SUMMARY
Using a functional approach to investigate the epigenetics of Type 2 Diabetes (T2D), we combine three lines of evidence – diet-induced epigenetic dysregulation in mouse, epigenetic conservation in humans, and T2D clinical risk evidence – to identify genes implicated in T2D pathogenesis through epigenetic mechanisms related to obesity. Beginning with dietary manipulation of genetically homogeneous mice, we identify differentially DNA-methylated genomic regions. We then replicate these results in adipose samples from lean and obese patients pre- and post-Roux-en-Y gastric bypass, identifying regions where both the location and direction of methylation change is conserved. These regions overlap with 27 genetic T2D risk loci, only one of which was deemed significant by GWAS alone. Functional analysis of genes associated with these regions revealed four genes with roles in insulin resistance, demonstrating the potential general utility of this approach for complementing conventional human genetic studies by integrating cross-species epigenomics and clinical genetic risk.
INTRODUCTION
Type 2 diabetes mellitus (T2D) is a metabolic disorder with a rapidly increasing worldwide prevalence. T2D affects 300 million adults worldwide and that number is predicted to rise to above 430 million by 2030 (Chen et al., 2012). Although T2D has a significant genetic risk component, as determined genome-wide association studies (McCarthy, 2010), the heritability estimate is only 21% when looking across all age groups (Almgren et al., 2011). These low heritability estimates, coupled with the rapid increase in worldwide prevalence, suggests a strong role for environmental risk factors. As an example, recent work on the efficacy of Roux-en-Y gastric bypass (RYGB) as a treatment for obesity has found that this procedure can have a profound positive effect on T2D-related metabolic indicators (Mingrone et al., 2012).
Epigenetics, the study of non-DNA sequence based information that is replicated during cell division, such as DNA methylation, has been suggested as a natural integrator of genetic susceptibility and environmental exposure in common disease (Bjornsson et al., 2004). Epigenetics has also attracted considerable scientific and lay attention due to its dynamic nature, association with common disease (Cui et al., 2003), and reversibility under targeted therapies (Sharma et al., 2010).
Most common human diseases are explained to a very limited degree by known individual common genetic variants, with ~3.4% of risk profile score explained for psychiatric disorders like schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics, 2014), and ~10.7% for T2D (Consortium et al., 2013). This combination of limited genetic causality, environmental influence and persistence over long time periods suggests a likely role for epigenetics in common human disease. However, epigenetic studies have their own limitations, including the need in most cases to use cells appropriate to the disease under study, confounding effects such as age, and the often considerable difficulty in designing replication sets, which are much easier in purely genetic studies because of the universality of the sample type (DNA from blood). A number of methodologies have been developed by our and other groups to adjust for cell type composition, confounding variables, and replication studies (which are typically much smaller) (Houseman et al., 2012; Liu et al., 2013; Montano et al., 2013).
There have been limited epigenetic analyses of T2D and associated metabolic traits thus far. Studies of pancreatic islets have found methylation differences between T2D patients and non-diabetic controls (Dayeh et al., 2014). Similar changes have also been found in peripheral blood leukocytes from obese humans early after RYGB (Kirchner et al., 2014). Two studies examining DNA methylation related to exercise and T2D status found epigenetic changes overlapping the TCF7L2 locus (Ronn et al., 2013) (Toperoff et al., 2012). Finally, one study that examined methylation in skeletal muscle from obese and lean subjects at 14 individually-selected loci found that methylation in obese subjects reverted to lean methylation levels after RYGB (Barres et al., 2013).
Here, we established an approach utilizing two species to identify candidate genes involved in obesity and T2D through epigenetic mechanisms. We first examined the epigenetic consequences of a high-fat diet in a carefully controlled experimental mouse obesity setting. We then replicated across species—in humans—by analyzing adipose tissue from a cohort that both reproduces and reverses a phenotype similar to the obese mouse. The use of samples from the same subjects pre- and post-RYGB allows a human isogenic comparison of the effect of obesity-induced metabolic disturbances. This cross-species approach exploits the power of evolutionary selection, whose mechanisms have survived the 50 million year separation between mouse and human, in a more comprehensive manner than simple replication from human set to human set, and may better identify functionally important environmental targets. We lastly stratified these cross-species obesity-associated regions using genetic association data from a large genome-wide association study (GWAS) for T2D to more directly link our obesity-derived phenotypes with human T2D. As a result of this approach, we are able to identify four genes with roles in insulin resistance, suggesting that this cross-species approach provides a powerful experimental system for identifying the genomic variation associated with common disease.
RESULTS
Alterations in DNA methylation in mouse adipocytes produced by high-fat diet
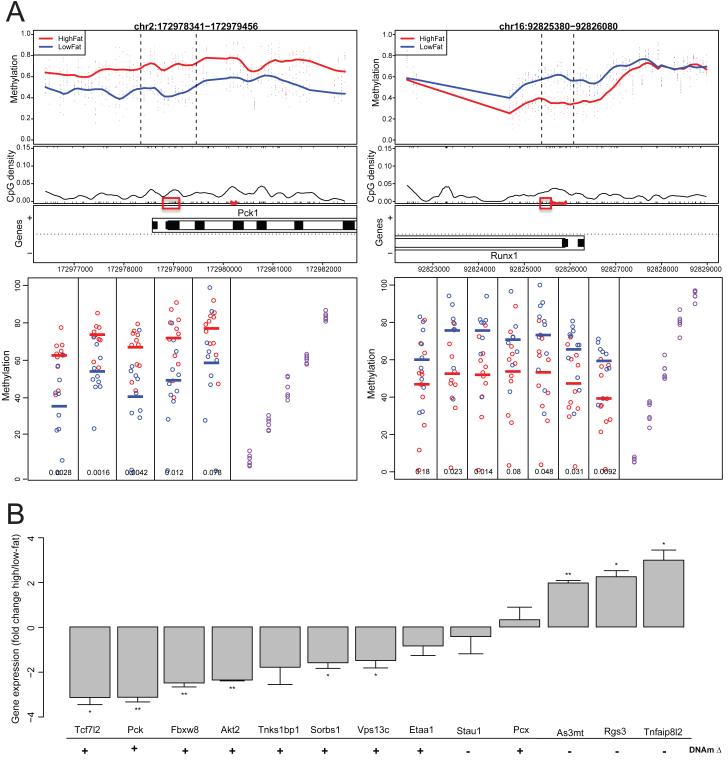
To detect DNA methylation differences, we used the Comprehensive High-throughput Array-based Relative Methylation (CHARM) method that in its current form can assay over 5 million CpG sites in mouse and 7.5 million CpG sites in human. In 12 adipocyte samples extracted from mouse adipose tissue, we find 232 differentially methylated regions (DMRs) correlated with diet status (Table 1). As an example, when comparing adipocytes from high-fat-fed mice versus low-fat-fed mice, we found hypermethylation overlying the promoter of phosphoenolpyruvate carboxykinase 1 (Pck1, Figure 1A). PEPCK, the product of Pck1, catalyzes a rate-limiting step in gluconeogenesis, is essential for lipid metabolism in adipose tissue, is known to be regulated by insulin, and has been linked to lipodystrophy and obesity in mice (Beale et al., 2004).
Table 1.
Genome-wide significant mouse DMRs
| Tissue | Analysis | Q-val < 0.05 | Q-val < 0.1 |
|---|---|---|---|
|
| |||
| Diet | 232 | 448 | |
| Weight | 183 | 288 | |
| Adipocytes | Fasting Glucose | 235 | 571 |
| GTT | 0 | 3 | |
| ITT | 294 | 419 | |
Q-values generated based upon comparison of observed DMR areas to areas generated by 1000 random permutations of phenotype/methylation associations. See also Table S1 for a full list of all mouse DMRs.
Figure 1. Genome-wide significant methylation changes related to diet-induced obesity in C57BL/6 mice.
(A) Two genome-wide significant DMRs are hypermethylated in adipocytes purified from mice raised on a high-fat diet. Each point represents the methylation level in adipocytes from an individual mouse at a specific probe, with smoothed lines representing group methylation averages. These points are colored blue for lean mice and red for obese mice. (B) Body weight (grams) and glucose tolerance (AUC) are associated with methylation in adipocytes at genome-wide significant levels. Each point in the top panels represents one probe, with the y-axis representing the Pearson correlation coefficients of the probes with the analyzed phenotype. Dotted lines represent the extent of the DMR as generated automatically via CHARM. The bottom panels display gene location information for the chromosomal coordinates on the x-axis.
In addition to the high-fat versus low-fat analysis, even more DMRs were detected when analyzing methylation differences related to the metabolic phenotypes of body weight, fasting glucose, and insulin and glucose tolerance test area-under-curve (ITT/GTT AUC) values (Table 1, Table S1). One example of a mouse GTT-associated DMR is in the Fasn gene, which produces fatty acid synthase. Most DMRs found were significantly associated with more than one trait, which is not entirely unexpected as the phenotypes themselves are highly correlated (Figure S1).
We additionally examined DNA methylation in pancreatic islets purified from whole mouse pancreata and hepatocytes extracted from mouse liver tissue. We found significant correlations between methylation and mouse diet and weight in pancreatic islets, and correlations between methylation and weight and ITT in hepatocytes (Table S1).
Pooling tissues together and surveying for DNA methylation changes in common across tissues yielded no significant results.
Gene ontology for mouse DMRs
We implemented gene set analyses to assess the overall biological importance of the DNA methylation changes we observed in mouse adipocytes. The genome-wide significant adipocyte DMRs were near genes that were significantly overrepresented in lipid metabolic and immune/inflammatory pathways compared to the background list of genes represented on our array, with enrichment Q-values < 9.7*10−3 (Table S2). Examining hyper- and hypo-methylated DMRs separately in high-fat-fed obese mice, we observed that the metabolic pathway enrichment was derived from genes near hypermethylated DMRs, while the inflammatory pathway enrichment was present mainly in genes near hypomethylated DMRs.
Inflammatory and immune related systems are known to be upregulated in adipocytes specifically in both obesity and T2D (Hotamisligil, 2010). Similarly, recent work has shown adipose de novo lipogenesis downregulation associated with metabolic dysfunction (Roberts et al., 2009). These pathways, however, have not previously been shown to be significantly associated with methylation changes in a diet-induced obesity phenotype.
Methylation replication in mice and associated gene expression studies
We then tested for replication of the methylation results at nine DMRs in adipocytes and three DMRs in pancreatic islets in an independent set of 18 mice (Figure 2A, Table S3). The 625 genome-wide significant adipocyte DMRs have FDR Q-values ranging from 0.004 to 0.05. In order to determine whether our results would replicate throughout this range, we examined a subset of DMRs with levels of statistical significance that spanned from the most significant to just below the 0.05 cutoff. Mice used in the replication set were also reared on a high-fat diet but were separate from those used for CHARM. Nine mouse adipocyte DMRs were assayed by bisulfite pyrosequencing. Eight of these regions had at least one CpG showing significant differential methylation in the same direction as detected by CHARM.
Figure 2. Replication of mouse methylation changes in additional mice, and associated gene expression changes.
(A) Methylation changes observed after CHARM analysis at two genome-wide significant DMRs are replicated using bisulfite pyrosequencing. Red boxes indicate CpGs assayed in pyrosequencing. For the lower pyrosequencing plots, the y-axis represents methylation, and individual CpGs are plotted along the x-axis. Purple dots represent control DNA artificially methylated to have 0, 25, 50, 75 and 100% methylation. (B) Gene expression changes for genes near genome-wide significant mouse adipocyte DMRs. RNA levels were normalized to same-sample 18S RNA measurements and are displayed as [CT (high-fat samples) – CT (low-fat samples)]2. Error bars represent standard error of the CT differences between groups. * p<0.05, ** p<0.005, *** p<0.0005. The direction of the genome-wide significant CHARM DMR closest to the gene is denoted below the gene names; + and − represent regions hyper- or hypomethylated in the high-fat samples, respectively. See also Figure S2 for whole-genome gene expression correlations, and Tables S4 and S5 for pyrosequencing and tissue purification, respectively.
Although these were fractionated cells under investigation, to further ensure that the results were not due to cell-type shifts in the high-fat-fed obese mice resulting from the infiltration of immune cells into adipose tissue, we used quantitative PCR to characterize the expression of multiple macrophage- and adipocyte-specific markers in our purified adipocyte samples from low-fat-fed and high-fat-fed mice. We saw no significant change in the levels of expression of the macrophage (inflammatory) markers F4/80, Cd14, or Cd68, and we did see the expected obesity-related within-adipocyte changes of the adipocyte markers AdipoQ and Ccl2 (Table S4).
To examine whether these methylation changes between high-fat- and low-fat-fed mice involved changes in the expression of nearby genes, we used quantitative PCR (qPCR) to examine the expression of thirteen genes near genome-wide significant DMRs (Figure 2B). We used qPCR to examine mRNA from the same adipocytes and mice that were analyzed by CHARM. Of the thirteen genes examined, nine showed significant changes in mRNA expression in the opposite direction as methylation changes (Figure 2B).
Furthermore, we assessed whether these DNA methylation changes correlated with previously published genome-wide gene expression data in a similar cohort (Xu et al., 2003). We saw significant inverse correlations between diet-related methylation changes and diet-related gene expression changes (Figures S2A and S2B). These results compare favorably to other functional analyses of discovered DMRs (Kim et al., 2010). Taken together, these data show that we find robustly significant DMRs in mice that correlate with metabolic traits, that these DMRs replicate in separate animals, and that methylation at many of these regions appears to have a functional effect on gene expression.
Mouse DMRs replicated evolutionarily in human adipose tissue
We reasoned that many functionally relevant DMRs in mice exposed to a high-fat diet serve an important metabolic function that would be conserved across species and often susceptible to similar environmental cues. Therefore, to determine whether the methylation changes observed in mouse adipocytes could be replicated in an evolutionarily divergent cohort, we performed CHARM analysis on human subcutaneous adipose tissues from 7 lean subjects and 14 obese sex-matched insulin resistant subjects of the same age range, as well as 8 obese subjects post-RYGB.
We first examined the replication of mouse adipocyte DMRs in human adipose tissue from obese versus lean. We observed very strong overlap between DMRs in human obese versus lean tissue and DMRs in high-fat-fed versus low-fat-fed mouse adipocytes (all p<10−15, Figure S3A, rightmost five bars), showing that there is a strong correlation between areas that are regulated by methylation in metabolic dysfunction in both mice and humans.
Next, in order to determine which mouse methylation changes would replicate in human, we determined that out of a total of 625 genome-wide significant mouse adipocyte DMRs, 576 had homologous regions on the human genome (hg19), calculated via the liftOver UCSC tool (Hinrichs et al., 2006), and 497 had human CHARM probes within 5kb. This is a remarkably high fraction (86.3%), suggesting that our assay method, CHARM, is highly comprehensive, and also that the location of CpG regions is strongly conserved in evolution. Of the 497 conserved DMRs, 249 (50.3%) showed significant differential methylation (p<0.05) between obese and lean people (Table S5). These numbers were similar when analyzing differential methylation before and after RYGB surgery (227 out of 497). As a final restrictive step in using human methylation to validate our mouse results, we determined that 170 (68%) of these regions had a consistent direction of methylation change between high-fat-fed obese mice and obese humans, such that if a particular region had higher methylation in high-fat-fed mice, that region would also have higher methylation in obese humans and vice versa.
When more restrictive human methylation significance cutoffs are used, the percentage of regions with consistent directionality (true positive rate) rises, but the total number of retained regions drops, with 67/77 (87%) directionally consistent at human obesity P-values <0.005, and 25/25 (100%) consistent at P-values < 0.0005 (Figure S3B). All 170 directionally conserved regions were associated with the metabolic phenotypes of fasting glucose, GTT, and/or ITT in addition to mouse diet status. Furthermore, 134 of these regions had a consistent effect directionality between obesity- and RYGB surgery-related methylation (e.g. higher in obesity and pre-surgery and vice versa), and a further 105 had post-surgery methylation values that were in between lean and pre-surgery methylation values, i.e., regions where methylation in obese subjects appeared to revert towards a lean phenotype after surgery (enrichment p=2.8x10−3).
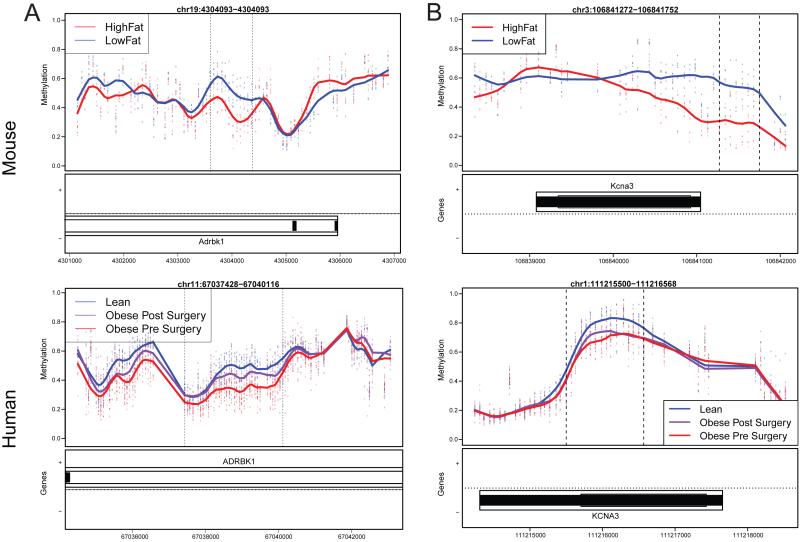
In Figure 3, we present two regions that have significant methylation changes in human adipose tissue, are in homologous regions of the genome as mouse DMRs, are directionally consistent with the mouse DMRs, and have human post-surgery methylation levels that have moved closer to the lean phenotype. These regions are over two genes; ADRBK1 (adrenergic, beta, receptor kinase 1, Figure 3A), and KCNA3 (potassium voltage-gated channel, shaker-related subfamily, member 3, Figure 3B).
Figure 3. Overlapping methylation changes in human and mouse adipose tissue.
Two genome-wide significant DMRs found in mouse adipocytes over Adrbk1 (A, top) and Kcna3 (B, top) are shown along with the corresponding methylation changes in human adipose tissue in (A, bottom), and (B, bottom). For the panels denoting methylation, each point represents the methylation level from an individual mouse or human at a specific genomic location, with smoothed lines representing group methylation averages. Y-axis – methylation values. Below each methylation plot is a panel showing genomic coordinates for the respective species and any genes at those coordinates. See also Figure S3 for tissue and species overlaps, and Tables S6 and S7 for conserved adipose mouse DMRs in human and for enrichment between DIAGRAM and conserved DMRs, respectively.
We also assessed whether the human adipose DNA methylation changes correlated with previously published human genome-wide gene expression data from obese and lean individuals (Arner et al., 2012). As with our mouse data, we saw a highly significant inverse correlation between obesity-related methylation changes and obesity-related gene expression changes (Figure S2A and S2B, right panels).
We performed a similar mouse-human comparison in pancreatic islets using published DNAm data from T2D and control subjects (Dayeh et al., 2014), showing that 67% (odds ratio= 7.2, p=7.2x10−6) of the mouse pancreatic islet DMRs that replicated in the human data had methylation change in the same direction, and that these probes were far more associated with human T2D status than the rest of the probes on the array (p = 1.18x10−9, Figure S3C), demonstrating that our mouse-derived islet DMRs are enriched for potential epigenetic alteration in human T2D. Finally, we also validated multiple mouse hepatocyte DMRs in human liver tissue, with 62.5% replicating (Table S3).
Genetic risk loci association with overlapping regions of human and mouse methylation changes
We incorporated data from human GWAS for T2D using two complementary approaches that allow further characterization of our candidate obesity-related DMRs. GWAS summary statistics were obtained from the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) T2D genome-wide association meta-analysis, comprising data from 12 separate GWAS studies totaling 12,171 T2D cases and 56,682 controls (diagram-consortium.org). We first directly explored the association between genes with obesity-related DMRs and genes conferring clinical genetic risk for T2D by calculating statistical enrichment of the GWAS regions overlapping our DMRs. We found marginally significant enrichment for adipose DMRs among at least marginally significant GWAS signals (GWAS p-value cutoffs starting with p<10−6, corresponding to enrichment p-values ranging from 0.0048 to 0.0165, Table S6). Given the small number of directly overlapping regions, these results are likely strongly influenced by the strength of the TCF7L2 signal. While much of the early literature on TCF7L2 focused on its role in pancreatic islets, there is growing evidence that extra-pancreatic effects may contribute to the T2D phenotype at this locus (Nilsson et al., 2014).
We further examined statistical enrichment in the context of regulatory networks involving genes implicated in GWAS. Genes at 23 genome-wide significant GWAS signals (usually the gene nearest to the lead SNP) were directly (one-step) connected to genes near DMRs either by transcriptional control or direct protein-protein interaction (Figure 4A). This amount of interaction represents significantly more than expected by random chance (p = 0.0206) (Figure S4), and demonstrates how genes implicated by methylation appear to be acting in the same pathways as genes implicated by GWAS. Similarly, expanding beyond one-step connections, many of the 30 regions implicated by both methylation data and GWAS are connected to genes identified by the mouse-only and human-mouse analyses and act in the same pathways (Figure 4B).
Figure 4. Diagrammatic representation of the interactions between epigenetically conserved and genetically-associated genes implicated in this study.
Generated using QIAGEN’s Ingenuity IPA (Ingenuity® Systems, www.ingenuity.com), these diagrams represent the connections between genes implicated in our analyses. A) Genes with genome-wide significant linkage to T2D in the DIAGRAM meta-analysis were connected to genes near directionally conserved cross-species DMRs. Genes with no connections were dropped. B) Starting with a set of 23 genes near T2D-associated directionally conserved cross-species DMRs, this network was grown by adding genes near species-conserved and mouse-only genome-wide significant DMRs in order to represent one potential regulatory network. Gene colors explained in within-figure legend. See also Figure S4 for the permutation analysis of the enrichment of interactions in Figure 4A.
Given these results, we sought to further filter our obesity-related DMRs down to the subset of genes likely associated with T2D. We hypothesize that DMRs that overlap associated marker SNPs for T2D can identify genes with epigenetic mechanisms of risk in adipose tissue. As many of the DMRs overlapping GWAS T2D loci with low p-values implicate genes already known to be involved in T2D, obesity and related phenotypes, we therefore selected the subset of DMRs within genetic loci that had at least marginal statistical association with T2D clinical risk.
This approach reduced the 170 regions of directionally consistent and evolutionarily conserved methylation change in adipose tissue using the SNP-level summary statistics of the DIAGRAM analysis. In all, 30 cross-species and directionally conserved adipose DMRs directly overlapped with 27 marker SNPs (or close proxies with linkage disequilibrium > 0.8) that had some evidence of association with T2D (at least p<0.01, Table 2; see Methods). We also identified ten regions where conserved pancreatic islet DMRs overlap with DIAGRAM SNPs (Table S7).
Table 2.
Mouse-human DMRs with genetic T2D risk loci association
| Gene Name | Relative location of DMR |
Distance to TSS |
RYGB Reversion |
DIAGRAM P-value |
|---|---|---|---|---|
| Tcf7l2 | inside intron | 43058 | − | 4.90E-68 |
| Tcf7l2 | inside intron | 77345 | − | 4.90E-68 |
| As3mt | overlaps 5' | 0 | + | 9.60E-06 |
| Etaa1 | inside intron | 618 | + | 4.70E-05 |
| Tnfsf8 | overlaps 5' | 0 | − | 0.00029 |
| Plekho1 | overlaps exon | 4965 | + | 0.00045 |
| Tnfaip8l2 | inside intron | 337 | + | 0.00045 |
| Akt2 | inside intron | 20427 | − | 0.00049 |
| DIAGRAM GWAS 0.001 cutoff | ||||
| Lhfpl2 | inside intron | 2490 | + | 0.001 |
| Mkl1 | overlaps 5' | 0 | + | 0.0014 |
| BC048644 (Car5a) | overlaps exon | 146 | + | 0.0015 |
| Rgs3 | downstream | 108842 | + | 0.0019 |
| Fgd3 | inside intron | 11100 | + | 0.002 |
| Stau1 | overlaps 5' | 0 | + | 0.0022 |
| Tmcc3 | inside intron | 43772 | + | 0.0025 |
| Tbx3 | inside exon | 12714 | − | 0.0029 |
| Gstz1 | inside intron | 10332 | + | 0.0029 |
| Taok3 | inside intron | 549 | + | 0.0036 |
| Bnip3 | inside intron | 1863 | − | 0.0039 |
| Dlst | overlaps 5' | 0 | + | 0.0053 |
| Kcna3 | close to 3' | 2192 | + | 0.0064 |
| Cln8 | inside intron | 3055 | + | 0.0065 |
| Cd37 | exon | 2687 | + | 0.0069 |
| Nfib | inside intron | 100380 | − | 0.0071 |
| Pck1 | promoter | 453 | + | 0.0072 |
| Pck1 | overlaps 5' | 0 | + | 0.0072 |
| Pcx | inside intron | 59049 | + | 0.0073 |
| Hoxd3 | inside intron | 7307 | + | 0.0084 |
| Cd33 | overlaps 5' | 0 | + | 0.0087 |
| Evl | exon | 157 | + | 0.0099 |
Shown are the names of the nearest gene to the mouse and human differential methylation, the position of the DMR relative to the gene, the distance to the transcriptional start site (TSS), whether the direction of methylation change (sign of smoothed effect statistic) post-RYGB surgery reverts toward lean subject methylation levels (RYGB Reversion), and the p-value of the T2D genetic association in the region. See also Table S7 for an analogous table with the pancreatic islet results instead and Table S8 for conserved adipose DMRs that overlap with adipose enhancers.
In these final 30 regions, not only have we connected methylation change to obesity-induced metabolic phenotypes across two species, but the association with T2D-associated SNPs also provides a candidate mechanism for the methylation changes observed in human obesity and RYGB surgery. These 27 identified SNPs could potentially explain up to 2.69% of genetic T2D liability, though only one of these loci reached genome-wide significance in DIAGRAM (Morris et al., 2012). Even excluding this GWAS-positive loci (TCF7L2), which explains 1.12% of the variance alone, the remaining regions could explain up to 1.57% of genetic variance in T2D susceptibility. These data suggest that for at least some of these loci, genetic variation underlies changes in methylation that are causal for T2D risk. It is also possible that these regions are also susceptible to environmental factors that influence local methylation and that they therefore serve to integrate genetic and epigenetic effects.
Note that this filtering-based approach is independent of assessing the statistical enrichment of T2D GWAS signal, either at SNP- or gene-level, within our cross-species obesity-associated DMRs, an approach commonly used with GWAS summary statistic data. This approach therefore does not diminish the potential function of genes with GWAS-positive statistical association for T2D or our DMRs that do not overlap with GWAS-associated SNPs for contributing epigenetically to obesity.
We hypothesized that one mechanism by which DNA methylation and genetic variation contribute to T2D risk may involve enhancer activity. Using publicly available human enhancer maps in 86 independent cell and tissue types (Hnisz et al., 2013), we found that a striking proportion of DMRs mapped to adipose nuclei enhancers and super-enhancers (which had the largest degree of overlap across all cell types). While the background proportion of overlap for CHARM was 17.2% for adipose enhancers and 3.8% for super enhancers, 40.6% (69 overlaps, p = 1.58x10−15) and 14.7% (25 overlaps, p=5.72x10−13) of the directionally consistent 170 regions, and 53.3% (16 overlaps, p=5.65x10−7) and 20% (6 overlaps, p=3.24x10−5) of the further 30 GWAS-associated regions above lie in adipose enhancers and super enhancers, respectively (Table S8). Thus, a major mechanism for methylation-mediated metabolic dysfunction is likely through epigenetic modification of enhancers. Note that most of these enhancers were not previously known to be related to T2D through conventional GWAS or other methods.
Functional analysis of genes implicated by cross-species methylation
In order to establish that our cross-species method can identify functional genes implicated in obesity, insulin resistance, T2D, and related research, we functionally assayed five genes. We selected genes with no prior association with metabolic phenotypes and that had methylation reversion after RYGB. As RYGB is a targeted, environmental therapy that improves multiple deleterious phenotypes including insulin sensitivity, we hypothesized that this subset of our results would be the most likely to have an effect on T2D- and obesity-related phenotypes. We then examined the physiological effect of altering the expression of these genes on adipocyte cell culture models using insulin-stimulated glucose uptake assays. This procedure can measure the responsiveness of adipocytes to insulin, a phenotype disrupted in obesity. We assayed seven 3T3-L1 adipocyte cell lines, each stably expressing shRNAs or expression plasmids corresponding to one of the five selected genes or a suitable control. In order to mimic the effects of a high-fat diet, genes hypermethylated in high-fat adipocytes were knocked down, and genes hypomethylated were overexpressed. Significant changes in glucose uptake were found for four of these five (Figure 5B). Potential roles for all of these genes in modulating insulin sensitivity and resistance are considered in the Discussion.
Figure 5. Overexpression and shRNA-mediated knock down of selected genes in 3T3-L1 adipocytes.
Selected genes from the set of 30 species conserved and T2D-SNP overlapping adipose DMRs were either stably overexpressed (A) or knocked down with shRNA (B). Glucose uptake is plotted as fold difference from normal, and significance was determined by two-way ANOVA modified by Bonferroni correction denoted as follows: * p<0.05, ** p<0.01, *** p<0.001. (C) DNA methylation and gene expression levels for high-fat-fed mice and obese human versus low-fat-fed mice and lean humans (e.g., “↓“ indicates hypomethylation / lower gene expression in high-fat-fed and obese compared to low-fat-fed and lean). Bold arrows indicate significant changes.
DISCUSSION
In mouse, we identified 625 genome-wide significant differentially methylated regions (DMRs) that correlate with diet-induced obesity phenotypes in adipocytes. Of these regions, 249 had significant conserved methylation changes in human obesity, and 170 of these had the same direction of methylation change in both species. Thirty of these DMRs also overlapped with SNPs or nearby proxies that have been associated with human T2D genetic risk. These data show for the first time that DNA methylation changes in metabolic disease are conserved across species and that this conservation overlaps genomic regions where genetic polymorphisms have been associated with T2D. Our approach combines three lines of evidence – epigenetic dysregulation following high fat diet in mouse, epigenetic directional consistency in humans, and some evidence for clinical risk of T2D – to identify genes likely functionally implicated in the pathogenesis of T2D specifically through epigenetic mechanisms related to obesity.
In the present study, while we use nominal P-value significance to identify human methylation and GWAS results, we first perform a multiple comparison correction in our initial set of mouse DMRs using a false discovery rate algorithm. As there is a growing awareness that the cumulative effect of common SNPs with low minor-allele frequency scores potentially explain large amounts of phenotypic variability beyond that of genome-wide significant SNPs identifiable by GWAS (Yang et al., 2010), approaches like ours that can use alternative methods to identify significant areas of potential genetic risk are necessary. The unique SNPs in these regions potentially account for 2.76% of T2D genetic variance, almost half of which is known by purely genetic analysis and may be epigenetically mediated.
We observed significant changes associated with four out of five genes assayed by insulin-stimulated glucose uptake assay, a common indicator of insulin resistance. Screens using this assay and performed on sample sets not enriched for genes in gluco-insulinemic pathways have found a far smaller percentage of genes that will alter glucose uptake (~10%) (Tang et al., 2006), indicating that our method can successfully select potential targets with a much higher than random probability of affecting insulin sensitivity.
Three of the genes that we found had altered glucose uptake fell into the classical inverse methylation-gene expression correlation: Mkl1, Plekho1 and Tnfaip8l2 were all hypomethylated in high-fat-fed mice and obese humans, had increased gene expression in corresponding subjects, and, when these genes were overexpressed in cell culture adipocytes, exhibited decreased glucose uptake in response to insulin, which would fit with the increased insulin resistance commonly observed in obesity and diabetes. While none of these genes have previously published roles in insulin resistance, several have suggestive links to metabolic phenotypes. Mkl1 is known to be a transcriptional coactivator of serum response factor (SRF), which been associated with insulin resistance in skeletal muscle (Jin et al., 2011). Similarly, PLEKHO1 has recently been shown to inhibit AKT/PI3K signaling (Zhang et al., 2014), a pathway known to be involved in insulin signaling. With regards to the direction of glucose uptake change, we note that insulin signaling induces both positive and negative feedback within affected cells (Gual et al., 2005), and without a methylation-gene expression candidate mechanism it is not possible to determine which feedback loop the methylation changes are involved with.
It is worth noting that as these genes did not contain common variants that passed the genome-wide significant GWAS threshold, they would not have been identified by GWAS alone. Similarly, only four out of these five genes had significant gene expression changes. This functional assay illustrates how our method of combining cross-species methylation data with GWAS results for common SNPs can implicate genes that would not have been detected otherwise.
Recent work in our laboratory has identified regions of the genome where DNA methylation acts to mediate a genetic effect on rheumatoid arthritis (Liu et al., 2013), and the methylation changes in obese humans could potentially act in an analogous role. Our results in obese and insulin-resistant mouse models, however, identify methylation differences even between inbred mice, and thus are definitively the result of environmental stimuli rather than a genetic underpinning. The fact that we see many of these same methylation changes in obese humans, and that these changes are located over regions with known genetic links to T2D, implies that DNA methylation levels could be integrating and mediating genetic and environmental causes of metabolic disease at specific genomic loci.
It is encouraging that many of the new genes described here show pathway relationships to known genetic associations (Figure 4). For example, PRC1, a regulator of cytokinesis, is associated with T2D by a genome-wide significant DIAGRAM result, but it has no known connection to any other gene implicated by genome-wide significant DIAGRAM loci. Its transcription, however, is regulated by FOXO1, an important transcription factor in gluconeogenesis, insulin signaling and adipocyte differentiation that we find to be differentially methylated in both mouse and human obesity. FOXO1 is in turn regulated by TCF7L2, one of the strongest GWAS results. Furthermore, combining genes from all levels of this study creates potential regulatory networks that include genes with known involvement in T2D but also incorporate closely connected genes with no previously known obesity or T2D association that are shown to be involved with obesity and insulin resistance in this story (Figure 4B). Some of these genes, such as FASN and APP, appear to be loci in this network, and could represent potentially important targets.
There are many approaches for, and important applications of, interrogating the association of functional and genetic elements using GWAS summary statistics (Consortium et al., 2012; Jostins et al., 2012; Nicolae et al., 2010), but our approach is unique in its leverage of carefully controlled biological systems to directly integrate cross-species functional epigenomics and clinical genetic risk by stratification. This work, of course, does not address or diminish the many GWAS associations that are not associated with methylation changes. Additionally, it is important to note that while we do not directly address the issue of methylation causality in this study, causality is, at the least, multi-tiered. Our new functional data certainly indicates that these epigenetic changes are functionally proximate to T2D-relevant phenotypes and therefore important for discovery and for clinical translation. Current systems biology literature challenges conventional notions of causality as there is both positive and negative feedback in most complex living systems (Noble, 2012).
The approach described in this study may have broad applicability to identify candidate genes that may better dissect mechanisms and potential routes of treatment in common human disorders, such as cancer and cardiovascular disease. The accessibility of a limited cohort of relevant patients with well characterized clinical materials before and after disease exposure is plausible for cross-species replication. This type of analysis can generate a reliable, functional candidate disease gene set that can be used to interrogate SNP datasets and lend additional support to specific targets that would not ordinarily pass the genome-wide correction threshold. The end result is a process that can integrate information from multiple complementary sources to identify potential targets essential for the pathogenesis of common diseases, such as obesity or T2D, that do not involve highly penetrant single genes, but rather arise from multiple defects along pathways that integrate genetic, epigenetic, and environmental cues.
EXPERIMENTAL PROCEDURES
For full details of all methods (and primer sequences), please see Supplemental Experimental Procedures.
Mouse Sample Preparation
All animal protocols were approved by the Institutional Animal Care and Use Committee of The Johns Hopkins University School of Medicine. Male C57BL/6 mice were purchased from Charles River. Mice were fed a high-fat diet ormatched control low-fat diet. Diet was provided for a period of 12 weeks, beginning at 4 weeks of age. At termination of the study, animals were fasted overnight, euthanized, and tissues were collected.
Intraperitoneal glucose and insulin tolerance tests
Cohorts of mice (between 20 and 24 weeks of age) were injected with glucose or insulin. Animals were fasted overnight (16 h) prior to the glucose tolerance test. For the insulin tolerance test, food was removed 2 h prior to insulin injection. Serum samples were collected and glucose concentrations determined at six time points after injections.
Mouse Hepatocyte Isolation
A protocol for primary hepatocyte isolation was performed using Collagenase (BD Biosciences) and gradient centrifugation as adapted from previously published methods (Berry and Friend, 1969; Li et al., 2010).
Mouse Primary Adipocyte Isolation
Mature adipocytes were isolated from mouse fat pads using Collagenase (Sigma) and resuspension washes as previously described (Stahl et al., 2002).
Pancreatic Islet Isolation
Pancreatic islets used for CHARM were isolated as previously described (Hussain et al., 2000). For the pancreatic islets used in the replication set, whole pancreases were obtained from high-fat-fed and low-fat-fed mice, stained for insulin, cryosectioned into 8μm sections, and then laser-capture microdissection was used to isolate pancreatic islets.
3T3-L1 transduction and transfection
3T3-L1 cells were transducted with Sigma Mission™ lentiviral particles (Sigma) and transfected with overexpression plasmids using Lipofectamine 3000 (Life Technologies) as per the respective manufacturers’ protocols.
Cell culture and glucose uptake assay
3T3-L1 cell lines (ATCC) were maintained and differentiated as per manufacturer’s protocol, and glucose uptake assays were performed on differentiated knock-down and over-expression lines.
Clinical Cohort
This study was approved by the Regional Ethics Committee of Stockholm. All participants provided informed oral and written consent. Clinical characteristics are shown for the obese men before and after RYGB surgery (n = 14, 8, respectively) and non-obese (normal weight) men with a similar age range (n = 7). Full information for human subjects can be found in Table S9.
Human Sample Surgery and Subcutaneous Adipose Tissue Biopsies
A standard laparoscopic RYGB with a 1 m Roux limb was performed. Subcutaneous abdominal adipose biopsies (50–100 mg) were obtained from the obese and non-obese (normal weight) subjects. Biopsies were obtained at the beginning of RYGB surgery (obese subjects) or elective laparoscopic cholecystectomy (lean subjects). Biopsies taken from the obese subjects 6 months after RYGB surgery were obtained after an overnight 12 hour fast from the same surgical incision as the initial biopsy.
CHARM DNA methylation analysis
Genomic DNA from all samples was purified with the MasterPure DNA purification kit (Epicentre) following the manufacturer’s protocol. Genomic DNA was fractionated, digested with McrBC, gel-purified, labeled and hybridized to a CHARM microarray as described (Ladd-Acosta et al., 2010). The array design specifications are freely available on our website (rafalab.jhu.edu). Subsequent technical pre-processing, normalization and correction for batch effects were performed as previously described (Jaffe et al., 2012).
Bisulfite Pyrosequencing
Genomic DNA from each replication sample was bisulfite treated and PCR amplified using nested primers. DNA methylation was subsequently determined by pyrosequencing with a PSQ HS96 (Biotage) as previously reported (Migheli et al., 2013). Artificially methylated control standards of 0, 25, 50, 75 and 100% methylated samples were created using mixtures of purified and SssI-treated whole genome amplified genomic DNA.
Quantitative PCR analysis
Validated primers for all genes were taken from PrimerBank (Wang and Seed, 2003) and synthesized by Integrated DNA Technologies (Coralville, IA, USA). RNA was extracted with Trizol reagent (Life Technologies, Carlsbad, CA, USA), cDNA was created with Quantitect Reverse Transcriptase Kit (Qiagen, Venlo, Netherlands), and quantitative-PCR was performed with Fast SYBR Green (Applied Biosystems, Foster City, CA, USA) on a 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). RNA levels were normalized to same-sample 18S RNA levels.
GO annotation
We analyzed GO annotation using the GOrilla tool (Eden et al., 2009). Enrichment was calculated by comparing genes identified from our analysis to a background of all genes detectable on the appropriate array.
Whole-genome gene expression analysis
Whole genome gene expression data for mouse and human analogues of our study was downloaded from GEO (Barrett et al., 2013). The mouse data was already pre-processed, and the human data was pre-processed using Robust Multi-array Averaging (RMA) from the Affy R library (Bioconductor). The gene expression data was then matched against the DMRs closest to corresponding genes, the log fold change (logFC) of the gene expression was plotted against the average value of the smoothed effect estimate within the DMR, and p-values were generated using t-tests based on Pearson’s correlation coefficient.
Enrichment between human and mouse DMRs
The liftOver tool from the UCSC genome browser transformed the coordinates from the human DMRs from the hg19 human genome to the mm9 mouse genome, as implemented in the rtracklayer Bioconductor package (Lawrence et al., 2009). For each pair of DMR lists, one from the two lifted-over human DMRs and another from the 25 mouse trait DMRs (Table S1), we calculated the number of DMRs at given within-specific p-value significance levels, and also the number that overlapped within 5kb across species. Enrichment tests were chi-squared tests based on the number of species-overlapping significant DMRs, then DMRs only significant within each species, and finally the number of lifted probe groups that were not significant in either species.
Cross-species statistical analysis
We combined significant adipocyte mouse DMRs (at FDR < 5%) across the five traits (glucose, GTT, ITT, weight, and diet) by retaining the maximal coordinates over overlapping cross-trait DMRs resulting in 625 independent DMRs associated with at least 1 trait in adipocytes in mouse. These regions were lifted over from the mouse mm9 genome build to the human hg19 genome build as implemented in the rtracklayer Bioconductor package (Lawrence et al., 2009). These DMRs were annotated to the nearest human charm probe group based on the annotation within 5kb. We then computed a difference and corresponding p-value in obese versus lean and then in obese humans pre-versus post RYGB surgery using linear regression, and retained the minimum p-value, number of probes with p < 0.05, and the slope at the smallest p-value, within each of the mapped DMRs.
DIAGRAM GWAS analysis
We integrated GWAS results into the mouse-human DMRs by obtaining publicly available results from the DIAGRAM meta-analysis (http://diagram-consortium.org/downloads.html; Stage 1 GWAS: Summary Statistics download).
We estimated the variance in disease susceptibility based on the algorithms provided in the Methods section of Morris et al (Morris et al., 2012) and from Wray et al (Wray et al., 2010) using 1000 Genomes-derived risk allele frequencies and assuming a disease prevalence of 8% for a given collection of risk SNPs.
We assessed potential enrichment between the DMRs and the GWAS results using two complementary approaches – the first assessed the enrichment in genome location between DMRs and the LD blocks from the GWAS (Collado-Torres and Jaffe, 2014), and the second assessed enrichment in gene symbols based on all genes directly connected (one-step) to genes linked to T2D with genome-wide significance by the DIAGRAM meta-analysis based on regulatory networks generated using QIAGEN’s Ingenuity IPA (Ingenuity® Systems, www.ingenuity.com)
Data availability
Both raw and processed microarray data has been uploaded to GEO, the Gene Expression Omnibus, as series record GSE63981.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH Director Pioneer Award DP1 ES022579 to A.P.F, by NIDDK R01 grant DK084171 to G.W.W, by the Novo-Nordisk Foundation and Stockholm County Council to EN and J.R.Z., and by the European Research Council to J.R.Z. We thank Arni Runarsson for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Almgren P, Lehtovirta M, Isomaa B, Sarelin L, Taskinen MR, Lyssenko V, Tuomi T, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54:2811–2819. doi: 10.1007/s00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- Arner E, Mejhert N, Kulyte A, Balwierz PJ, Pachkov M, Cormont M, Lorente-Cebrian S, et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61:1986–1993. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Kirchner H, Rasmussen M, Yan J, Kantor FR, Krook A, Naslund E, et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3:1020–1027. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale EG, Hammer RE, Antoine B, Forest C. Disregulated glyceroneogenesis: PCK1 as a candidate diabetes and obesity gene. Trends Endocrinol. Metab. 2004;15:129–135. doi: 10.1016/j.tem.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Berry MN, Friend DS. High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J. Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus--present and future perspectives. Nat. Rev. Endocrinol. 2012;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- Collado-Torres L, Jaffe AE. enrichedRanges: Identify enrichment between two sets of genomic ranges. GitHub. 2014 [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GLG, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Dayeh T, Volkov P, Salo S, Hall E, Nilsson E, Olsson AH, Kirkpatrick CL, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS genetics. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87:99–109. doi: 10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MA, Daniel PB, Habener JF. Glucagon stimulates expression of the inducible cAMP early repressor and suppresses insulin gene expression in pancreatic beta-cells. Diabetes. 2000;49:1681–1690. doi: 10.2337/diabetes.49.10.1681. [DOI] [PubMed] [Google Scholar]
- Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Goldfine AB, Boes T, Henry RR, Ciaraldi TP, Kim EY, Emecan M, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J. Clin. Invest. 2011;121:918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner H, Nylen C, Laber S, Barres R, Yan J, Krook A, Zierath JR, et al. Altered promoter methylation of PDK4, IL1 B, IL6, and TNF after Roux-en Y gastric bypass. Surg. Obes. Relat. Dis. 2014 doi: 10.1016/j.soard.2013.12.019. [DOI] [PubMed] [Google Scholar]
- Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Curr Protoc Hum Genet. 2010 doi: 10.1002/0471142905.hg2001s65. Chapter 20, Unit 20 21 21-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Ralphs KL, Tosh D. Isolation and culture of adult mouse hepatocytes. Methods Mol. Biol. 2010;633:185–196. doi: 10.1007/978-1-59745-019-5_13. [DOI] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI. Genomics, type 2 diabetes, and obesity. N. Engl. J. Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- Migheli F, Stoccoro A, Coppede F, Wan Omar WA, Failli A, Consolini R, Seccia M, et al. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS One. 2013;8:e52501. doi: 10.1371/journal.pone.0052501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingrone G, Panunzi S, De Gaetano A, Guidone C, Iaconelli A, Leccesi L, Nanni G, et al. Bariatric surgery versus conventional medical therapy for type 2 diabetes. N. Engl. J. Med. 2012;366:1577–1585. doi: 10.1056/NEJMoa1200111. [DOI] [PubMed] [Google Scholar]
- Montano CM, Irizarry RA, Kaufmann WE, Talbot K, Gur RE, Feinberg AP, Taub MA. Measuring cell-type specific differential methylation in human brain tissue. Genome Biol. 2013;14:R94. doi: 10.1186/gb-2013-14-8-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS genetics. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, Poulsen P, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- Noble D. A theory of biological relativity: no privileged level of causation. Interface focus. 2012;2:55–64. doi: 10.1098/rsfs.2011.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Hodson L, Dennis AL, Neville MJ, Humphreys SM, Harnden KE, Micklem KJ, et al. Markers of de novo lipogenesis in adipose tissue: associations with small adipocytes and insulin sensitivity in humans. Diabetologia. 2009;52:882–890. doi: 10.1007/s00125-009-1300-4. [DOI] [PubMed] [Google Scholar]
- Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS genetics. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl A, Evans JG, Pattel S, Hirsch D, Lodish HF. Insulin causes fatty acid transport protein translocation and enhanced fatty acid uptake in adipocytes. Dev. Cell. 2002;2:477–488. doi: 10.1016/s1534-5807(02)00143-0. [DOI] [PubMed] [Google Scholar]
- Tang X, Guilherme A, Chakladar A, Powelka AM, Konda S, Virbasius JV, Nicoloro SM, et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, Wainstein J, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum. Mol. Genet. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS genetics. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang Y, Xiao F, Wang S, Xing G, Li Y, Yin X, et al. CKIP-1 regulates macrophage proliferation by inhibiting TRAF6-mediated Akt activation. Cell Res. 2014;24:742–761. doi: 10.1038/cr.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Both raw and processed microarray data has been uploaded to GEO, the Gene Expression Omnibus, as series record GSE63981.