Abstract
Pseudomonas aeruginosa genus bacteria are well known for their increased drug resistance (phenotypic ang genotypic resistance). The most important resistance mechanisms are: enzyme production, reduction of pore expression, reduction of the external membrane proteins expression, efflux systems, topoisomerase mutations. These mechanisms often accumulate and lead to multidrug ressitance strains emergence. The most frequent acquired resistance mechanisms are betalactamase-type enzyme production (ESBLs, AmpC, carbapenemases), which determine variable phenotypes of betalactamines resistance, phenotypes which are associated with aminoglycosides and quinolones resistance. The nonenzymatic drug resistance mechanisms are caused by efflux systems, pore reduction and penicillin-binding proteins (PBP) modification, which are often associated to other resistance mechanisms. Phenotypic methods used for testing these mechanisms are based on highlighting these phenotypes using Kirby Bauer antibiogram, clinical breakpoints, and “cut off” values recommended by EUCAST 2013 standard, version 3.1.
Keywords: ESBLs, AmpC, Carbapenemase, genotypic resistance, phenotypic resistance
Introduction
Pseudomonas aeruginosa produces infections at patients in special conditions and at hospitalized patients (bladder or tracheobronchial cateters), being a rare infection at healthy patients. The rate of P.aeruginosa colonization increases at hospitalized patients. P.aeruginosa is a bacillus that poses serious treatment issues because of the high intrinsic resistance level due to the very poor permeability of this bacillus (P.aeruginosa has moth of its pores closed) and to the reduction of an external membrane protein expression. [1]
Multidrug resistance is increasing and spreading rapidly among nosocomial bacteria, health care assistance bacteria, and community bacteria. Natural (genotypic) resistance represents the wild-type phenotype and defines the action spectrum of an antibiotic, having cromosomial support. Acquired resistance emerges in a sensitive population, has variable frequency and has cromosomial or extracromosomial support (plasmid, bacteriophage, transposons). Knowing the resistance phenotypes is a very important step in choosing the correct antibiotic treatment. [2, 3]
Pseudomonas aeruginosa betalactamines resistance phenotypes
The involved mechanisms are: impermeability, efflux, hydrolyzing enzymes production or target modification. Many of these can be associated, making the phenotypes difficult to interpret.
Natural resistance. Wild-type phenotype
Natural resistance is linked to the production of cephalosporinase, induced by aminopenicillins and first generation cephalosporins, doubled by variable impermeability and efflux systems. This phenotype presents resistance to aminopenicillins, aminopenicillins associated to betalactamase inhibitor, first and second generation cephalosporins. This phenotype remains susceptible to carboxypenicillins (ticarcillin, cabenicillin), ureidopenicillins (piperacillin, mezlocillin), to third and fouth generation cephalosporins, to carbapenems and monobactams.
Acquired resistance
Is linked to enzymatic mechanisms (panicillinases, cephalosporinases, extended-spectrum betalactamases, metallo-betalactamases) or nonenzymatic mechanisms (impermeability, efflux and target modification).
Enzymatic acquired resistanceis linked to betalactamases production and represents the main resistance mechanism to betalactamines. These enzymes are grouped into four classes (according to Ambler classification), depending on the aminoacid sequence. A,C and D classes take action through a serine-based mechanism, while B class require zinc to take action. The most important betalactamases are: penicillinases, ESBLs, AmpC and carbapenemases.
a.1) Penicillinases. Provide resistance to carboxypenicillins, ureidopenicillins, cefoperazone and sensitivity to ceftazidime, cefepime, imipenem and betalactamase inhibitors.
a.2) Extended-spectrum betalactamases (ESBLs). Represent betalactamases that hydrolyze extended-spectrum cephalosporins (cefotaxime, ceftriaxone, ceftazidime), monobactams (aztreonam) and that are not affected by betalactamase and carbapenemase inhibitors. Second generation cephalosporins are not hydrolyzed by ESBLs, but are hydrolyzed by ESLB/AmpC association. The most important ESBLs are TEM, SHV, PER, VEB (class A): sensitivity to imipenem, carboxypenicillins +betalactamase inhibitors, ureidopenicillin +betalactamase inhibitors. Until 2000, the ESBLs were TEM (Temoniera) and SHV (Sulphydril variable) and they were frequently associated to nosocomial infections. Since 2000, CTX-M (cefotaximase) has become the main ESBL and has rapidly spread [4, 5].
a.3) AmpC betalactamases (class C). Are constitutive or inducible cephalosporinases, chromosomally or plasmid mediated. AmpC betalactamases are inhibited by aztreonam, but not by clavulanic acid, tazobactam, sulbactam. Second generation cephalosporins are hydrolyzed by AmpC, but fourth generation cephalosporines are not hydrolyzed [6].
a.4) Carbapenemases. Excessive use of carbapenems (because of ESBLs spread) has lead to the emergence of carbapenemases.
Class A – NMC (non-metalloenzyme carbapenemases), which hydrolyze the imipenem, but are not inhibited by EDTA. They are represented by plasmid carbapenemases: KPC (Klebsiella pneumonia carbapenemase), GES (Guyana extended spectrum 1-12). Their substrate is represented by penicillins, cephalosporins, carbapenems. They are not inhibited by EDTA and they do not hydrolyze aztreonam, piperacillin or piperacillin/tazobactam. [7,8]
ms and cephalosporins of any generation, not the aztreonam. VIM is oftel present in P.aeruginosa strains and it is spreading rapidly around the globe [9, 10].
Class C – CMY, described in 2006 in a very aggressive Enterobacter aerogenes strain, but it was transmitted through a plasmid path to other Gram negative bacteria.
Class D – OXA-carbapenemases (OXA-2, OXA-10): sensitivity only to imipenem; this class represents a great therapeutic concern.
Nimish Patel et.all. showed in a study published in the 48th Annual Reunion of IDSA (2010) that the use of ertapenem in Pseudomonas aeruginosa infections has reduced the Pseudomonas aeruginosa resistance to imipenem. The authors believe that this is possible because of the reducing of ciprofloxacin use and the decrease of the activity of the ciprofloxacin – induced efflux pump, which is linked to imipenem resistance. Authors suggest that the decrease of ciprofloxacin use could control the nosocomial infections with Pseudomonas aeruginosa resistant to cabapenems.
Nonenzymatic acquired resistance
b.1) Efflux, including three mechanisms:
- MexA-MexB-OprM (membrane proteins which mediate the expression of a natural efflux); resistance to carboxypenicillins and aztreonam
- MexC-MexD-OprJ – resistance to cefepime and cefpirome
- MexE-MexF-OprN (mechanism often liked to D2 pores decrease) – resistance to imipenem
b.2) Lack of pores (D2 pores loss) – variable resistance to carbapenems
b.3) Penicillin-binding proteins (PBP) modification: PBP 2 or PBP 4 – resistance to imipenem; PBP 3 – resistance to all betalactamines, but not to imipenem [11,12,13].
A summary of this data is shown in Table 1.
Table 1.
P.aeruginosa betalactamines resistance phenotypes [11]
| Drug | Wild-type phenotype | Penicillinase | AmpC | BLSE | Carbapenemase | Efflux | D2 | |||||
| CTX-M | PER | A | B | D | OprM | OprJ | OprN | |||||
| Ticarcillin | S | R | R | R | R | R | R | R | R | S | S | S |
| Ticarcillin – clavulanic acid | S | S | R | S | S | R | R | R | R | S | S | S |
| Piperacillin | S | R | R | R | R | S | R | R | S | S | S | S |
| Piperacillin – tazobactam | S | S | R | S | S | S | R | R | S | S | S | S |
| Cefoperasone | S | R | R | R | R | R | R | R | S | S | S | S |
| Ceftazidime | S | S | R | R | R | R | R | R | S | S | S | S |
| Cefepime | S | S | S | S | R | R | R | R | S | R | S | S |
| Cefpirome | S | S | S | S | R | R | R | R | S | R | S | S |
| Aztreonam | S | S | S | S | R | S | S | R | R | S | S | S |
| Imipenem | S | S | S | S | S | R | R | S | S | S | R | R |
1.Pseudomonas aeruginosa aminoglycosides resistance phenotypes
The wild-type phenotype is sensitive to all aminoglycosides.
Acquired resistance is produced through many mechanisms: impermeability, efflux, enzymatic inactivation, respiratory mutants or different combination of these mechanisms.
Aminoglycoside resistance through impermeability is frequent at P.aeruginosa; enzymatic resistance is also frequent; three enzyme classes are responsible for this kind of resistance: aminozid-phosphotransferase (APH), aminozid-nucleotidyltransferase (ANT) and aminozid-acetyltransferase (AAC) [11,14].
P.aeruginosa resistance phenotypes are as follows:
- G: resistance to gentamicin; sensitive to tobramicin, netilmicin, amykacin, isepamicin
- GNt: resistance to gentamicin, netilmicin; sensitive to tobramicin, amykacin, isepamicin
- GT: resistance to gentamicin, tobramicin; sensitive to netilmicin, amykacin, isepamicin
- GNtT: resistance to gentamicin, tobramicin, netilmicin; sensitive to amykacin, isepamicin
- TNtA: resistance to tobramicin, netilmicin, amykacin; sensitive to gentamicin, isepamicin (?)
- GTNtA: resistance to gentamicin, tobramicin, netilmicin, amykacin; sensitive to isepamicin (?)
- impermeability phenotype: resistance to gentamicin, tobeamicin, netilmicin, amykacin, isepamicin
2.Pseudomonas aeruginosa quinolones resistance phenotypes
The wild type phenotype is sensitive in vitro to norfloxacin, pefloxacin, ofloxacin, ciprofloxacin, levofloxacin. In practice only ciprofloxacin is used.
Acquired resistance appears through different mechanisms:
Impermeability: pores and LPS (pores represent the main pathway for quinolones through the external membrane)
Target affinity modification: submits A and B of the DNA-gyrase and subunits C and D of the topoisomerase
Active efflux: OprM, OrpJ, OprN (low resistance)
P.aeruginosa resistance phenotypes are as follows:
- I (wild-type phenotype): sensitive to norfloxain, pefloxacin, ofloxacin/levofloxacin, ciprofloxacin
- II: sensitive to ciprofloxacin; intermediary to norfloxacin, pefloxacin, ofloxacin/levofloxacin
- III: sensitive to ciprofloxacin; resistant to norfloxacin, pefloxacin, ofloxacin/levofloxacin
- IV: resistant to ciprofloxacin, norfloxacin, pefloxacin, ofloxacin/levofloxacin
- efflux phenotype: resistant to norfloxacin, ciprofloxacin; sensitive to pefloxacin, ofloxacin/levofloxacin
3.Pseudomonas aeruginosa resistance to trimethoprim-sulfamethoxazole and to nitrofurantoin is intrinsic and is caused by decreased access to the target enzyme.
4.Pseudomonas aeruginosa resistance to colistin is very low, fact that made some authors believe that it is no longer appropriate to use carbapenems as first line therapy in infections with this germ. These authors propose as first line therapy in Pseudomonas aeruginosa infections the colistin, in association with other active antibiotics, in order to prevent the emergence of colistin – resistant strains.[15]
I.ESBLs highlighting tests
Phenotypic tests are the most used and consist of antibiotic susceptibility testing by Kirby Bauer method, using clinical break-points and cut-off values, recommended by EUCAST standard. [16]
Screening tests – resistance testing with cephalosporin and monobactam discs (ceftriaxone<25mm, cefotaxime<27mm, ceftazidime<22mm, cefpodoxime<17mm, aztreonam<27mm)
Confirmation tests – are based on the synergy between cephalosporins and clavulanic acid.
a.Double disk method (synergy test): a disc containing amoxicillin+clavulanic acid (20 μg+10 μg) is placed in the center of a Mueller-Hinton gelose medium plate and at about 20-35 mm from this central disc, other discs containing ceftazidime, ceftriaxone, cefotaxime are placed (champagne stopper method) (Fig. 1).
Fig.1.
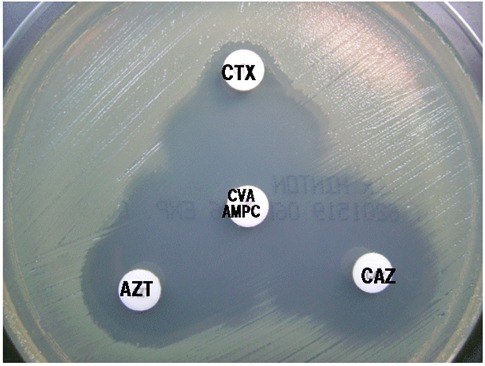
Double disc method
b.Combined disc method: consists in the comparison between the diameters of the inhibition areas of a cephalosporin disc and a cephalosporin+clavulanic acid disc (cefotaxime 30 μg or ceftazidime 30 μg). If the strain produces ESBLs, the inhibition area of the disc containing clavulanic acid is with at least 5 mm larger than the inhibition area of the disc without clavulanic acid. (Fig.2)
Fig.2.
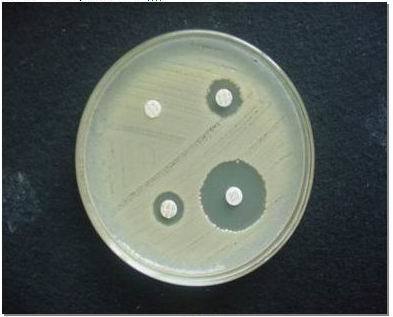
Combined disc method
c.E-test: uses two plastic strips which contain ceftazidime/cefotaxime on one half and ceftazidime/cefotaxime + clavulanic acid on the other half. (Fig.3)
Fig.3.
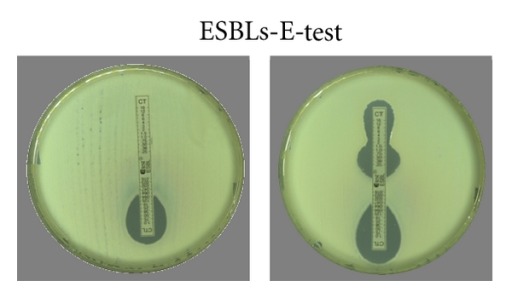
E-test
d.ESBL-AGAR medium: the plates contain on the left half MacConkey selective medium supplemented with ceftazidime 2 mg (red uncolonized) and on the right half Drigalski medium, supplemented with cefotaxime 1.5 mg (green uncolonized). This way resistance to ceftazidime and cefotaxime is tested simultaneously. Double detection increases the test sensitivity. (Fig.4)
Fig.4.

ESBL AGAR medium (Microgen Bioproducts LTD)
II.AmpC highlighting tests
The phenotypic non-standard screening method is represented by the cefoxitin sensitivity reduction. Another method uses AmpC-inhibiting enzymes, such as cloxacillin and boronic acid. These inhibitors are incorporated in the cefoxitin 30 μg or cefpodoxime 10 μg discs. The inhibition area growth around the disc with inhibitor is considered to be a positive test, confirming that the strain produces AmpC. [17,8, 9]
III.Carbapenemase highlighting tests
Screening tests: according to CLSI, a strain that produces carbapenemases presents at least 21 mm diameter to meropenem, imipenem or ertapenem. [18]
Confirmation tests
Chrom ID CARBA is a BioMerieux medium on which only carbapenemase-producing strains grow.
E-test is used for the determination of the minimum inhibitory concentration of the carbapenemase-producing strains.
Modified Hodge recommended by CLSI (Fig. 5). On a Mueller-Hinton plate inoculated with an E.coli ATCC 25922 reference strain, three strains are linearly seeded: K.pneumoniae ATCC BAA 1705 (carbapenemase +; 1 in Fig.5), K.pneumoniae ATCC BAA 1706 (carbapenemase -; 2 in Fig.5) and the isolated strain (3 in Fig.5). In the center of the plate the ertapenem disc is placed. [17,19]
Fig.5.
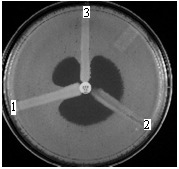
Modified Hodge
CARBA NP test. Is based on a color reaction produced by the medium acidification caused by the degradation of imipenem: phenol red indicator turns yellow. This method is 100% sensitive in comparison to molecular techniques and fast (in about 2 hours the result is ready and can be adapted to any laboratory). [20]
All of these confirmation tests determine the carbapenemase production without the carbapenemases classification. The following methods allow the carbapenemases classification.
Class A carbapenemases confirmation tests
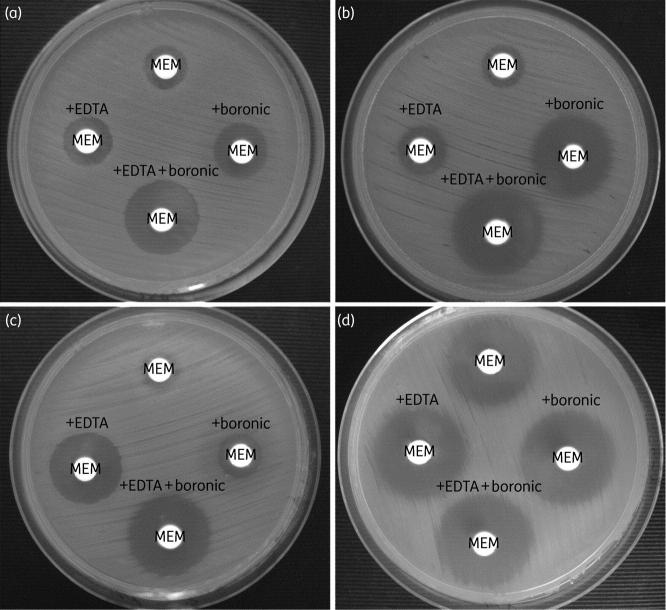
The method uses meropenem 10 μg discs and meropenem 10 μg + boronic acid 600 μg discs. A 5 mm or larger diameter between the meropenem disc and the meropenem+boronic acid disc proves that the strain produces excessive chromosomial or plasmid carbapenemase (Fig.6). [21]
Fig.6.
(a) P.aeruginosa strain with class A and class B carbapenemases production. (b) P.aeruginosa strain with class A carbapenemase production. (c) P.aeruginosa strain with class B carbapenemase production. (d) P.aeruginosa strain without carbapenemase production.
Class B carbapenemases confirmation tests
Metallo-betalactamases highlighting is based on the synergy between the metallo-betalactamases inhibitors (such as EDTA or dipicollinic acid) and the carbapenems (imipenem or meropenem). The MBL detection is based on the 5 mm or larger difference between the meropenem disc and the meropenem+EDTA disc (Fig.6). [22]
Class D carbapenemases confirmation tests
Chrom ID OXA-48 on the chromogenic medium produced by BioMerieux. First evaluation of Chrom ID-OXA media has been performed during the European Society of Clinical Microbiology and Infectious Diseases meeting in Berlin, held between the 27th and the 30th of April 2013. The carbapenemase detection on chromogenic media was compared to the PCR molecular method for carbapenemase detection, the correspondence being 98% (Fig.7). [23, 24,25]
Fig.7.
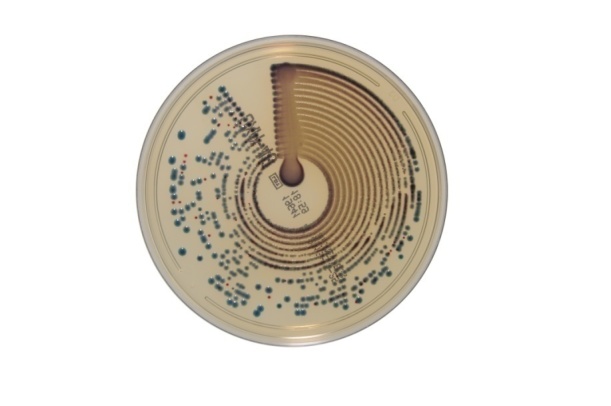
Chrom ID OXA-48 medium (BioMerieux)
Conclusions
Pseudomonas aeruginosa is one of the most resistant bacteria to antibiotics, therefore acknowledging the resistance phenotypes is very useful to the clinician in applying the targeted therapy and the successful resolution of the infection.
Microbiology Laboratory plays an important role in finding the most eloquent methods for detecting multidrug-resistance mechanisms of the bacteria, therefore the tight collaboration between the laboratory and the clinician is essential for the patient first, but also for the clinician and the microbiologist.
References
- 1.Barlow G., Nathwani D. Is Antibiotic Resistance a Problem? A Practical Guide for Hospital Clinicians. Postgraduate Medical Journal. 2005;81(April):680–692. doi: 10.1136/pgmj.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Nosocomial Inflections Surveillance System (NNISS) CDC definitions for nosocomial infections. 2004 [Google Scholar]
- 3.Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. Emergence of antibiotic resistant Pseudomonas aeruginosa: comparison of risks associted with different antipseudomonadal agents. Amtimicrob. Agents Chemother. 1999;43(6):1379–1382. doi: 10.1128/aac.43.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas ME, Karageorgopoulos DE. Extended-spectrum beta-lactamase-producing organisms. Journal of Hospital infection. 2009;73:345–354. doi: 10.1016/j.jhin.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin microbiol and Infect. 2008;14:90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Ronco E, Naas T, Nordmann P. Extended-spectrum ß-lactamase TEM-4 in Pseudomonas aeruginosa. Clin Microbiol Infect. 1999;5:651–652. doi: 10.1111/j.1469-0691.1999.tb00425.x. [DOI] [PubMed] [Google Scholar]
- 7.Carmeli Y. The Role of Carbapenems: The Predictive Factors for Multi-Drug Resistant Gram-Negatives. http://www.invanz.co.il 2006 [Google Scholar]
- 8.Studemeister AE, Quinn JP. Selective imipenem resistance in Pseudomonas aeruginosa associated with diminished outer membrane permeability. Antimicrob Agents Chemother. 1988;32:1267–1268. doi: 10.1128/aac.32.8.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picao RC, Andrade SS, Nicoletti AG, et al. Metallo-ß-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP, GIM, SIM, SPM, or VIM producing organisms. Clin Microbiol. 2008;46:2028–2037. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh TR, Weekers J, Livermore DM, Toleman MH. Dissemination of NDM-1-positive bacteria in the New Delhi environment and its implications for human health:an enveronmental point prevalence study. Lancet Infect.Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 11.Francois Jehl, Monique Chomarat, Michele Weber, et al. De la antibiograma la prescriptie. Bucuresti: Ed. Orizonturi; 2003. Antibioticele: clasificare, spectru si mecanisme de actiune. pp. 12–30. [Google Scholar]
- 12.Nobuhisa Masuda, Eiko Sakagawa, Satoshi Ohya, et al. Substrate Specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM Efflux Pumps in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2000;44(12):3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole K, Gotoh N., Tsujimoto H., et al. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 14.Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin AS, Barone AA, Penco J, et al. Intravenous Colistin as Therpy for Nosocomial Infections Caused by Multidrug-Resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clinical Infections Deseases. 1999;28(5):1008–1011. doi: 10.1086/514732. [DOI] [PubMed] [Google Scholar]
- 16.EUCAST. Clinical breakpoints – bacteria, version 3.1. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_3.1.pdf. 2013 [Google Scholar]
- 17.Clinical and Laboratory Standards Institute (CLSI) . Performance Standards for Antimicrobial Disk Susceptibility Tests. CLSI Twentieth Informational Supplement, M100-S20. 2010 [Google Scholar]
- 18.Jacoby GA, Walsh KE, Walker VJ. Identification of Extended-Spectrum, AmpC, and Carbapenem-Hydrolyzing beta-Lactamases in Escherichia coli and Klebsiella pneumoniae by Disk Tests. Journal of Clinical Microbiology. 2006;44:1971–1976. doi: 10.1128/JCM.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC . Modified Hodge Test for Carbapenemase Detection in Enterobacteriaceae . http://www.ndhealth.gov/microlab/Uploads/HodgeTest.pdf. 2010 [Google Scholar]
- 20.Nordman P., Poirel L., Dortet L. Rapid Detection of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasteran F., Mendez T., Guerriero L. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J.Clin.Microbiol. 2009;47:1631–1639. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miriagou V, Cornaglia G, Edelstein M, et al. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clinical Microbiology and Infection. 2010;16:112–122. doi: 10.1111/j.1469-0691.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- 23.Devigne L, . Pantel. A., Bourguignon M.P., Celier M., Vignon V., Zambardi G., Lavigne J.P. First evaluation of chromID OXA-48, a new chromogenic medium for detection of E nterobacteriaceae producing OXA-48 carbapenemase. ECCMID; 2013 Apr 27-30 ; Berlin, Germany. [Google Scholar]
- 24.Girlch D, Anglade C, Zambardi G, Nordmann P. Comparative evaluation of a novel chromogenic medium(chrom ID OXA-48) for detection of OXA-48 producing Enterobacteriaceae. Diagn. Microbiol.Infect.Dis. 2013 Dec.77(4):296–230. doi: 10.1016/j.diagmicrobio.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 25.WHO. WHONET Software. http://www.who.int/drugresistance/whonetsoftware/en/ 2007 [Google Scholar]