Background: Different transcription factor combinations may control distinct or overlapping cellular functions.
Results: Intestines lacking tissue-restricted CDX2 and broadly expressed GATA4 or HNF4A show unique defects.
Conclusion: Combined with CDX2, GATA4 controls crypt cell replication, whereas HNF4A regulates enterocyte maturation and a cohort of functional enterocyte genes.
Significance: Combinatorial mechanisms for intestine-specific gene regulation may apply generally to other tissues.
Keywords: Cell Differentiation, Cell Proliferation, GATA Transcription Factor, Gene Regulation, Intestinal Epithelium, Lipid Metabolism, CDX2 Transcription Factor, HNF4A Transcription Factor
Abstract
Distinct groups of transcription factors (TFs) assemble at tissue-specific cis-regulatory sites, implying that different TF combinations may control different genes and cellular functions. Within such combinations, TFs that specify or maintain a lineage and are therefore considered master regulators may play a key role. Gene enhancers often attract these tissue-restricted TFs, as well as TFs that are expressed more broadly. However, the contributions of the individual TFs to combinatorial regulatory activity have not been examined critically in many cases in vivo. We address this question using a genetic approach in mice to inactivate the intestine-specifying and intestine-restricted factor CDX2 alone or in combination with its more broadly expressed partner factors, GATA4 and HNF4A. Compared with single mutants, each combination produced significantly greater defects and rapid lethality through distinct anomalies. Intestines lacking Gata4 and Cdx2 were deficient in crypt cell replication, whereas combined loss of Hnf4a and Cdx2 specifically impaired viability and maturation of villus enterocytes. Integrated analysis of TF binding and of transcripts affected in Hnf4a;Cdx2 compound-mutant intestines indicated that this TF pair controls genes required to construct the apical brush border and absorb nutrients, including dietary lipids. This study thus defines combinatorial TF activities, their specific requirements during tissue homeostasis, and modules of transcriptional targets in intestinal epithelial cells in vivo.
Introduction
Tissue-specific gene expression reflects the coordinated activities of transcription factors (TFs)3 that are restricted to one or a few cell types and others that are expressed more broadly. In tissues of endodermal origin, TFs such as CDX2, PTF1, and PDX1 are restricted to individual organs (1–3), whereas others such as HNF4A and GATA are expressed in many endoderm-derived tissues. It is unclear if the latter TFs control separate and distinct programs and cellular function or merely support the activity of master regulators, such as the intestine-restricted CDX2.
In adult mammals, the small intestine contains abundant proliferative cells in submucosal crypts and mature post-mitotic cells along the villi. The homeodomain TF CDX2 is expressed exclusively in the epithelium of the small intestine and colon in both replicating crypt cells and differentiated villus cells (4–7). CDX2 is required for proper specification of the intestine during development (8) and is considered a master regulator of intestinal identity because its ectopic expression in the stomach or esophagus activates intestine-restricted genes (9, 10). In adult mice, absence of CDX2 dysregulates genes involved in terminal cell differentiation, and fatal malnutrition ensues over ∼3 weeks (11–13). Simultaneous inactivation of its homolog CDX1 results in nearly complete arrest of intestinal crypt cell proliferation (14). Together, these findings demonstrate diverse requirements for CDX2 in embryonic tissue specification, cell differentiation, and maintenance of the adult intestinal epithelium. It is unclear how CDX2 partners with various TFs to mediate these diverse functions.
One way to identify the TFs important in any tissue is through DNA sequence motifs that are highly enriched among active cis-regulatory sites. Enhancers active in the intestinal epithelium show few recurring sequence motifs, including the one preferred by CDX2; the GATA motif, especially in replicating cells; and the consensus motif for HNF4A, mainly in differentiated villus cells (11). This select group of TFs thus seems particularly important in intestinal gene regulation. Indeed, knock-out mice lacking single factors show diverse, subtle, and nonlethal defects. GATA4 and GATA6 show regional (proximal 4/5) and global intestinal expression, respectively, and the corresponding mutant mice have subtle defects in crypt cell replication, secretory cell differentiation, and control of selected enterocyte genes (15–17). Loss of HNF4A perturbs colon development (18), but Hnf4a−/− adult mouse intestines are overtly normal, with modestly perturbed gene expression (19). Coupled with the frequent co-occurrence of their specific sequence motifs near CDX2-binding sites, the limited defects in Gata and Hnf4a mutant mice led us to postulate that they may regulate intestinal genes in combination with CDX2. These TFs might act in several different ways: (i) simply support CDX2 activity at CDX2-dependent genes, (ii) partner with CDX2 in distinct combinations to regulate different cellular functions, or (iii) serve additional, CDX2-independent functions.
To evaluate these possibilities, we generated inducible compound-mutant mice that lack Cdx2 and either Gata4/6 or Hnf4a in the adult intestine. Distinct defects in each compound-mutant strain revealed unambiguous joint requirements for CDX2 and GATA4 in crypt cell proliferation and for CDX2 and HNF4A in differentiated villus enterocytes. CDX2 and HNF4A, in particular, co-regulate genes necessary to absorb dietary lipids. Thus, the lineage-restricted factor CDX2 functions in obligate partnerships with different broadly expressed factors to regulate distinct aspects of intestinal epithelial structure and function.
EXPERIMENTAL PROCEDURES
Mice
Gata4Fl/Fl, Gata6Fl/Fl, Cdx2Fl/Fl, Hnf4aFl/Fl, and transgenic Villin-CreERT2 mice were described previously (11, 20–23). Gata4Fl/Fl, Gata6Fl/Fl, Cdx2Fl/Fl, and Villin-CreERT2 mice were crossed to generate conditional Gata4Fl/Fl;Gata6Fl/Fl;Cdx2Fl/Fl;Villin-CreERT2 compound-mutant mice. Cdx2Fl/Fl, Hnf4aFl/Fl, and Villin-CreERT2 mice were mated to generate Hnf4aFl/Fl;Cdx2Fl/Fl;Villin-CreERT2 mice. Genotypes were verified using previously published protocols for each mutant strain (11, 20–23). To activate Cre, mice received intraperitoneal injections of 1 mg of tamoxifen (Sigma) in sunflower oil (Sigma) daily for 4–5 days. Mice were weighed daily and euthanized when the first mouse of a particular genotype became moribund (4 days for Gata4delCdx2del mice and 7 days for Hnf4adelCdx2del mice). Controls were injected with tamoxifen but lacked Villin-CreERT2. The Animal Care and Use Committees at our institutions approved and monitored animal use.
Motif Enrichment in ChIP-seq Data
CDX2 ChIP-seq data on intestinal villus cells (Gene Expression Omnibus (GEO) accession number GSM851117) (24) were analyzed using SeqPos (25) to identify enriched TF motifs within the 5000 most significant CDX2 peaks.
Histochemistry and Immunohistochemistry
The proximal (duodenum), central (jejunum), or distal (ileum) third of the small intestine was fixed overnight in 4% paraformaldehyde; embedded in paraffin; and sectioned at 5-μm thickness. Some mice were injected intraperitoneally with BrdU (1 mg/ml; Sigma) 1 h before euthanasia. H&E, Alcian blue, and periodic acid-Schiff staining followed standard procedures. To detect alkaline phosphatase, slides were incubated in 2 mm NaCl, 10 mm Tris-HCl (pH 9.5), and 5 mm MgCl2 for 5 min, followed by nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP ready-to-use tablets, Roche Applied Science) solution for 15–30 min, and washed with PBS.
Immunohistochemistry was performed as described previously (11) using the following primary antibodies: rabbit anti-Ki67 (1:200), goat anti-HNF4A (1:1000), and goat anti-GATA4 (1:400) (Santa Cruz Biotechnology); rat anti-BrdU (1:300; AbD Serotec); mouse anti-CDX2 (1:20; BioGenex); rabbit anti-cleaved caspase-3 (1:1000; Cell Signaling); goat anti-CRS4C1 (1:1000; gift of Andre Ouellette, University of Southern California, Los Angeles, CA); and goat anti-GATA6 (1:50; R&D Laboratories). Representative images of histology and immunohistochemistry from at least five mice of each genotype were obtained using an Olympus BX40 light microscope. The addition of scale bars and adjustment of brightness and contrast were performed in Photoshop.
To quantify morphological changes, at least 50 crypts or villi from each mouse (n = 3) were measured and averaged. For Ki67 and goblet cell counts, at least 10 crypts or villi from at least three mice were counted and averaged. For BrdU-positive cell counts, 25 crypts were counted and averaged. Significance was determined by t test using GraphPad Prism software. p values <0.05 were considered significant and are indicated in each figure.
Analysis of Microarray Data
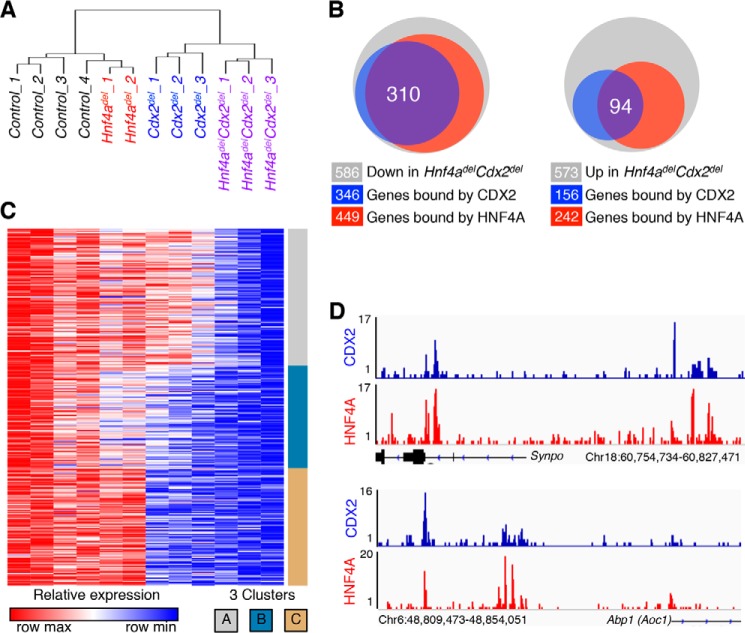
Raw microarray data from the Hnf4a/Cdx2 knock-out genetic series (GEO accession number GSE34567) (24) were reanalyzed using oneChannelGUI (26). Background correction and normalization were performed using the robust multi-array average method (27). Significant differential gene expression was determined using Limma (28), with p value adjustment (Q value) using the Benjamini-Hochberg correction for multiple testing (29). Fold changes in probes targeting the same gene were averaged together, so each gene is represented in the list only once. GENE-E software was used to perform hierarchical clustering of samples based on Pearson correlation and to create heat map images. The genes displayed in the heat map (see Fig. 5C) were bound by CDX2 (see methods discussed below) and significantly down-regulated in Hnf4adelCdx2del intestines compared with controls. Each row displays the relative expression value in that row from the minimum (blue) to maximum (red). Rows were then clustered using k-means clustering into three groups containing genes with similar expression patterns across the genotypes.
FIGURE 5.
mRNA analysis of single- and compound-mutant intestinal epithelia reveals dependences on single or multiple TFs. A, hierarchical cluster analysis of global mRNA expression in a series of Hnf4a;Cdx2 mutant intestinal epithelia revealed that transcripts were more profoundly affected in Hnf4adelCdx2del than in either single mutant. B, Venn diagram depicting dysregulated genes in Hnf4adelCdx2del mice (gray) and their overlap with CDX2 (blue) and HNF4A (red) binding. Gene numbers represented in each circle are indicated below. C, heat map showing expression levels in each genotype of the 310 down-regulated and co-occupied genes. The patterns of change fall into three k-means clusters (A–C). D, data traces for CDX2 and HNF4A ChIP-seq at the Abp1 (Aoc1) and Synpo loci, demonstrating TF co-occupancy at selected sites. The y axis indicates the density of mapped sequence tags, and genome coordinates are shown on the x axis, Chr, chromosome.
RNA Expression Analysis
Mouse intestinal epithelium was harvested by incubating fresh jejuna in 5 mm EDTA solution for 45 min as described previously (24). RNA was isolated using TRIzol reagent (Invitrogen) and the RNeasy kit (Qiagen). For quantitative RT-PCR, RNA was reverse-transcribed (SuperScript III, Invitrogen) and assessed using FastStart Universal SYBR Green Master Mix (Roche Applied Science) and specific primers for Cdx2 (5′-TCACCATCAGGAGGAAAAGTG-3′ and 5′-GCAAGGAGGTCACAGGACTC-3′), Gata4 (5′-TTTGAGCGAGTTGGG-3′ and 5′-GAATGCGGGTGTGC-3′), Gata6 (5′-CAGCAAGCTGTTGTGGTC-3′ and 5′-GTCTGGTACATTTCCTCCG-3′), and Hnf4a (5′-GGTCAAGCTACGAGGACAGC-3′ and 5′-ATGTACTTGGCCCACTCGAC-3′). Data were normalized for abundance of Gapdh (5′-GCCTTCCGTGTTCCTACCC-3′ and 5′-TGCCTGCTTCACCACCTTC-3′) or HPRT (5′-AAGCTTGCTGGTGAAAAGGA-3′ and 5′-TTGCGCTCATCTTAGGCTTT-3′) mRNA and expressed relative to control tissues.
Global assessment of RNA levels was performed on isolated jejunal epithelia from two control and two Hnf4adelCdx2del mice. RNA was isolated using TRIzol reagent and the RNeasy kit, followed by treatment with the Turbo DNA-free kit (Ambion) to remove genomic DNA. The RNA integrity number for each sample was ≥9.8. RNA-seq libraries were prepared with the TruSeq RNA sample preparation kit (Illumina) according to the manufacturer's instructions. 75-bp single-end reads were sequenced on an Illumina NextSeq 500 instrument. Sequence tags were aligned with the Mus musculus reference genome build 9 (mm9), and the Tuxedo software package was used to align reads, assemble transcripts, and determine differences in transcript levels using a false discovery rate of 0.05 (30). The Integrative Genomics Viewer was used to visualize aligned reads (31).
Association of TF Binding with Nearby Genes
Binding sites for CDX2 and HNF4A from ChIP-seq experiments (GEO accession number GSE34568) (24) were associated with the nearest gene within 30 kb using GREAT software (32). Genes with at least one binding site for each TF within this range were considered in our further analysis. BioVenn was used to generate Venn diagrams (33).
Gene Ontology Analysis
DAVID functional annotation clustering analysis was performed using medium classification stringency and default options (34). Clusters with significant enrichment scores (>1.3) were considered (34). When similar annotation clusters recurred in the list, we selected a representative gene ontology term from the cluster and listed it with the cluster enrichment score for that group of terms.
Electron Microscopy
Mouse ilea were flushed with PBS, fixed overnight or longer in EM fixative (2% formaldehyde and 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 7.4)), and embedded in TAAB 812 resin (Marivac Ltd., Nova Scotia, Canada). 80-nm sections were cut, stained with 0.2% lead citrate, viewed, and imaged with a Philips Tecnai BioTwin Spirit electron microscope at an accelerating voltage of 80 kV.
Analysis of Intestinal Lipid Absorption
Mice were placed on a diet (D12331, Research Diets) containing 58% calories from fat (the regular diet, Prolab IsoPro RMH 3000, contains 14% calories from fat) starting 5 days before the first dose of tamoxifen until euthanasia. Intestines were fixed overnight at 4 °C, equilibrated in 20% sucrose overnight at 4 °C, embedded in O.C.T. compound (Tissue-Tek), and frozen on dry ice. Lipid accumulation was visualized in 10-μm tissue sections incubated in 0.6% Oil Red O (Sigma) in propylene glycol at 60 °C for 8 min.
RESULTS
cis-Element Features Identify Candidate Partner TFs in Intestinal Epithelial Cells
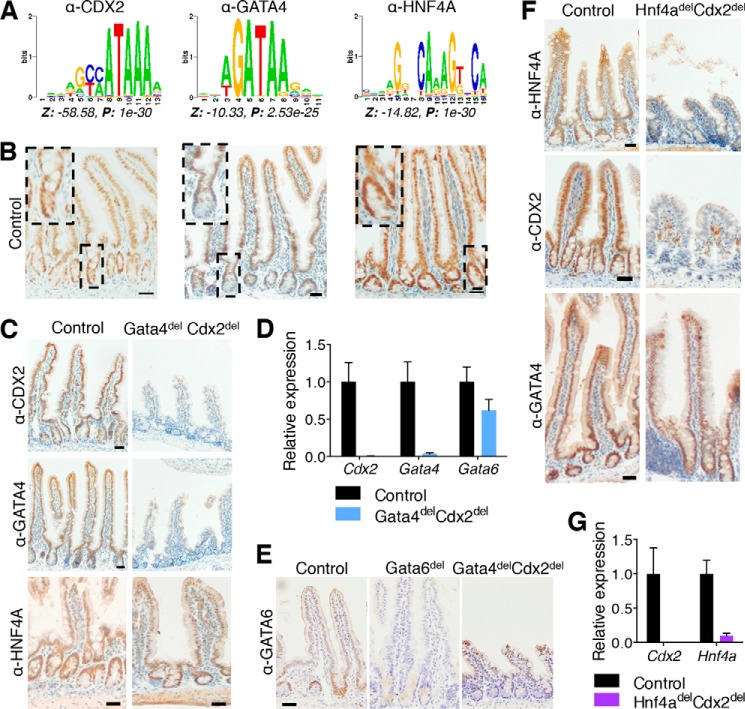
To assess the potential for coordinate regulation among intestine-specific and pan-endodermal TFs, we searched for overrepresented sequence motifs in CDX2 ChIP-seq data from mouse intestinal villus cells (24). Sequences corresponding to GATA and HNF4A were significantly enriched near CDX2-bound sites (Fig. 1A), indicating that these endoderm-restricted TFs may recurrently join CDX2 at intestinal enhancers in vivo. Indeed, ChIP-seq for GATA4 and HNF4A in the intestinal villus indicate co-occupancy with CDX2 at thousands of sites (24, 35). Interestingly, all three TFs are expressed in crypt and villus epithelial cells (Fig. 1B), suggesting the possibility of overlapping requirements that might be revealed in compound-mutant mice. If CDX2 and these partner TFs largely overlap in function, then the compound-mutant mice should phenocopy one another. Alternatively, distinct phenotypes would indicate that TF pairs control distinct programs. Moreover, transcripts perturbed upon combined TF loss should identify genes that require more than one TF for optimal expression.
FIGURE 1.
Recurrence of DNA sequence motifs at CDX2-binding sites in mouse intestinal villus cells and efficient gene deletion in conditional mutant mice. A, motifs for CDX2, GATA, and HNF4A are enriched near the 5000 strongest CDX2-binding sites in wild-type mouse intestinal villus cells. Position weight matrices, with the corresponding Z-score and p value, are indicated for each motif. B, immunohistochemistry for each TF showed nuclear staining throughout the crypt-villus axis in wild-type mice. Dashed boxes and insets show crypt details. C, CDX2 (upper panels) and GATA4 (middle panels) immunohistochemistry in control and Gata4delCdx2del jejuna confirmed the absence of the targeted TF proteins. Immunohistochemistry for HNF4A (lower panels) revealed similar levels in Gata4delCdx2del and control mice. D, quantitative RT-PCR for Cdx2 and Gata4 revealed complete loss of these mRNAs in the Gata4delCdx2del jejunum compared with controls (n = 2 each). However, Gata6 mRNA levels were only mildly reduced (n ≥ 4 each). E, GATA6 immunohistochemistry showed persistent protein in the Gata4delCdx2del jejunum after 4 days of tamoxifen. At the same time, mice with Gata6del alone recombined the allele efficiently and lost GATA6. F, immunohistochemistry for HNF4A (upper panels) and CDX2 (middle panels) verified the absence of both proteins in the ileum of compound-mutant intestines. Staining for GATA4 (lower panels) in the duodenum of Hnf4adelCdx2del mice (GATA4 is not expressed in the ileum) was similar to the control. G, quantitative RT-PCR for Cdx2 and Hnf4a showed complete and nearly complete loss of mRNA levels (n = 3 each), respectively. Scale bars = 30 μm. Graphs show means ± S.E.
Efficient Depletion of TFs in Mouse Intestinal Epithelium
As GATA4 and CDX2 are implicated in the control of selected villus cell genes in vivo (15, 36) and in replication of cultured progenitor cells (11), we first considered the intestinal GATA factors and crossed mice to obtain conditional Gata4Fl/Fl;Gata6Fl/Fl;Cdx2Fl/Fl;Villin-CreERT2 mutants. Tamoxifen treatment activated Cre recombinase and achieved intestinal loss of GATA4 and CDX2 mRNA and protein (Fig. 1, C and D), although Gata6 loss was incomplete due to inefficient recombination at the Gata6Fl allele (Fig. 1, D and E) (17). Thus, we refer to these mice as Gata4delCdx2del and note that HNF4A expression was preserved (Fig. 1C).
CDX2 and HNF4A co-occupy thousands of intestinal enhancers, where loss of CDX2 perturbs chromatin structure, resulting in decreases in or loss of HNF4A binding (24). The absence of HNF4A alone has little consequence on intestinal function (19) or chromatin structure, but the combined absence of CDX2 and HNF4A affects more transcripts than loss of either TF alone (24). To determine the functional consequences of combined TF loss, we produced Hnf4adelCdx2del intestines, which showed total or nearly total loss of CDX2 and HNF4A mRNA and protein throughout (Fig. 1, F and G), without affecting GATA4 (Fig. 1F).
Loss of Cdx2 and Gata4 Impairs Intestinal Crypt Cell Replication
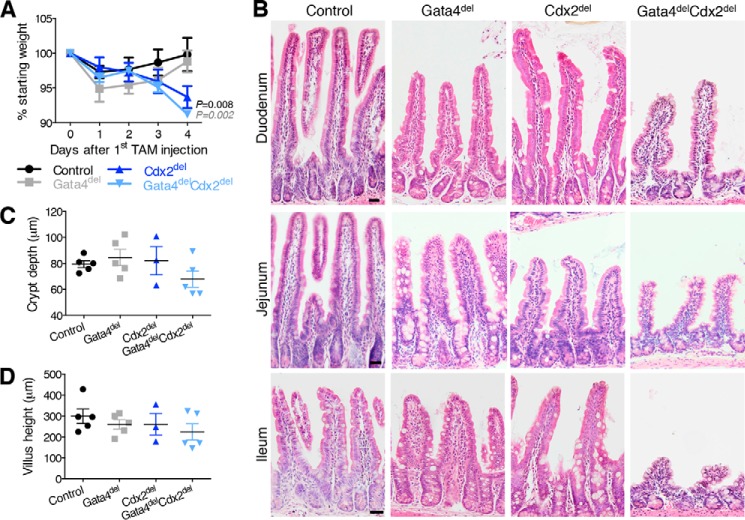
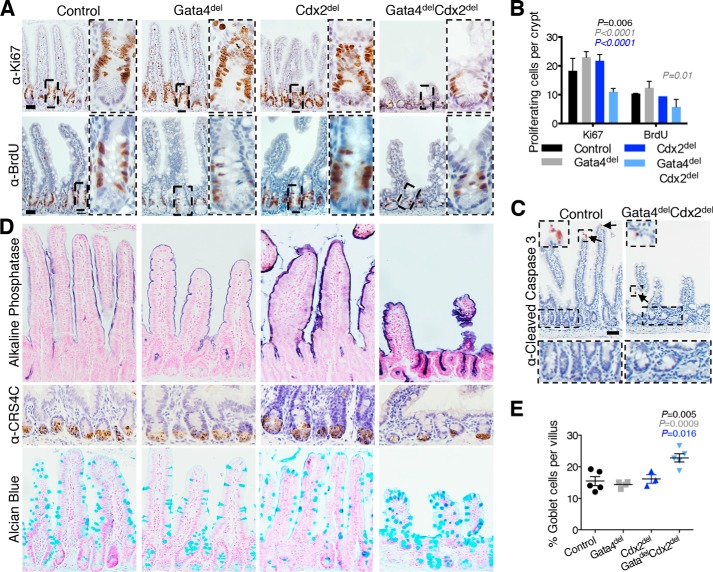
Gata4delCdx2del mice lost weight rapidly (Fig. 2A), became moribund, and required euthanasia within days of induced gene recombination. GATA4 is not expressed in the distal ileum, where CDX2 levels are the highest and the defects in Cdx2del intestines are the most severe (37). In the duodenum and jejunum, where GATA4 is abundant in wild-type mice, villi were slightly stunted in Gata4del mutants. In contrast, Gata4delCdx2del mice showed shallow crypts and short villi throughout the small intestine (Fig. 2, B–D). As the effects of the Cdx2del mice are most severe distally, we focused analysis on the most distal portion of the intestine that normally expresses Gata4, i.e. the jejunum. Here, significantly fewer crypt cells expressed the proliferation marker Ki67 (Fig. 3, A and B). In addition, the number of cells in S-phase, as marked by incorporation of BrdU during a 1-h pulse, was also reduced in Gata4delCdx2del mice (Fig. 3, A and B). The reduced crypt and villus heights did not reflect increased apoptosis in addition to the proliferation deficit (Fig. 3C), but ectopic alkaline phosphatase expression (enterocytes) and Alcian blue staining (goblet cells) in Gata4delCdx2del crypts indicated precocious cell maturation (Fig. 3D), probably reflecting the cell cycle arrest. Villus alkaline phosphatase expression and Alcian blue staining verified the presence of mature cells (Fig. 3D), and although cells retained a columnar morphology, they varied in shape, size, and nuclear morphology. The fraction of goblet cells, but not of enterocytes or Paneth cells, was modestly increased over intestines lacking only GATA4 or CDX2 (Fig. 3, D and E). These data show that intestinal crypt cell replication is a prominent shared function for CDX2 and GATA4, with a significantly larger defect in the compound-mutant mouse than in either single-mutant mouse. The villus defects may reflect this poor crypt cell turnover or indicate additional joint functions in cell maturation.
FIGURE 2.
Morbidity and morphologic defects with combined loss of Gata4 and Cdx2. A, mouse body weights each day after the start of tamoxifen (TAM) injection revealed weight loss in Cdx2del and Gata4delCdx2del mice. Significance was calculated between Gata4delCdx2del mice and other genotypes on day 4 (n ≥ 5). B, duodenal (upper panels), jejunal (middle panels), and ileal (lower panels) tissue sections stained with H&E showed reduced villus height and crypt depth in Gata4delCdx2del intestines compared with loss of either TF alone. Scale bars = 30 μm. C and D, quantitation of crypt (C) and villus (D) length in the Gata4delCdx2del jejunum compared with single mutants and controls (n ≥ 3). Graphs show means ± S.E. Statistical significance was assessed using Student's t test; p values are indicated when significant and are color-coded to match the sample with which Gata4delCdx2del samples were compared.
FIGURE 3.
Effects of combined loss of Gata4 and Cdx2 on proliferating and differentiated intestinal cells. A, immunohistochemistry of proliferative markers Ki67 and BrdU (administered 1 h before euthanasia) confirmed significantly reduced proliferation in Gata4delCdx2del jejuna compared with single-mutant and control littermates (quantified in B). Dashed boxes outline the areas magnified to the right. Scale bars = 50 μm. C, cleaved caspase-3 staining revealed that apoptosis was not increased in Gata4delCdx2del intestines compared with control villi or crypts (areas in dashed boxes are magnified below). Arrows indicate caspase-3-positive cells (an example is magnified in the dashed box on the upper left. D, histochemistry and immunohistochemistry of the jejuna for alkaline phosphatase and CRS4C1 in Gata4delCdx2del and control mice showed no differences in mature enterocytes and Paneth cells, respectively. Alcian blue staining revealed an increased fraction of villus goblet cells in Gata4delCdx2del intestines (quantified in E). Of note, Gata4delCdx2del mice had ectopic alkaline phosphatase and Alcian blue staining in crypts. Graphs show means ± S.E. Significant changes between Gata4delCdx2del and other genotypes were calculated using Student's t test. Results are color-coded by genotype; insignificant differences are not marked. Scale bars = 50 μm (A) and 30 μm (C).
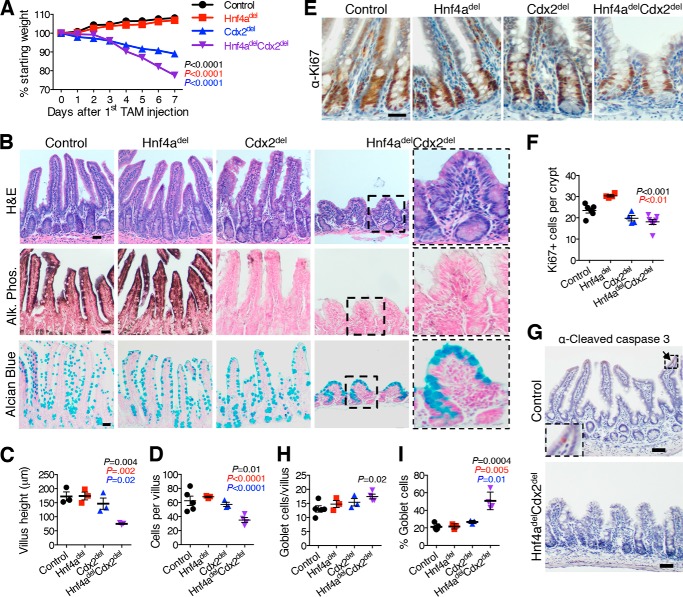
Combined Loss of CDX2 and HNF4A Compromises Enterocyte Differentiation without Affecting Crypt Cell Replication
Swift weight loss in Hnf4adelCdx2del mice (Fig. 4A) warranted euthanasia 2 weeks earlier than in Cdx2del littermates, with severe diarrhea and steatorrhea occurring during the course of tamoxifen administration. Duodenal and jejunal villus heights were similar to those in Cdx2del intestines, and enterocytes in these regions showed little cellular atypia (data not shown). By contrast, villi in the ileum were dysplastic and significantly stunted (Fig. 4, B and C), with many cells showing pyknosis, and total villus cell numbers were significantly lower compared with Hnf4adel or Cdx2del villi (Fig. 4D). Importantly and in contrast to Gata4delCdx2del mice, this was not a result of reduced crypt cell proliferation (Fig. 4, E and F) or increased apoptosis (Fig. 4G) but rather of aberrant epithelial cell differentiation. A total absence of alkaline phosphatase in Cdx2del and Hnf4adelCdx2del villi in the ileum (Fig. 4B) implicated the enterocyte compartment. Indeed, Alcian blue staining revealed a predominance of goblet cells (Fig. 4B), whereas total goblet cell numbers were similar to those control mice (Fig. 4H). Thus, the fraction of goblet cells per villus was significantly increased in the Hnf4adelCdx2del ileum (Fig. 4I), indicating a lack of mature enterocytes. Weight loss leading to rapid demise is likely due to this enterocyte deficit, which was greatest in the ileum, distinct from the absent (Hnf4adel) or subtle (Cdx2del) defects observed in single-mutant mice. The rapid and dramatic consequences of combined loss of CDX2 and HNF4A indicate that the two TFs control, cooperatively or in parallel, transcriptional programs specific to mature intestinal villus cells.
FIGURE 4.
Combined loss of Hnf4a and Cdx2 accelerates death and results in reduction of absorptive enterocytes. A, weight loss in Hnf4delCdx2del mice was accelerated compared with single mutants or controls, requiring euthanasia within 7 days of induced gene deletion. Significance was calculated between Hnf4adelCdx2del mice and other genotypes on day 7 (n = 6). TAM, tamoxifen. B, H&E staining of the ileum in the Hnf4a/Cdx2 genetic series showed significant shortening of Hnf4adelCdx2del villi (quantified in C). Both Cdx2del and Hnf4adelCdx2del mice lacked alkaline phosphatase (Alk. Phos.) in ileal villus cells, indicating defective enterocyte maturation. Alcian blue staining revealed goblet cell abundance in Hnf4adelCdx2del villi. Dashed boxes outline areas magnified to the right. D, the total number of ileal villus cells was reduced in Hnf4adelCdx2del mice. E, Ki67 immunohistochemistry showed that combined loss of Hnf4a and Cdx2 affected proliferation no more than loss of Cdx2 alone (quantified in F). G, cleaved caspase-3 staining revealed no significant apoptosis in the Hnf4adelCdx2del ileal epithelium. A few apoptotic cells were present at the villus tips in control mice (arrow). A caspase-3-positive cell is magnified in dashed box at the lower left. H, quantitation of total goblet cell numbers per villus in the ileum showed no increase in Hnf4adelCdx2del intestines compared with single-mutant intestines. I, Hnf4adelCdx2del mice exhibited an increased proportion of ileal goblet cells. Scale bars = 30 μm (B and E) and 50 μm (G). Graphs show means ± S.E. Significant changes between Hnf4adelCdx2del and other genotypes were calculated using Student's t test. Results are color-coded by genotype as indicated; insignificant results are not indicated.
Identification of Genes Co-regulated by CDX2 and HNF4A
To investigate transcriptional programs that depend on CDX2 and/or HNF4A in maturing cells, we first compared transcriptomes in mature villi 7 days after tamoxifen-induced deletion of Hnf4a, Cdx2, or both genes (24). Because abnormalities in the ileum could represent direct consequences of TF loss or the indirect effects of epithelial dysfunction, we examined jejunal transcriptomes, reasoning that the absence of a strong phenotype in this region would highlight the primary effects, free of confounding considerations. Compound-mutant Hnf4adelCdx2del samples clustered separately from single Hnf4adel or Cdx2del mutants, indicating increased severity of global changes in gene expression (Fig. 5A). Compared with the controls, 573 transcripts were significantly increased and 586 transcripts were reduced in Hnf4adelCdx2del intestines (Q < 0.05).
To identify likely direct transcriptional targets, we selected genes showing CDX2 and HNF4A co-occupancy in jejunal villus epithelial cells (<30 kb from the nearest gene). More than half (52.9%) of the genes reduced in Hnf4adelCdx2del intestines were bound by both TFs, compared with 16.4% of genes with increased levels (Fig. 5B), consistent with a role for these factors primarily in gene activation. Among the 310 genes down-regulated in Hnf4adelCdx2del mice compared with controls and showing TF co-occupancy, k-means clustering identified three prominent patterns with roughly equal frequency (Fig. 5C and supplemental Table 1): genes equally reduced compared with controls in Cdx2del and Hnf4adelCdx2del intestines, but not in Hnf4adel intestines (cluster C); genes modestly perturbed in each single mutant and further dysregulated in the compound mutant (cluster B); and transcripts that barely changed in either single mutant compared with controls, but were significantly reduced in Hnf4adelCdx2del intestines (cluster A). Selected examples of genes from clusters A (Synpo) and C (Abp1/Aoc1) (Fig. 5D) showed binding of both CDX2 and HNF4A. This analysis indicates that CDX2 alone is required for proper expression of genes in cluster C, whereas additional loss of HNF4A is necessary to affect genes in clusters A and B.
To examine these changes in gene expression with greater confidence and quantitative accuracy, we performed RNA-seq on epithelial cells isolated from control and Hnf4adelCdx2del jejuna. We confirmed disruption of Cdx2 and Hnf4a by RNA-seq, noting an absence of transcript reads from exon 2 in Cdx2 and a nearly complete absence of transcript reads from exons 4 and 5 in Hnf4a (data not shown).
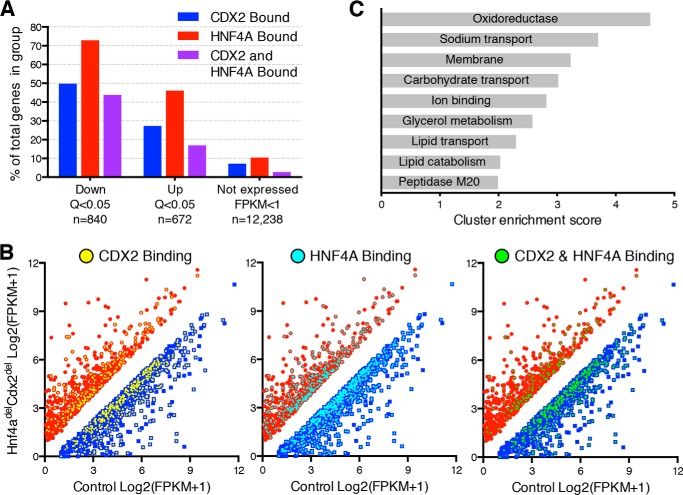
RNA-seq results corroborated our microarray data and expanded the number of candidate direct transcriptional targets, identifying 840 down-regulated and 642 up-regulated genes (Q < 0.05) in Hnf4adelCdx2del intestines. Integration of RNA-seq and ChIP-seq data indicated that both CDX2 and HNF4A were bound within 30 kb of 44% (368) of the down-regulated genes, 17% of the up-regulated or unaffected genes, and 3% of the genes not expressed in intestinal villi (Fig. 6, A and B). These findings further support the idea that CDX2 and HNF4A function mainly as activators. Although the two TFs could, in principle, affect intestinal genes independently, their binding near genes that respond to loss of both TFs is a good indication of co-regulation.
FIGURE 6.
RNA-seq analysis further identifies CDX2- and HNF4A-co-regulated intestinal genes. A and B, integration of CDX2 and HNF4A binding data from ChIP-seq with mRNA expression data from RNA-seq analysis of control and Hnf4adelCdx2del intestines. A, genes significantly down- or up-regulated (Q < 0.05) are shown in relation to those not expressed (fragments/kb of transcript/million mapped reads (FPKM) < 1) in either sample. The fraction of genes in each group bound by CDX2 (blue), HNF4A (red), and both TFs (purple) indicates again that these TFs activate mainly target genes. B, FPKM graphs showing RNA-seq expression of significantly up- and down-regulated genes are overlaid with binding for CDX2 (left panel, yellow), HNF4A (middle panel, aqua), and both (right panel, green), demonstrating a higher distribution of bound down-regulated genes. C, gene ontology annotation clusters from DAVID analysis of the 362 genes significantly down-regulated in Hnf4adelCdx2del intestines and bound by both TFs.
CDX2 and HNF4A Co-regulate Genes for Specific Aspects of Digestive Physiology
To determine the physiologic consequences of gene co-regulation, we performed gene ontology analysis on the 368 genes that RNA-seq identified as reduced in Hnf4adelCdx2del intestines and for which ChIP-seq showed binding of both TFs (supplemental Table 2). Clustering of significantly enriched gene ontology terms highlighted genes central to enterocyte functions, including oxidation-reduction; transport of ions, carbohydrates, and lipids; and components of the plasma membrane (Fig. 6C). Thus, combined loss of CDX2 and HNF4A in vivo produced attrition of enterocytes in the ileum and significantly altered gene expression in jejunal enterocytes, albeit with few overt histologic manifestations. Together, these anomalies can account for the rapid uniform lethality in Hnf4adelCdx2del mice.
Because defects in cell membranes and in nutrient absorption should be objectively observable, we focused efforts on identifying these deficits in Hnf4adelCdx2del mice. Apical microvilli in Hnf4adel mice are grossly intact (19). In contrast, microvilli on the apical membranes of Cdx2del enterocytes were moderately scant and stunted, whereas those in Hnf4adelCdx2del cells were sparse, shortened, and disarrayed (Fig. 7A). Beyond this significant brush-border defect, the Hnf4adelCdx2del enterocytes showed additional intracellular anomalies, including an accumulation of light droplets or vacuoles near the cell apex and dark, possibly membrane-bound vesicles within mitochondria. The contribution of these defects to cell attrition is not presently clear. However, together with reduced expression of peptidases, other hydrolases, and genes involved in symporter activity and intestinal absorption, the brush-border anomalies undoubtedly contribute to the severe malnutrition.
FIGURE 7.
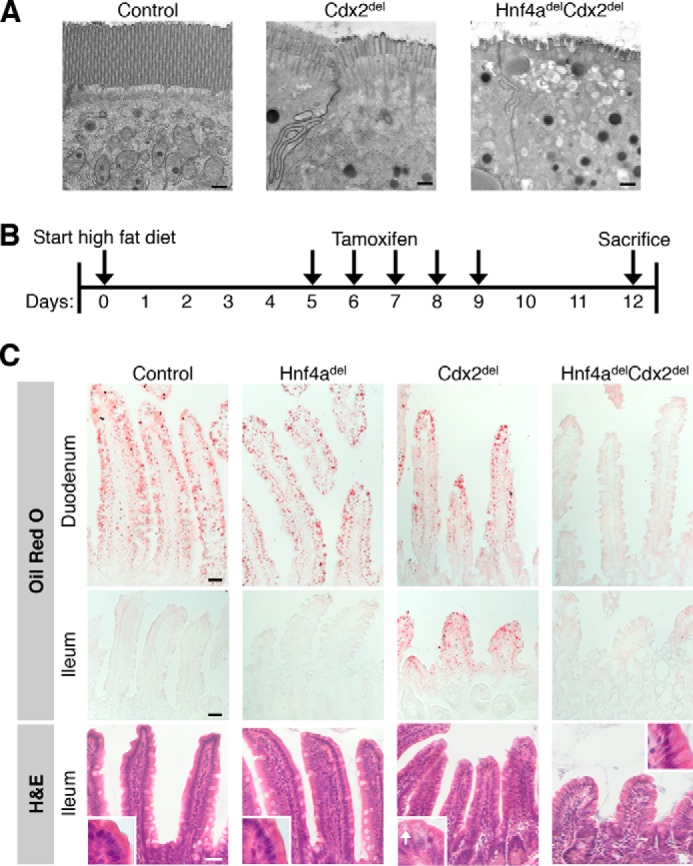
HNF4A and CDX2 co-regulate enterocyte apical microvilli and absorption of dietary lipids. A, electron micrographs of ileal enterocyte apices showing an intact brush border in control mice, shortened microvilli in Cdx2del mice, and severely depleted and stunted microvilli in Hnf4adelCdx2del intestines. Scale bars = 500 nm. B, experimental schema for maintaining mice on a high-fat diet before, during, and after inducing gene deletion. C, Oil Red O staining for neutral lipids revealed ectopic lipid absorption in Cdx2del ileal enterocytes and a total lack of lipid absorption in Hnf4delCdx2del intestines. H&E staining of the ileum revealed light lipid droplets in the Cdx2del epithelium (inset, white arrow), but not in other genotypes. Scale bars = 30 μm.
In addition, we made particular note of lipid metabolism because intestinal absorption of dietary lipids is highly relevant to human health, and HNF4A is known to regulate hepatic lipid metabolism (23). To test the hypothesis that co-regulation of genes related to lipid absorption might also underlie the profound rapid malnutrition in Hnf4adelCdx2del mice, we placed mice on a high-fat diet starting 5 days before TF gene disruption (Fig. 7B) and stained intestines 12 days later with Oil Red O to detect lipid deposits. Enterocytes accumulate lipids as a result of fatty acid and cholesterol transport as well as de novo biosynthesis (38). In the presence of bile salts and pancreatic lipase, enterocytes in the proximal intestine (duodenum) absorb dietary lipids efficiently, leaving no luminal lipid residue for absorption by distal (ileal) enterocytes (39). Accordingly, Cre− and Hnf4adel control mice showed prominent Oil Red O staining in duodenal cells and virtually none in the ileum (Fig. 7C). In contrast, Cdx2del single-mutant mice showed reduced lipid in duodenal enterocytes and significant levels in the ileum, indicating that CDX2-null cells absorb dietary lipids inefficiently, leaving a residual amount for ileal absorption. Hnf4adelCdx2del intestines showed minimal Oil Red O staining in any region (Fig. 7C), revealing a global and profound defect in lipid absorption. Importantly, this was not a trivial consequence of enterocyte depletion because the duodenum, where enterocytes were plentiful, showed no lipid uptake. Moreover, we observed no lipid uptake in ileal villi that contained both goblet cells and a few enterocytes. Taken together, these findings reveal that CDX2 and HNF4A co-regulate genes necessary for dietary lipid metabolism, which is more severely impaired when both TFs are absent than when either TF alone is lost. This striking defect likely also contributes to rapid weight loss in Hnf4adelCdx2del mice.
DISCUSSION
Selected lineage-restricted TFs exert considerable control over each cell type's unique transcriptional program. For example, GATA1, TAL1, EKLF, and NF-E2 together regulate most erythroid blood cell genes (40), and a few basic helix-loop-helix TFs control much of the muscle cell-specific transcriptome (41). Several lines of evidence implicate CDX2, HNF4A, and GATA4/6 in control of intestinal genes. First, functional cis-elements for individual intestinal genes repeatedly reveal these and few other TF activities (42, 43). Second, the corresponding sequence motifs recur frequently at intestine-active enhancers identified by histone marks and nucleosome depletion (11, 44) or differential DNA methylation (45). Here, we have shown that additional loss of CDX2 dramatically unmasks GATA4 and HNF4A requirements in knock-out mice. Defects in both compound-mutant intestines are rapidly lethal, include distinct components, and help delineate functional TF hierarchies in the intestine.
Arrested crypt cell replication in Gata4delCdx2del intestines indicates collaboration of these TFs in intestinal crypts and contrasts sharply with the lack of replication deficits in Hnf4adelCdx2del intestines. Although our data do not address whether CDX2 and GATA control the same genes additively or different genes, the GATA sequence motif and binding of a closely related factor, GATA6, are highly enriched near CDX2-binding sites in replicating human intestinal cells in culture (11), suggesting possible regulation of the same genes. CDX2 and GATA proteins likely have additional, joint roles in villus cell maturation, as evidenced by a large overlap of their respective binding sites in the intestinal villus (35). It was difficult to address this point unequivocally in our analysis because the deficit in crypt cell replication may explain the reduced villus cell height whether cell maturation is intact or defective. In the future, identification of GATA4-binding sites and GATA4-dependent genes in intestinal crypt cells may uncover co-regulated genes in this compartment, much like this study uncovered CDX2/HNF4A-co-regulated genes in villus cells.
Suppression of crypt cell replication and the increase in goblet cell numbers in Gata4delCdx2del intestines may reflect reduced Notch signaling or an effect on some other shared pathway. Whether the phenotype is caused by defects in stem cells or transit-amplifying progenitors also warrants further investigation. By contrast, because Hnf4adelCdx2del crypt cells proliferate normally, we can categorically attribute these phenotypes to failures in cell maturation and in enterocytes, in particular.
CDX2 loss disrupts chromatin structure and binding of other TFs such as HNF4A (24), implying that CDX2 maintains chromatin access in intestinal cells. In a simple hierarchy, where other TFs depend on CDX2 wholly, compound-mutant and Cdx2del intestines should largely phenocopy one another, but our studies on HNF4A indicated otherwise. Hnf4adelCdx2del mice fared profoundly worse than Cdx2del littermates, with accelerated demise, near absence of ileal enterocytes, significant gene dysregulation in jejunal villus cells, and lack of dietary fat absorption. Many more genes are dysregulated in Hnf4adelCdx2del than in either single mutant. One likely reason is that HNF4A regulates genes in addition to those where CDX2 enables chromatin access, but CDX2 loss is necessary to expose that dependence. A second reason is that HNF4A provides necessary additive activity at regulatory sites that both TFs co-occupy. Indeed, the genes most affected in Hnf4adelCdx2del intestines are enriched for such co-occupancy, indicating that loss of single TFs may preserve some enhancer function, but the absence of both TFs is necessary to abrogate transcription completely.
In summary, combinatorial control allows TFs to elicit diverse transcriptional outcomes in both embryos and adult tissues, and our data highlight this aspect of gene control in vivo. The intestinal “master regulator” CDX2 participates in various TF combinations. Specific pairings with partners such as GATA proteins drive cell replication, whereas separate pairings with HNF4A and other TFs regulate genes in terminally differentiated cells. The contributions of each TF no doubt vary at different cis-regulatory sites. At some sites, the absence of a single factor has profound effects, as occurs commonly with loss of CDX2, but not GATA4 or HNF4A. At other sites, gene expression suffers only when more than one TF is absent. Our studies reveal many examples of such cooperativity, and the scope of gene dysregulation in Hnf4adelCdx2del (compared with Cdx2del) intestines matched the greater severity of tissue defects and malnutrition. Particularly at enterocyte genes necessary to construct the microvillus brush border or absorb nutrients, including dietary fat, both CDX2 and HNF4A are necessary. These TFs co-occupy the corresponding enhancers, and loss of both TFs affects mRNA levels more than the absence of either factor alone. These findings illuminate the transcriptional basis for vital intestinal functions.
Supplementary Material
Acknowledgments
We thank Sylvie Robine and William Pu for sharing Villin-CreERT2 and conditional Gata mutant mice, respectively; Justina Chen and Kelly Stapleton for assistance with mouse dissections and immunostaining; Marian Neutra for insights into intestinal phenotypes; Andre Ouellette for the gift of anti-CRS4C1 antibody; Pere Puigserver for providing high-fat mouse chow; the Electron Microscopy Core Facility at Harvard Medical School; the Center for Functional Cancer Epigenetics for preparing RNA-seq libraries; and the Molecular Biology Core Facilities at Dana-Farber Cancer Institute for next-generation sequencing.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK08229 (to R. A. S.) and R01DK061382 (to S. D. K.) and Fellowships F31CA180784 (to A. K. S. R.) and K01DK088868 (to M. P. V.). This work was also supported by a National Science Foundation graduate research fellowship (to A. K. S. R.); the Human Genetics Institute of New Jersey (to M. P. V.); the Nutricia Research Foundation, the European Society for Pediatric Research, KWF Kankerbestrijding, and Prins Bernhard Cultuurfonds (to B. E. A.); and Specialized Program of Research Excellence in Gastrointestinal Cancer Award P50CA127003 from the NCI, National Institutes of Health.
RNA-seq data have been deposited in the Gene Omnibus Expression Database with GEO accession number GSE62633.

This article contains supplemental Tables 1 and 2.
- TF
- transcription factor
- GEO
- Gene Expression Omnibus.
REFERENCES
- 1. James R., Erler T., Kazenwadel J. (1994) Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J. Biol. Chem. 269, 15229–15237 [PubMed] [Google Scholar]
- 2. Krapp A., Knöfler M., Frutiger S., Hughes G. J., Hagenbüchle O., Wellauer P. K. (1996) The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 15, 4317–4329 [PMC free article] [PubMed] [Google Scholar]
- 3. Ohlsson H., Karlsson K., Edlund T. (1993) IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12, 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. German M. S., Wang J., Chadwick R. B., Rutter W. J. (1992) Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 6, 2165–2176 [DOI] [PubMed] [Google Scholar]
- 5. James R., Kazenwadel J. (1991) Homeobox gene expression in the intestinal epithelium of adult mice. J. Biol. Chem. 266, 3246–3251 [PubMed] [Google Scholar]
- 6. Suh E., Chen L., Taylor J., Traber P. G. (1994) A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol. 14, 7340–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin T., Drucker D. J. (1996) Activation of proglucagon gene transcription through a novel promoter element by the caudal-related homeodomain protein Cdx-2/3. Mol. Cell. Biol. 16, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao N., White P., Kaestner K. H. (2009) Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu T., Zhang X., So C. K., Wang S., Wang P., Yan L., Myers R., Chen Z., Patterson A. P., Yang C. S., Chen X. (2007) Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis 28, 488–496 [DOI] [PubMed] [Google Scholar]
- 10. Silberg D. G., Sullivan J., Kang E., Swain G. P., Moffett J., Sund N. J., Sackett S. D., Kaestner K. H. (2002) Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology 122, 689–696 [DOI] [PubMed] [Google Scholar]
- 11. Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., Montgomery R. K., Fleet J. C., Brown M., Liu X. S., Shivdasani R. A. (2010) Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hryniuk A., Grainger S., Savory J. G., Lohnes D. (2012) Cdx function is required for maintenance of intestinal identity in the adult. Dev. Biol. 363, 426–437 [DOI] [PubMed] [Google Scholar]
- 13. Stringer E. J., Duluc I., Saandi T., Davidson I., Bialecka M., Sato T., Barker N., Clevers H., Pritchard C. A., Winton D. J., Wright N. A., Freund J. N., Deschamps J., Beck F. (2012) Cdx2 determines the fate of postnatal intestinal endoderm. Development 139, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verzi M. P., Shin H., Ho L. L., Liu X. S., Shivdasani R. A. (2011) Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31, 2026–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bosse T., Piaseckyj C. M., Burghard E., Fialkovich J. J., Rajagopal S., Pu W. T., Krasinski S. D. (2006) Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol. Cell. Biol. 26, 9060–9070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beuling E., Baffour-Awuah N. Y., Stapleton K. A., Aronson B. E., Noah T. K., Shroyer N. F., Duncan S. A., Fleet J. C., Krasinski S. D. (2011) GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology 140, 1219.e1–2-1229.e1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beuling E., Aronson B. E., Tran L. M., Stapleton K. A., ter Horst E. N., Vissers L. A., Verzi M. P., Krasinski S. D. (2012) GATA6 is required for proliferation, migration, secretory cell maturation, and gene expression in the mature mouse colon. Mol. Cell. Biol. 32, 3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garrison W. D., Battle M. A., Yang C., Kaestner K. H., Sladek F. M., Duncan S. A. (2006) Hepatocyte nuclear factor 4α is essential for embryonic development of the mouse colon. Gastroenterology 130, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Babeu J.-P., Darsigny M., Lussier C. R., Boudreau F. (2009) Hepatocyte nuclear factor 4α contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G124–G134 [DOI] [PubMed] [Google Scholar]
- 20. Sodhi C. P., Li J., Duncan S. A. (2006) Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pu W. T., Ishiwata T., Juraszek A. L., Ma Q., Izumo S. (2004) GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 275, 235–244 [DOI] [PubMed] [Google Scholar]
- 22. el Marjou F., Janssen K.-P., Chang B. H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. (2004) Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 [DOI] [PubMed] [Google Scholar]
- 23. Hayhurst G. P., Lee Y. H., Lambert G., Ward J. M., Gonzalez F. J. (2001) Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verzi M. P., Shin H., San Roman A. K., Liu X. S., Shivdasani R. A. (2013) Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 33, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu T., Ortiz J. A., Taing L., Meyer C. A., Lee B., Zhang Y., Shin H., Wong S. S., Ma J., Lei Y., Pape U. J., Poidinger M., Chen Y., Yeung K., Brown M., Turpaz Y., Liu X. S. (2011) Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanges R., Cordero F., Calogero R. A. (2007) oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics 23, 3406–3408 [DOI] [PubMed] [Google Scholar]
- 27. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 28. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 29. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 [Google Scholar]
- 30. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P. (2011) Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hulsen T., de Vlieg J., Alkema W. (2008) BioVenn–a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 35. Aronson B. E., Rabello Aronson S., Berkhout R. P., Chavoushi S. F., He A., Pu W. T., Verzi M. P., Krasinski S. D. (2014) GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27. Biochim. Biophys. Acta 1839, 1273–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benoit Y. D., Paré F., Francoeur C., Jean D., Tremblay E., Boudreau F., Escaffit F., Beaulieu J. F. (2010) Cooperation between HNF-1α, Cdx2, and GATA-4 in initiating an enterocytic differentiation program in a normal human intestinal epithelial progenitor cell line. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G504–G517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Wering H. M., Bosse T., Musters A., de Jong E., de Jong N., Hogen Esch C. E., Boudreau F., Swain G. P., Dowling L. N., Montgomery R. K., Grand R. J., Krasinski S. D. (2004) Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G899–G909 [DOI] [PubMed] [Google Scholar]
- 38. Sturley S. L., Hussain M. M. (2012) Lipid droplet formation on opposing sides of the endoplasmic reticulum. J. Lipid Res. 53, 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamada T., Alpers D. H. (2009) Textbook of Gastroenterology, 5th Ed., Wiley-Blackwell, Hoboken, NY [Google Scholar]
- 40. Orkin S. H., Zon L. I. (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tapscott S. J. (2005) The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 42. Gregory P. A., Lewinsky R. H., Gardner-Stephen D. A., Mackenzie P. I. (2004) Coordinate regulation of the human UDP-glucuronosyltransferase 1A8, 1A9, and 1A10 genes by hepatocyte nuclear factor 1α and the caudal-related homeodomain protein 2. Mol. Pharmacol. 65, 953–963 [DOI] [PubMed] [Google Scholar]
- 43. Mitchelmore C., Troelsen J. T., Spodsberg N., Sjöström H., Norén O. (2000) Interaction between the homeodomain proteins Cdx2 and HNF1α mediates expression of the lactase-phlorizin hydrolase gene. Biochem. J. 346, 529–535 [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu J., Adli M., Zou J. Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P. L., Bennett D. A., Houmard J. A., Muoio D. M., Onder T. T., Camahort R., Cowan C. A., Meissner A., Epstein C. B., Shoresh N., Bernstein B. E. (2013) Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sheaffer K. L., Kim R., Aoki R., Elliott E. N., Schug J., Burger L., Schübeler D., Kaestner K. H. (2014) DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 28, 652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.