
In support of this idea, we found significant up-regulation of miR-638 during PMA- or ATRA-induced myeloid differentiation of primary AML blasts, compared with that of untreated (vehicle) control (Fig. 1C). Furthermore, miR-638 up-regulation was also detected during PMA- or ATRA-induced myeloid differentiation of leukemic cell lines in vitro. THP-1, derived from an acute monocytic leukemia patient, can be differentiated into monocytic/macrophage-like cells by PMA treatment (29), whereas, as a kind of promyelocytic leukemia cell line, HL-60 can be induced by either PMA or ATRA to differentiate into a monocytic/macrophage- or granulocytic-like phenotype, respectively (30). As expected, up-regulation of miR-638 coincided with increased myeloid-specific surface markers CD14 and CD11b in THP-1 and HL-60 cells induced by PMA or ATRA, respectively (Fig. 2D). In addition, elevated miR-638 expression was also observed in NB4 cells, an acute promyelocytic leukemia cell line (31), undergoing granulocytic differentiation induced by ATRA. These findings demonstrate that myeloid differentiation and miR-638 expression may correlate with each other, suggesting a regulatory role of miR-638 in myeloid differentiation.
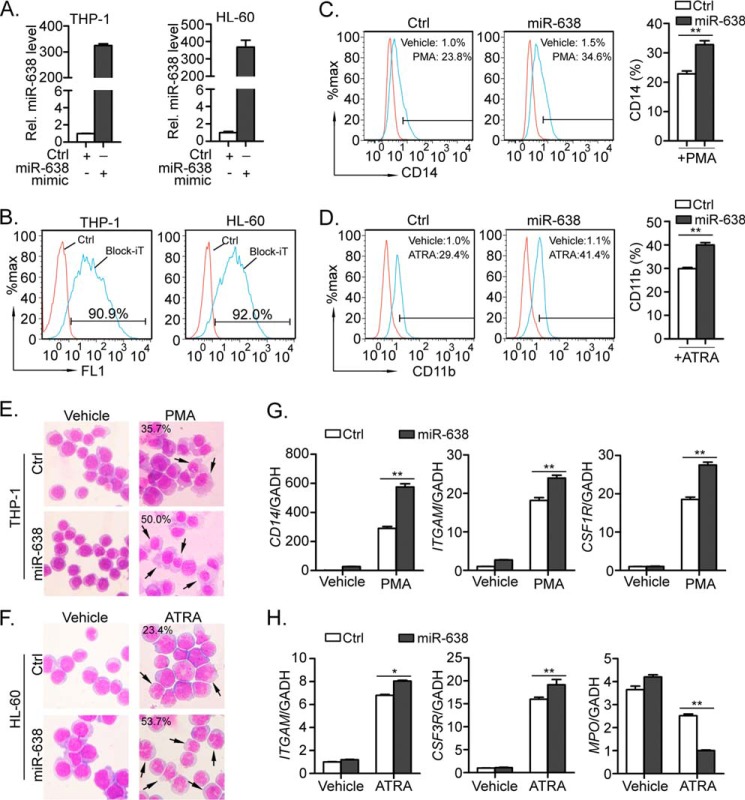
FIGURE 2.
Ectopic expression of miR-638 promotes myeloid differentiation in THP-1 and HL-60 cells induced by PMA or ATRA. A, THP-1 and HL-60 cells were transfected with control mimic (Ctrl) or miR-638 mimic. Levels of miR-638 in the transfected cells were measured by quantitative RT-PCR. Comparative real-time PCR was performed in triplicate and normalized to U6 snRNA. B, miRNA transfection efficiency was determined by co-transfecting cells with BLOCK-iT Oligo (Invitrogen) and detected (24 h after transfection) by flow cytometry. C and D, PMA-treated THP-1 cells or ATRA-treated HL-60 cells were further stained with a fluorescence-conjugated antibody against CD14 or CD11b, and the expression of CD14 or CD11b was detected by flow cytometry (left). Numbers in the graphs represent the percentages of positively stained cells after PMA or ATRA treatment for 72 h (blue histograms) compared with vehicle-treated cells (red histograms). A representative experiment of three is presented (left). The bar graph shows statistics for flow cytometry (right). E and F, the morphology of the treated cells was examined by staining cells with May-Grünwald-Giemsa, and the representative images are shown at ×200 magnification. The black arrows indicate mature macrophages or segmented neutrophils after treatment for 72 h, and numbers indicate their percentages. G and H, PMA-treated THP-1 cells or ATRA-treated HL-60 cells were collected, and the expression levels of myeloid differentiation markers (CD14, ITGAM, CSF1R/CSF3R, and MPO) were measured by quantitative RT-PCR. Comparative real-time PCR was performed in triplicate and normalized to GAPDH mRNA. Error bars, S.D. *, p < 0.05; **, p < 0.01.
miR-638 Promotes Forced Myeloid Terminal Differentiation of Leukemic Cells
To investigate the role of miR-638 in leukemic cell differentiation, we used THP-1 and HL-60 cells as models of myeloid differentiation. We transiently transfected these cells with either synthetic miR-638 mimics or scrambled mimics as a control. The expression levels of mature miR-638 in THP-1 and HL-60 cells were confirmed by quantitative RT-PCR (Fig. 2A). The transfection efficiency was determined by BLOCK-iT and was more than 90% in both groups (Fig. 2B). The transfected cells were then treated with PMA or ATRA for 72 h, and monocytic or granulocytic differentiation was measured. When compared with the cells treated by vehicle, the proportions of CD14 (monocyte specific marker)- and CD11b (granulocyte-specific)-positive cells were both significantly higher in miR-638-transfected THP-1 cells (34.6% for miR-638 versus 23.8% for control; Fig. 2C) and HL-60 cells (41.4% for miR-638 versus 29.4% for control; Fig. 2D), respectively, as measured by flow cytometry. During forced myeloid differentiation, PMA- or ATRA-treated leukemic cells always show significant morphological changes. As revealed by May-Grünwald-Giemsa staining, a greater number of differentiated monocytes was observed in THP-1 cells transfected with miR-638 mimics compared with control cells (Fig. 2E). Similarly, ATRA-treated HL-60 cells exhibited the typical characteristics of granulocytes with multiple nuclear lobules, whereas miR-638-transfected HL-60 cells showed greater numbers of mature granulocytes than did control cells (Fig. 2F). Moreover, elevated expression levels of CD14 mRNA and monocytic markers CD11b (ITGAM) and M-CSF receptor (CSF1R) were confirmed in miR-638-transfected THP-1 cells when compared with control cells (Fig. 2G). When treated with ATRA, the similar results were also observed for the two markers of granulocyte differentiation, ITGAM and G-CSF receptor (CSF3R), in miR-638-transfected HL-60 cells. On the contrary, MPO, a marker for myeloblasts and promyelocytes (32), was much lower in miR-638-overexpressed cells than in control cells (Fig. 2H).
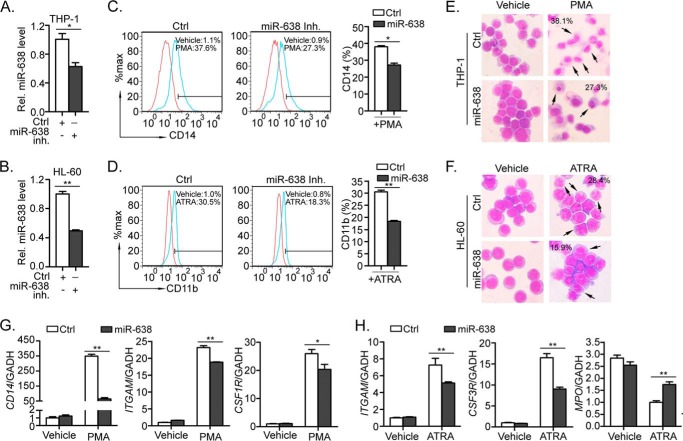
To further verify the role of miR-638 in forced myeloid terminal differentiation, miR-638 was knocked down by transiently transfecting cells with miR-638-specific inhibitors in leukemic cells, and miR-638 inhibitor efficiently knocked down miR-638 expression in THP-1 and HL-60 cells (Fig. 3, A and B). After PMA treatment, the miR-638 inhibitor-transfected THP-1 cells had fewer differentiated monocytes (Fig. 3, C and E) than did control cells with reduced expression of CD14, ITGAM, and CSF1R (Fig. 3G). Similarly, knockdown of miR-638 in HL-60 cells impaired granulocytic differentiation (Fig. 3, D and F) reduced expression of CD11b and CSF3R and increased MPO expression compared with control cells (Fig. 3H). Taken together, these findings suggest that miR-638 may promote PMA- or ATRA-induced myeloid differentiation in leukemic cells.
FIGURE 3.
miR-638 inhibition impairs myeloid differentiation in THP-1 and HL-60 cells induced by PMA and ATRA. A and B, THP-1 and HL-60 cells were transfected with control (Ctrl) or miR-638 inhibitors (miR-638 inh.). Expression of miR-638 in the transfected cells was measured by quantitative RT-PCR. C and D, transfected THP-1 and HL-60 cells were treated with (PMA) or without (Vehicle) PMA for 72 h. Cells were further stained with a fluorescence-conjugated antibody against CD14 or CD11b, and the expression of CD14 or CD11b was detected by flow cytometry (left). The bar graph shows statistics for flow cytometry (right). E and F, the morphology of the treated cells was examined by staining cells with May-Grünwald-Giemsa, and the representative images are shown at ×200 magnification. The black arrows indicate mature macrophages or segmented neutrophils after treatment for 72 h, and numbers indicate their percentages. G and H, the expression levels of myeloid differentiation markers (CD14, ITGAM, CSF1R/CSF3R and MPO) were measured by quantitative RT-PCR in PMA-treated THP-1 or ATRA-treated HL-60 cells. *, p < 0.05; **, p < 0.01, compared with control with Student's t test. Error bars, S.D.
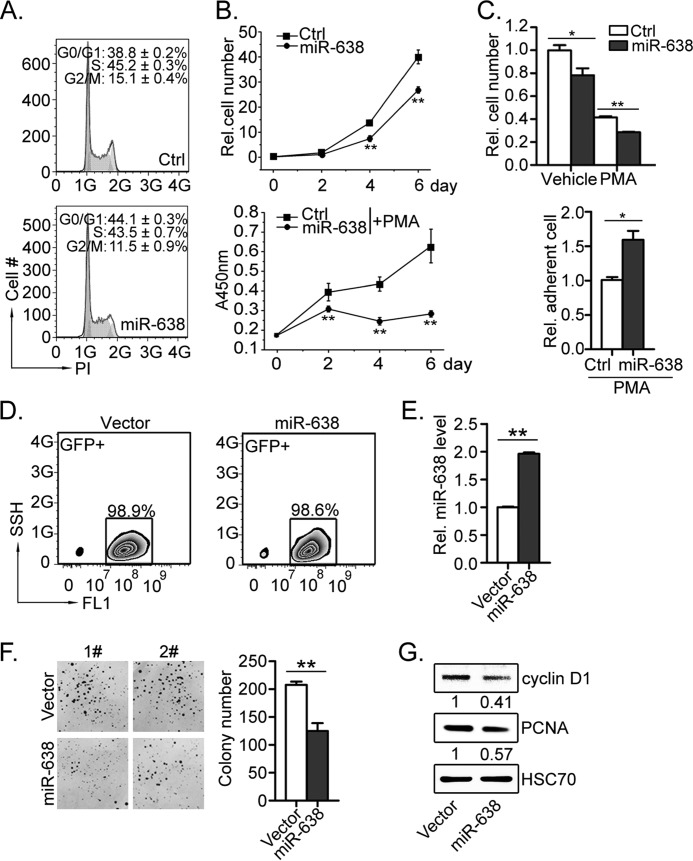
miR-638 Causes Cell Cycle Arrest, Reduces Cell Proliferation, and Impairs Colony Formation in HL-60 Cells
Most differentiation-promoting factors affect cell survival and/or proliferation. Therefore, we further investigated whether miR-638 affects cell cycle and proliferation of leukemic cells. We found that introduction of miR-638 mimic into HL-60 cells by transient transfection caused cell cycle arrest at the G1 phase. The percentage of cells in G1 phase increased in response to miR-638 overexpression compared with control cells (44.1% for miR-638 versus 38.8% for control, p < 0.01) (Fig. 4A). Moreover, cell numbers were reduced for cells overexpressing miR-638 compared with the number of control cells after 6 days of cell culture (Fig. 4B, top). Upon PMA treatment, the reduced proliferation was even more pronounced measured by CCK-8 (Fig. 4B, bottom). Similarly, significantly lower cell numbers were also observed for miR-638-transfected HL-60 cells compared with control (Fig. 4C, top). In addition, overexpression of miR-638 significantly increased the number of HL-60 cells adherent to the bottom of dishes after PMA treatment compared with control cells (Fig. 4C, bottom), which is a typical characteristic of differentiated monocytes. These observations suggest that miR-638 may cause cell cycle arrest at the G1 phase, resulting in reduced cell proliferation.
FIGURE 4.
Ectopic expression of miR-638 induced cell cycle arrest, inhibited proliferation, and impaired colony formation of HL-60 cells. A, HL-60 cells were transfected with control mimics (Ctrl) or miR-638 mimics (miR-638). Cells were stained with propidium iodide, and cell cycle profiles were analyzed by flow cytometry. Data in histograms are shown as means ± S.D. B, transfected cells were replated, and the cell numbers were counted on the day indicated (top). Transfected cells were also treated with PMA, and the growth curve was measured by CCK-8 at the indicated time points (bottom). Data are presented as a line chart. **, Student's t test p < 0.01. C, after transfection, equal numbers of transfected cells were plated, and the cell numbers (including cells adherent to the bottom) of PMA-treated or -untreated (Vehicle) cells on day 3 were further counted and presented as bar graphs (top). Cells adherent to the bottom after PMA treatment were also determined (bottom). *, Student's t test, p < 0.05; **, p < 0.01. D, HL-60 cells were infected with retrovirus overexpressing miR-638 (miR-638) or the control (Vector) retrovirus. The retroviral vector carried a GFP reporter gene, and GFP-positive HL-60 cells were purified by sorting GFP+ cells. The numbers shown in histograms represent the percentage of GFP-positive cells after sorting. E, expression of miR-638 was measured by quantitative RT-PCR. F, after sorting, the infected cells were seeded in soft agar and then cultured for 2 weeks. Representative dishes are shown (left); the bar graph shows statistics for the colony formation assay. **, p < 0.01 compared with control. G, expression of cyclin D1 and PCNA was measured by Western blot. HSC70 served as loading control. The numbers indicate the densitometric ratio of cyclin D1 or PCNA in miR-638 cells normalized to HSC70, which was further normalized to control cells. Error bars, S.D.
Forming colonies in soft agar is a characteristic of transformed tumor cells. Thus, we tested whether miR-638 affected the colony-forming capacity of HL-60. We overexpressed mature miR-638 by retrovirus infection. HL-60 cells infected by retrovirus expressed GFP and were purified by cell sorting (up to 98% purity; Fig. 4D). Quantitative RT-PCR showed that miR-638 expression was increased by about 2-fold (Fig. 4E). As expected, miR-638 overexpression impaired colony formation of HL-60 cells in soft agar. We found that miR-638-overexpressing HL-60 cells formed fewer colonies with a smaller size (Fig. 4F). Consistent with the results of cell cycle profiling experiments, miR-638 overexpression also reduced expression of cyclin D1 and PCNA, two factors that promote proliferation and cell cycle progression to S phase (Fig. 4G). Taken together, these findings suggest that down-regulation of miR-638 may promote proliferation and contribute to the transformation of leukemic cells.
Cyclin-dependent Kinase 2 Is a Novel Target of miR-638
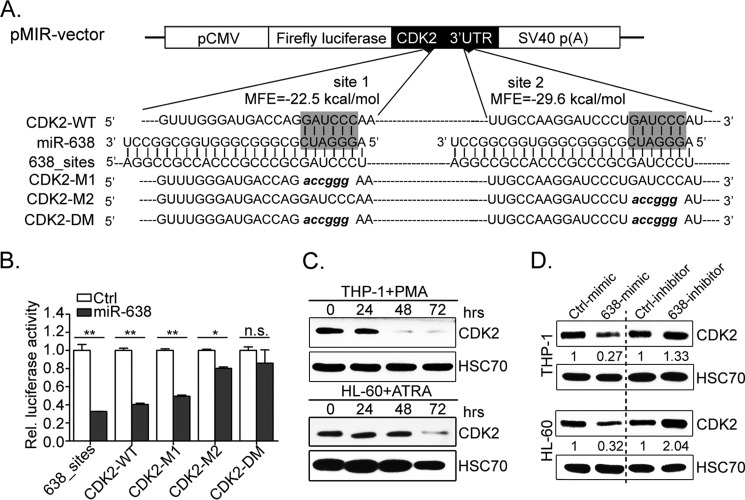
miRNAs function primarily by fine tuning the expression of specific target genes. Various computational algorithms have been developed to predict putative miRNA targets based on the rule that miRNAs commonly attach to the 3′-UTR of the target mRNA (33). Among the numerous putative target genes of miR-638 predicted by TargetScan (34), cyclin-dependent kinase 2 (CDK2), which was reported as a contributor in leukemia (35), was selected for further verification. Two putative binding sites (site 1 and site 2) in the 3′-UTR of CDK2 (NM_001798.3 as referential sequence) were predicted to have favorable hybridization minimum free energy values of −22.5 and −29.6 kcal/mol (Fig. 5A), respectively, as calculated by RNAhybrid (57). To prove whether CDK2 was a miR-638 target gene, we constructed luciferase reporter vectors harboring the wild-type 3′-UTR of CDK2 (CDK2-WT), mutant 3′-UTR of CDK2 at site 1 or site 2 (CDK2-M1 and CDK2-M2 with sequences in lowercase letters, respectively), or mutant 3′-UTR of CDK2 at both sites (CDK2-DM with sequences in lowercase letters) (Fig. 5A). As a positive control, miR-638 mimics caused ∼70% reduction in luciferase activity from the 638_sites vector compared with that of control mimics (Fig. 5B). As expected, 60% reduction in luciferase activity was observed for the wild-type 3′-UTR vector (CDK2-WT). Mutation of site 1 in the 3′-UTR (CDK2-M1) slightly impaired the inhibitory effect of miR-638. This effect was more significant in the CDK2-M2 vector. Notably, the double mutation completely abolished the repressive effect of miR-638 (Fig. 5B). These observations suggest that miR-638 may regulate CDK2 by binding both of two sites in the 3′-UTR.
FIGURE 5.
miR-638 suppresses CDK2 expression. A, diagram illustrates the vector structure used for target validation. The two putative sites (site 1 and site 2, seed sequences) potentially targeted by miR-638 in the CDK2 3′-UTR mRNA are shaded. CDK2-WT contains two putative wild-type binding sites (seed regions) complementary to miR-638. CDK2-M1 or CDK2-M2 has a mutation at site 1 or site 2, respectively. CDK2-DM has mutations at both sites. 638_sites contains two completely complementary sequences of miR-638. B, firefly luciferase activities were measured and normalized by Renilla luciferase expression. Data are presented as relative luciferase activity. *, p < 0.05; **, p < 0.01; n.s., not significant. C, Western blot analysis of CDK2 protein expression during PMA-induced monocytic differentiation in THP-1 and ATRA-induced granulocytic differentiation of HL-60 cells at the indicated time points. HSC70 served as a loading control. D, Western blot analysis of CDK2 levels in THP-1 and HL-60 cells transfected with control/miR-638 mimics or miR-638 inhibitors. Representative Western blots are shown. The numbers indicate the density ratio of CDK2 to HSC70, which was further normalized by the control. Error bars, S.D.
To further confirm CDK2 as a novel target gene of miR-638 in vivo, we measured the expression of the CDK2 protein in THP-1 and HL-60 cells undergoing myeloid terminal differentiation. These cells showed significant up-regulation of miR-638 (Fig. 1D). Expectedly, the CDK2 protein was significantly down-regulated in THP-1 and HL-60 cells after 72 h of induction (Fig. 5C). In addition, ectopic expression of miR-638 noticeably decreased CDK2 protein levels in THP-1 and HL-60 (reductions of 73 and 68%, respectively). Knockdown of miR-638 resulted in significantly increased CDK2 levels (up-regulated levels to 133 and 204%, respectively). These findings demonstrate an inverse correlation between miR-638 expression and CDK2 protein levels, suggesting that CDK2 is a direct target of miR-638.
miR-638 Functions in Part through Inhibition of CDK2
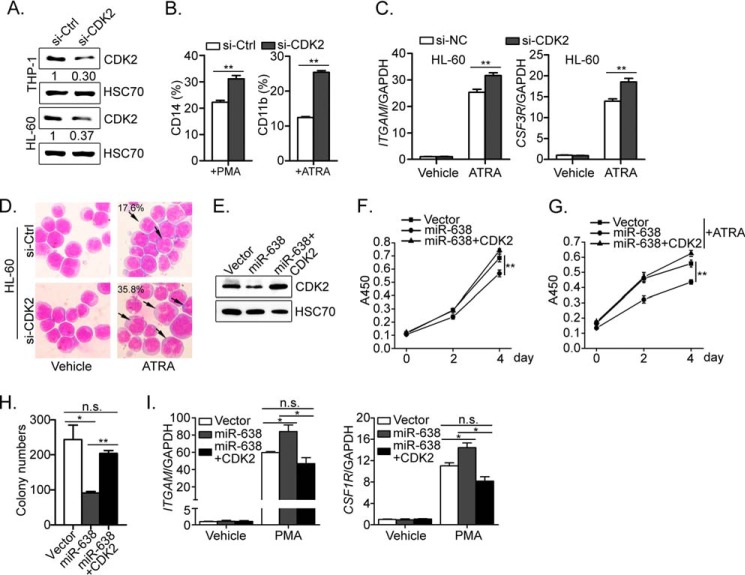
To investigate whether miR-638 promotes myeloid differentiation by suppressing CDK2, we first knocked down CDK2 expression in leukemic cells. Transient transfection of THP-1 and HL-60 cells with siRNA specific to CDK2 (si-CDK2) abolished expression of endogenous CDK2 protein (Fig. 6A). Upon PMA or ATRA treatment, CDK2 knockdown increased the percentage of CD14- and CD11b-positive cells (Fig. 6B), respectively, along with much higher levels of ITGAM and CSF3R mRNA expression in ATRA-treated cells (Fig. 6C). In the meantime, ATRA-treated HL-60 cells with lower CDK2 expression showed a greater proportion of mature granulocytes based on May-Grünwald-Giemsa staining (Fig. 6D).
FIGURE 6.
CDK2 partially mediates the effect of miR-638 in leukemic cells. A, Western blot analysis of CDK2 levels in THP-1 and HL-60 cells transiently transfected with control siRNA (Ctrl) or CDK2 siRNA (si-CDK2). Representative Western blots are shown. B, the transfected cells were further treated with or without PMA or ATRA as indicated for 72 h. Cells were further stained with FITC-conjugated CD14 or CD11b antibody and analyzed by flow cytometry. Each group was tested in triplicate, and data are shown as bar graphs. Error bars, S.D. **, p < 0.01, Student's t test. C, the expression of ITGAM and CSF3R in the transfected cells was measured by quantitative RT-PCR after ATRA treatment for 72 h. D, the morphology of the treated cells was examined by May-Grünwald-Giemsa staining, and the representative images are shown at ×200 magnification. The black arrows indicate mature neutrophils, and the numbers indicate their percentages. E, HL-60 cells were infected with control virus (Vector) or retrovirus expressing miR-638 or expressing miR-638 and CDK2 (miR-638 + CDK2). After GFP and puromycin selection, the expression of CDK2 in these cells was measured by Western blot. F and G, the resultant cells were plated in the same number, and the growth curves of untreated (F) or ATRA-treated (G) were measured by CCK-8 at the indicated time points. *, Student's t test, p < 0.05; **, p < 0.01. H, the resultant cells were also seeded in soft agar and cultured for 2 weeks. Colony numbers were counted and are presented as bar graphs. *, Student's t test p < 0.05; **, p < 0.01; n.s., not significant. I, after PMA treatment for 72 h, the expression of ITGAM and CSF1R in the infected cells was measured by quantitative RT-PCR. *, Student's t test, p < 0.05; n.s., not significant. Error bars, S.D.
To further test whether CDK2 miR-638 functions by inhibiting CDK2 expression, we restored CDK2 expression in HL-60 cells overexpressing miR-638. As shown in Fig. 6E, the CDK2 protein level was significantly suppressed by miR-638 overexpression and could be restored by the introduction of CDK2 (CDS region of CDK2 cDNA without 3′-UTR). Consistently, CDK2 overexpression eliminated the inhibitory effect of miR-638 in HL-60 cells (Fig. 6F), and this effect was more significant upon ATRA treatment, as evidenced by the increased cell proliferation rate compared with that of control cells (Fig. 6G). In addition, miR-638 alone significantly reduced colony numbers compared with the colony number of vector control, whereas restoration of CDK2 expression in miR-638-overexpressing cells significantly rescued the phenotype, comparable with that of control cells (Fig. 6H). When treated with PMA, CDK2 restoration significantly suppressed the expression of monocytic markers (ITGAM and CSF1R), and there was no significant difference between the vector and CDK2 overexpression group (Fig. 6I). These findings demonstrate that CDK2 overexpression counteracts the inhibitory effect of miR-638 on the proliferation ability of HL-60 cells. These data strongly support the idea that miR-638 inhibits proliferation and promotes forced differentiation of leukemic cells by suppressing CDK2 expression.
Ectopic Expression of miR-638 or CDK2 Knockdown Promotes Forced Myeloid Terminal Differentiation in Primary AML Blasts
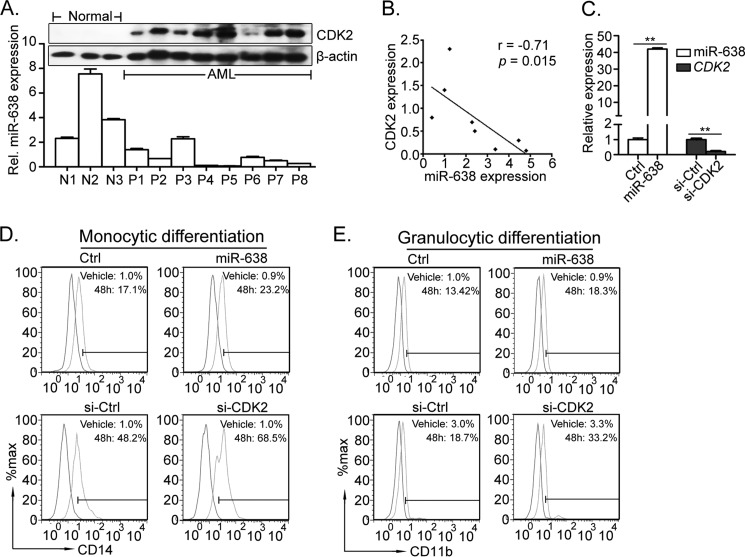
To determine the functions of the miR-638/CDK2 axis in acute myeloid leukemia, we further investigated the expression correlation of miR-638 and CDK2 in AML blasts. As shown in Fig. 7A, the expression of CDK2 protein was significantly higher in primary AML blasts than in MNCs from healthy donors. Pearson's correlation analysis showed that CDK2 expression inversely correlated with expression of miR-638 in primary AML blasts (r = −0.71, p = 0.015; Fig. 7B). Furthermore, we collected primary AML blasts from a newly diagnosed M1 patient with an initial percentage of circulating blasts exceeding 80%. These blasts were transfected with miR-638 mimic or siRNA specific for CDK2 and induced to undergo myeloid differentiation by PMA or ATRA. The expression level of the mature miR-638 was confirmed (40-fold higher than control cells) (Fig. 7C). Upon PMA treatment, miR-638 overexpression increased the expression of monocyte marker CD14 compared with that of control cells (Fig. 7D, top). Similarly, ATRA treatment pushed AML blasts to differentiate into mature granulocytes, and miR-638-overexpressing cells exhibited a greater number of mature granulocytes (Fig. 7E, top). In parallel, si-CDK2 transfection mimicked the phenotype of miR-638 overexpression, and CDK2 knockdown (Fig. 7C) promoted forced myeloid differentiation in primary AML blasts (Fig. 7, D and E, bottom).
FIGURE 7.
Enforced expression of miR-638 or CDK2 knockdown in AML blasts promotes forced myeloid differentiation. A, the expression levels of miR-638 (analyzed by quantitative RT-PCR) and CDK2 (analyzed by Western blot) in MNCs from each AML patient and healthy donor are shown. B, the correlation of miR-638 expression and CDK2 protein expression in AML patients was analyzed by Pearson's correlation analysis (two-tailed, r = −0.71, p = 0.015). C, analysis of miR-638 or CDK2 mRNA expression in primary AML blasts transfected with miRNA mimics or siRNAs. D, mononuclear cells isolated from AML patients were transfected with miR-638 mimics (top) or si-CDK2 (bottom) and treated with PMA (blue histograms) or vehicle (red histograms) for 48 h. Expression of CD14 was measured by flow cytometry. E, the transfected cells were also treated with ATRA (blue histograms) or vehicle (red histograms) for 48 h. The expression of CD11b was measured by flow cytometry. Error bars, S.D.
In conclusion, the effect of miR-638 on myeloid differentiation may function in part by suppressing CDK2. Dysregulation of the miR-638/CDK2 axis may contribute to acute myeloid leukemia.
DISCUSSION
Previously, miR-638 has been proposed to regulate human development (16) and tumorigenesis (36–38) and to play roles in hematopoiesis or leukemogenesis (39, 40). In this study, we revealed novel features of miR-638 and provided evidence that miR-638 is an important regulator of hematopoiesis and that its dysregulation contributes to leukemogenesis.
miR-638 appears to be developmentally regulated in hematopoiesis. Our data showed that miR-638 was specifically up-regulated in cells of the myeloid lineage but not in the lymphoid lineage (Fig. 1A). Multiple microRNAs have been shown to regulate myeloid (41–44) and lymphoid (45, 46) lineage commitment. Considering that there is substantial expression of miR-638 in the CD34+ population with sustained up-regulation in cells of the myeloid lineage, it is very tempting to postulate that miR-638 may promote myeloid commitment or play a role in hematopoietic stem cell differentiation into common myeloid progenitors.
miR-638 dysregulation may contribute to acute myeloid leukemia. We found significantly lower levels of miR-638 in AML blasts compared with normal MNCs, and restoration of miR-638 in leukemic cells promoted forced myeloid differentiation. However, miR-638 mimics or inhibitors did not alter CD14 or CD11b expression without PMA or ATRA induction. We speculated that miR-638 alteration alone was not strong enough to overcome the differentiation arrest in leukemic cells. Rather, it reduced the threshold for leukemic cells to undergo forced myeloid differentiation. Other evidence supporting miR-638 as a differentiation factor was derived from the report that miR-638 was induced in AML cells by C/EBPα (42), which is crucial for normal granulopoiesis and is frequently disrupted in AML (47). In addition, miR-638 is a p53-induced miRNA in malignant peripheral nerve sheath tumors (48), and p53 is frequently inactivated in numerous cancers. Thus, miR-638 may interplay with other oncogenes or tumor suppressor genes, dependent on different cellular context, to affect leukemogenesis or tumorigenesis. It is essential to determine the regulatory network associated with miR-638 before we can fully elucidate how miR-638 contributes to myeloid leukemia.
We identified CDK2 as a miR-638 target gene in myeloid differentiation. Down-regulation of CDK2 is a common event in end stage differentiated cells (49, 50), and irreversible down-regulation of three G1 phase cyclin-dependent kinases (CDK2, CDK4, and CDK6) at the mRNA and protein levels occurs during granulopoiesis (51). Herein, we demonstrated that up-regulation of miR-638 directly repressed CDK2 during myeloid commitment. Moreover, an RNAi assay proved that knocking down CDK2 promoted induced myeloid differentiation in both the cell line model and primary AML blasts. Studies in double-knock-out mice indicate that loss of CDK2 in neural stem cells promotes spontaneous neuronal differentiation (52, 53). In addition, other genuine genes targeted by miR-638 might also be involved in myeloid differentiation. We noticed that cyclin D1, which was an unverified target of miR-638 predicted by TargetScan and regarded to be involved in controlling the G1 to S transition, was conversely influenced by miR-638 levels in HL-60 (Fig. 4G). In consideration of cyclin D1 as a common oncogene in various cancers, including lymphomas, gliomas, and leukemia (54–56), it is likely that CDK2 might act in concert with cyclin D1 to impose a particular effect in myeloid differentiation.
In conclusion, the present study identified miR-638 as a novel regulator of myeloid differentiation in leukemic cells in part through suppressing its target gene CDK2. Considering the relationship of miR-638 and CDK2 expression with leukemic cell differentiation, the miR-638/CDK2 axis may serve as a marker for prognosis or treatment response.
Acknowledgments
We thank Dr. Beiyan Zhou (Texas A&M University) and Dr. Xuejun Zhu (Jiangsu Province Hospital of TCM) for sharing critical reagents.
This work was supported by National Natural Science Foundation of China Grants 31371481 (to Z. H.), 81000201 (to L. L.), 31271511 (to M. G.), and 81272627 (to S. L.); Ph.D. Programs Foundation of the Ministry of Education of China Grant 20110141110016 (to Z. H.); New Century Excellent Talents in University (NCET) of the Ministry of Education of China Grant NCET-12-0422 (to Z. H.), and Fundamental Research Funds for the Central Universities Grant 2042014KF0243 (to M. G.).
- AML
- acute myeloid leukemia
- miRNA
- microRNA
- PMA
- phorbol 12-myristate 13-acetate
- ATRA
- all-trans-retinoic acid
- MNC
- mononuclear cell
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. Palma C. A., Tonna E. J., Ma D. F., Lutherborrow M. A. (2012) MicroRNA control of myelopoiesis and the differentiation block in acute myeloid leukaemia. J. Cell Mol. Med. 16, 978–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mrózek K., Heerema N. A., Bloomfield C. D. (2004) Cytogenetics in acute leukemia. Blood Rev. 18, 115–136 [DOI] [PubMed] [Google Scholar]
- 3. Hackanson B., Bennett K. L., Brena R. M., Jiang J., Claus R., Chen S. S., Blagitko-Dorfs N., Maharry K., Whitman S. P., Schmittgen T. D., Lübbert M., Marcucci G., Bloomfield C. D., Plass C. (2008) Epigenetic modification of CCAAT/enhancer binding protein alpha expression in acute myeloid leukemia. Cancer Res. 68, 3142–3151 [DOI] [PubMed] [Google Scholar]
- 4. Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C. E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., Havelange V., Volinia S., Blum W., Rush L. J., Perrotti D., Andreeff M., Bloomfield C. D., Byrd J. C., Chan K., Wu L. C., Croce C. M., Marcucci G. (2009) MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 113, 6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 6. Mineno J., Okamoto S., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Kato I. (2006) The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 34, 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X. S., Gong J. N., Yu J., Wang F., Zhang X. H., Yin X. L., Tan Z. Q., Luo Z. M., Yang G. H., Shen C., Zhang J. W. (2012) MicroRNA-29a and microRNA-142–3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 119, 4992–5004 [DOI] [PubMed] [Google Scholar]
- 8. Tsukamoto Y., Nakada C., Noguchi T., Tanigawa M., Nguyen L. T., Uchida T., Hijiya N., Matsuura K., Fujioka T., Seto M., Moriyama M. (2010) MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 70, 2339–2349 [DOI] [PubMed] [Google Scholar]
- 9. Garzon R., Pichiorri F., Palumbo T., Iuliano R., Cimmino A., Aqeilan R., Volinia S., Bhatt D., Alder H., Marcucci G., Calin G. A., Liu C. G., Bloomfield C. D., Andreeff M., Croce C. M. (2006) MicroRNA fingerprints during human megakaryocytopoiesis. Proc. Natl. Acad. Sci. U.S.A. 103, 5078–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mukai H. Y., Motohashi H., Ohneda O., Suzuki N., Nagano M., Yamamoto M. (2006) Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol. Cell. Biol. 26, 7953–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukao T., Fukuda Y., Kiga K., Sharif J., Hino K., Enomoto Y., Kawamura A., Nakamura K., Takeuchi T., Tanabe M. (2007) An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell 129, 617–631 [DOI] [PubMed] [Google Scholar]
- 12. Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C. M. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pulikkan J. A., Dengler V., Peramangalam P. S., Peer Zada A. A., Müller-Tidow C., Bohlander S. K., Tenen D. G., Behre G. (2010) Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood 115, 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salvatori B., Iosue I., Djodji Damas N., Mangiavacchi A., Chiaretti S., Messina M., Padula F., Guarini A., Bozzoni I., Fazi F., Fatica A. (2011) Critical Role of c-Myc in Acute Myeloid Leukemia Involving Direct Regulation of miR-26a and Histone Methyltransferase EZH2. Genes Cancer 2, 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Favreau A. J., Cross E. L., Sathyanarayana P. (2012) miR-199b-5p directly targets PODXL and DDR1 and decreased levels of miR-199b-5p correlate with elevated expressions of PODXL and DDR1 in acute myeloid leukemia. Am. J. Hematol. 87, 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Y., Zeng Y., Zhang F., Xue L., Huang Z., Li W., Guo M. (2013) Characterization of MicroRNA Expression Profiles and the Discovery of Novel MicroRNAs Involved in Cancer during Human Embryonic Development. PLoS One 8, e69230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li P., Liu Y., Yi B., Wang G., You X., Zhao X., Summer R., Qin Y., Sun J. (2013) MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc. Res. 99, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li D., Wang Q., Liu C., Duan H., Zeng X., Zhang B., Li X., Zhao J., Tang S., Li Z., Xing X., Yang P., Chen L., Zeng J., Zhu X., Zhang S., Zhang Z., Ma L., He Z., Wang E., Xiao Y., Zheng Y., Chen W. (2012) Aberrant expression of miR-638 contributes to benzo(a)pyrene-induced human cell transformation. Toxicol. Sci. 125, 382–391 [DOI] [PubMed] [Google Scholar]
- 19. Tanaka M., Oikawa K., Takanashi M., Kudo M., Ohyashiki J., Ohyashiki K., Kuroda M. (2009) Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One 4, e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M., Zhou X., Yuan Z. (2013) Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 14, 793–803 [DOI] [PubMed] [Google Scholar]
- 21. Jaiswal R., Luk F., Gong J., Mathys J. M., Grau G. E., Bebawy M. (2012) Microparticle conferred microRNA profiles: implications in the transfer and dominance of cancer traits. Mol. Cancer 11, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lal A., Pan Y., Navarro F., Dykxhoorn D. M., Moreau L., Meire E., Bentwich Z., Lieberman J., Chowdhury D. (2009) miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., Xiang G., Mitchelson K., Zhou Y. (2011) Microarray profiling of monocytic differentiation reveals miRNA-mRNA intrinsic correlation. J. Cell. Biochem. 112, 2443–2453 [DOI] [PubMed] [Google Scholar]
- 24. Kharfan-Dabaja M., Ayala E., Lindner I., Cejas P. J., Bahlis N. J., Kolonias D., Carlson L. M., Lee K. P. (2005) Differentiation of acute and chronic myeloid leukemic blasts into the dendritic cell lineage: analysis of various differentiation-inducing signals. Cancer Immunol. Immunother. 54, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubois C., Schlageter M. H., de Gentile A., Guidez F., Balitrand N., Toubert M. E., Krawice I., Fenaux P., Castaigne S., Najean Y. (1994) Hematopoietic growth factor expression and ATRA sensitivity in acute promyelocytic blast cells. Blood 83, 3264–3270 [PubMed] [Google Scholar]
- 26. Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M. L., Nervi C., Bozzoni I. (2005) A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell 123, 819–831 [DOI] [PubMed] [Google Scholar]
- 27. Benedetti L., Grignani F., Scicchitano B. M., Jetten A. M., Diverio D., Lo Coco F., Avvisati G., Gambacorti-Passerini C., Adamo S., Levin A. A., Pelicci P. G., Nervi C. (1996) Retinoid-induced differentiation of acute promyelocytic leukemia involves PML-RARα-mediated increase of type II transglutaminase. Blood 87, 1939–1950 [PubMed] [Google Scholar]
- 28. Liu L., Wen Q., Gong R., Gilles L., Stankiewicz M. J., Li W., Guo M., Li L., Sun X., Li W., Crispino J. D., Huang Z. (2014) PSTPIP2 dysregulation contributes to aberrant terminal differentiation in GATA-1-deficient megakaryocytes by activating LYN. Cell Death Dis. 5, e988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., Tada K. (1980) Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171–176 [DOI] [PubMed] [Google Scholar]
- 30. Gallagher R., Collins S., Trujillo J., McCredie K., Ahearn M., Tsai S., Metzgar R., Aulakh G., Ting R., Ruscetti F., Gallo R. (1979) Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood 54, 713–733 [PubMed] [Google Scholar]
- 31. Duprez E., Tong J. H., Dérré J., Chen S. J., Berger R., Chen Z., Lanotte M. (1997) JEM-1, a novel gene encoding a leucine-zipper nuclear factor upregulated during retinoid-induced maturation of NB4 promyelocytic leukaemia. Oncogene 14, 1563–1570 [DOI] [PubMed] [Google Scholar]
- 32. Weil S. C., Rosner G. L., Reid M. S., Chisholm R. L., Farber N. M., Spitznagel J. K., Swanson M. S. (1987) cDNA cloning of human myeloperoxidase: decrease in myeloperoxidase mRNA upon induction of HL-60 cells. Proc. Natl. Acad. Sci. U.S.A. 84, 2057–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 35. Schmitz N. M., Leibundgut K., Hirt A. (2005) CDK2 catalytic activity and loss of nuclear tethering of retinoblastoma protein in childhood acute lymphoblastic leukemia. Leukemia 19, 1783–1787 [DOI] [PubMed] [Google Scholar]
- 36. Wang K., Xu L., Pan L., Xu K., Li G. (2014) The functional BRCA1 rs799917 genetic polymorphism is associated with gastric cancer risk in a Chinese Han population. Tumour Biol. 10.1007/s13277-014-2655-9 [DOI] [PubMed] [Google Scholar]
- 37. Ma K., Pan X., Fan P., He Y., Gu J., Wang W., Zhang T., Li Z., Luo X. (2014) Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Mol. Cancer 13, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao L. Y., Yao Y., Han J., Yang J., Wang X. F., Tong D. D., Song T. S., Huang C., Shao Y. (2014) miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig. Dis. Sci. 59, 1743–1753 [DOI] [PubMed] [Google Scholar]
- 39. Jiang X., Huang H., Li Z., He C., Li Y., Chen P., Gurbuxani S., Arnovitz S., Hong G. M., Price C., Ren H., Kunjamma R. B., Neilly M. B., Salat J., Wunderlich M., Slany R. K., Zhang Y., Larson R. A., Le Beau M. M., Mulloy J. C., Rowley J. D., Chen J. (2012) MiR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc. Natl. Acad. Sci. U.S.A. 109, 19397–19402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu D. X., Zhu W., Fang C., Fan L., Zou Z. J., Wang Y. H., Liu P., Hong M., Miao K. R., Liu P., Xu W., Li J. Y. (2012) miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 33, 1294–1301 [DOI] [PubMed] [Google Scholar]
- 41. Johnnidis J. B., Harris M. H., Wheeler R. T., Stehling-Sun S., Lam M. H., Kirak O., Brummelkamp T. R., Fleming M. D., Camargo F. D. (2008) Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129 [DOI] [PubMed] [Google Scholar]
- 42. Eyholzer M., Schmid S., Wilkens L., Mueller B. U., Pabst T. (2010) The tumour-suppressive miR-29a/b1 cluster is regulated by CEBPA and blocked in human AML. Br. J. Cancer 103, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seca H., Almeida G. M., Guimarães J. E., Vasconcelos M. H. (2010) miR signatures and the role of miRs in acute myeloid leukaemia. Eur. J. Cancer 46, 1520–1527 [DOI] [PubMed] [Google Scholar]
- 44. Wang X., Gocek E., Liu C. G., Studzinski G. P. (2009) MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle 8, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 46. O'Connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., Paquette R. L., Baltimore D. (2008) Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Radomska H. S., Huettner C. S., Zhang P., Cheng T., Scadden D. T., Tenen D. G. (1998) CCAAT/enhancer binding protein α is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18, 4301–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Subramanian S., Thayanithy V., West R. B., Lee C. H., Beck A. H., Zhu S., Downs-Kelly E., Montgomery K., Goldblum J. R., Hogendoorn P. C., Corless C. L., Oliveira A. M., Dry S. M., Nielsen T. O., Rubin B. P., Fletcher J. A., Fletcher C. D., van de Rijn M. (2010) Genome-wide transcriptome analyses reveal p53 inactivation-mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J. Pathol. 220, 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tikoo R., Casaccia-Bonnefil P., Chao M. V., Koff A. (1997) Changes in cyclin-dependent kinase 2 and p27kip1 accompany glial cell differentiation of central glia-4 cells. J. Biol. Chem. 272, 442–447 [DOI] [PubMed] [Google Scholar]
- 50. Martinez L. A., Chen Y., Fischer S. M., Conti C. J. (1999) Coordinated changes in cell cycle machinery occur during keratinocyte terminal differentiation. Oncogene 18, 397–406 [DOI] [PubMed] [Google Scholar]
- 51. Klausen P., Bjerregaard M. D., Borregaard N., Cowland J. B. (2004) End-stage differentiation of neutrophil granulocytes in vivo is accompanied by up-regulation of p27kip1 and down-regulation of CDK2, CDK4, and CDK6. J. Leukoc. Biol. 75, 569–578 [DOI] [PubMed] [Google Scholar]
- 52. Lim S., Kaldis P. (2012) Loss of Cdk2 and Cdk4 induces a switch from proliferation to differentiation in neural stem cells. Stem Cells 30, 1509–1520 [DOI] [PubMed] [Google Scholar]
- 53. Dobashi Y., Shoji M., Kitagawa M., Noguchi T., Kameya T. (2000) Simultaneous suppression of cdc2 and cdk2 activities induces neuronal differentiation of PC12 cells. J. Biol. Chem. 275, 12572–12580 [DOI] [PubMed] [Google Scholar]
- 54. Aref S., Mabed M., El-Sherbiny M., Selim T., Metwaly A. (2006) Cyclin D1 expression in acute leukemia. Hematology 11, 31–34 [DOI] [PubMed] [Google Scholar]
- 55. Wessendorf S., Schwaenen C., Kohlhammer H., Kienle D., Wrobel G., Barth T. F., Nessling M., Möller P., Döhner H., Lichter P., Bentz M. (2003) Hidden gene amplifications in aggressive B-cell non-Hodgkin lymphomas detected by microarray-based comparative genomic hybridization. Oncogene 22, 1425–1429 [DOI] [PubMed] [Google Scholar]
- 56. Büschges R., Weber R. G., Actor B., Lichter P., Collins V. P., Reifenberger G. (1999) Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 9, 435–442; discussion 432–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rehmsmeier M., Steffen P., Höchsmann M., Giegerich R. (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]