Synopsis
The myelodysplastic syndromes are a diverse group of clonal stem cell disorders characterized by ineffective hematopoiesis, peripheral cytopenias, and an increased propensity to evolve to acute myeloid leukemia. The molecular pathogenesis of these disorders is poorly understood, but recurring chromosomal abnormalities occur in ~50% of cases, and are the focus of much investigation. The availability of newer molecular techniques has allowed the identification of additional genetic aberrations, including mutations and epigenetic changes of prognostic and potential therapeutic importance. This review will focus on the key role of cytogenetic analysis in MDS in the context of the diagnosis, prognosis, and pathogenesis of these disorders.
Keywords: myelodysplastic syndromes, cytogenetics, molecular genetics, diagnosis, prognosis, classification
Introduction
The myelodysplastic syndromes (MDS) include a large spectrum of clonal hematopoietic stem cell disorders that are characterized by peripheral cytopenia(s), morphologic dysplasia, ineffective hematopoiesis, and a variable propensity to transform to acute myeloid leukemia (AML).1,2 The cytopenia can be limited to a single cell line resulting in anemia, thrombocytopenia, or neutropenia, and can be chronic and somewhat indolent, or profound involving all three lineages with life-threatening consequences. The morphologic dysplasia associated with the ineffective hematopoiesis may be subtle and difficult to recognize but, in some cases, it can be impressive and evident in both the bone marrow and peripheral blood. The variable increase in blasts relates, in part, to the risk for transformation to AML, although progression is not solely dependent on the blast percentage. MDS is a disease of older adults with a median age at diagnosis of ~70 years3. Other risk factors for the development of MDS include tobacco use and exposure to solvents, such as benzene and agricultural chemicals. In ~10–15% of cases, the disease arises as a late complication of cytotoxic therapy (radiotherapy and/or chemotherapy) for a prior disorder, and is referred to as a therapy-related myeloid neoplasm (t-MN). MDS is frequently associated with clonal cytogenetic abnormalities, of significant prognostic4,5 and emerging therapeutic importance. An in-depth analysis of some of these is providing significant insights into underlying molecular alterations, which may hold clues to unraveling the pathogenesis of these disorders. This review will focus on the key role of cytogenetic analysis in MDS, in the context of the diagnosis, prognosis and molecular pathobiology of these disorders.
Diagnostic Considerations
MDS Classification
The earliest recognition of myelodysplastic disorders came with the identification of an anemia that was long-standing and refractory, followed by the recognition that these were sometimes preleukemic.6 The classic myelodysplastic syndrome is exemplified in Figure 1, which illustrates pancytopenia with multilineage dysplasia. Although such a case is classic, it belies the wide pathologic spectrum of MDS, which includes cases that are diagnostically challenging and difficult to distinguish on one hand from other benign causes of cytopenia and, on the other hand, from AML and other more aggressive clonal myeloid neoplasms.
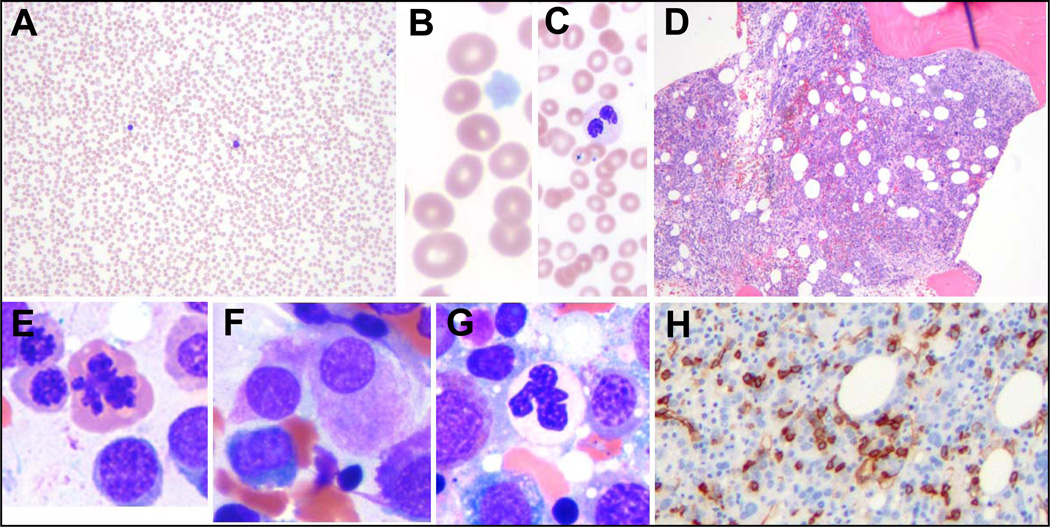
Fig 1.
Morphological features of a typical case of MDS. (A–C) The peripheral smear shows severe pancytopenia (A), with a macrocytic anemia (B), neutropenia with dysplastic neutrophils and thrombocytopenia with pale platelets (C). (D) The marrow is typically hypercellular, indicating ineffective hematopoiesis. (E–H) The marrow cytology reveals dysplasia in the erythroid (E), megakaryocytic (F), and granulocytic lineage (G), as well as increased blasts, which can sometimes be detected by CD34 immunostaining (H).
The disease spectrum was expanded with the first classification proposed by the French American British (FAB) group in 1982.7 In this classification scheme the myelodysplastic disorders were divided into 4 subtypes with increasing blast percentage, or as chronic myelomonocytic leukemia (CMML). The 4 entities included refractory anemia (RA), refractory anemia with ring sideroblasts (RARS), refractory anemia with excess of blasts (RAEB), and refractory anemia with excess of blasts in transformation (RAEB-T). These entities differed mainly by the percentage of blasts seen in the bone marrow (Table 1). In the FAB scheme, AML was defined by the presence of ≥30% blasts in the blood or marrow.
Table 1.
The Evolving Classification of MDS
| FAB 1982 | WHO 2001 | WHO 2008 | Blasts | Other Findings | Cytogenetic abnormalities |
|---|---|---|---|---|---|
| RA | RA |
RCUD RA RN RT |
<1% PB** <5% BM |
Uni or bi-lineage dysplasia* <15% RS |
25% |
| RARS | RARS | RARS | None in PB, <15% BM | At least 15% RS | 10% |
| RCMD RCMD-RS |
RCMD (−RS) | <1% PB*** <5% BM |
Dysplasia >10% in 2 cell lines, +/− RS, no AR**** |
50% | |
| RAEB | RAEB-1 | RAEB-1 | <5% PB 5–9% BM |
Cytopenia, <1K/ul mono; dysplasia in 1 or more lines, no AR**** |
50–70% |
| RAEB-2 | RAEB-2 | 5–19% 10–19 BM |
Cytopenia, <1K/uL mono, +/− AR; dysplasia in 1 or more lines |
50–70% | |
| MDS-U | <1% PB; <5% BM |
Cytopenia, <10% dysplasia; cytogenetic abnormalities present |
50% | ||
| MDS with 5q | <1% PB <5% BM |
Anemia, nml or incr. platelets, isolated del(5q), hypo-lobated megakaryocytes, no AR |
100% | ||
| RAEB-T | 20–29% BM | ||||
| CMML |
Stipulations:
If pancytopenia present, change to MDS-U;
If 1% blasts present, change to MDS-U;
if 2–4% PB blasts present, upgrade to RAEB-1;
if AR present, upgrade to RAEB-2.
Abbreviations: FAB, French American British classification; WHO, World Health Organization classification; RA, Refractory Anemia; RARS, Refractory Anemia with Ring Sideroblasts; RAEB, Refractory Anemia with Excess Blasts; RAEB-T, Refractory Anemia with Excess Blasts in Transformation; CMML, Chronic Myelomonocytic Leukemia; AML, Acute Myeloid Leukemia; RCMD, Refractory Cytopenia with Multi-lineage Dysplasia; RCUD, Refractory Cytopenia with Uni-lineage Dysplasia; MDS-U, Myelodysplastic syndrome-unclassified; AR, Auer Rods; PB, peripheral blood; BM, bone marrow; RS, ring sideroblasts (% indicates percent RS of total nucleated erythroid precursors).
Bold text indicates the current classification.
CMML was included by the FAB in the MDSs, although it was well recognized that CMML differed in that it had a proliferative component with increased circulating and marrow monocytes.8 At times, the peripheral leukocytosis was particularly elevated in the so-called myeloproliferative type, whereas the process was considered to be a dysplastic type and included in the MDS category when the count was less than 13K/uL.9
The classification of MDS presented by the WHO committee in 2001 resulted in significant changes to the classification of both MDS and AML (Table 1).10 Most notable was the reduction in the blast percentage required for a diagnosis of AML from 30% to 20% leading to the elimination of RAEB-T. The new classification also included a new subtype of MDS that, despite the lack of increased blasts (less than 5%), had a more aggressive course, probably owing to the presence of more pronounced multi-lineage dysplasia.11 This category was called refractory cytopenia with multi-lineage dysplasia (RCMD), and it comprised a substantial proportion of cases previously grouped in the low grade RA and RARS categories. Although the recognition of RCMD as a relatively more aggressive MDS served to de-emphasize the importance of the blast percentage for prognosis, the new classification sub-divided the RAEB category into 2 types, with 5–9% blasts (RAEB-1) and 10–19% blasts (RAEB-2), paradoxically emphasizing the prognostic significance of blast percentage in this category.12
A further significant change in the WHO 2001 classification scheme for MDS included the exclusion of CMML from the MDS category, and the development of a separate nosologic group for CMML and other diseases in which there were features of both myelodysplasia and myeloproliferation.13 These “overlap” disorders are called myelodysplastic syndromes/myeloproliferative neoplasms (MDS/MPN) and include CMML, “atypical CML”, and a unclassifiable category, which includes a provisional entity called RARS-T, a disorder that resembles RARS, but has associated thrombocytosis.
Further refinements in the WHO classification scheme for MDS were made most recently in 2008 (Table 1).14,15 These included expanding low grade MDSs from refractory anemia to refractory cytopenia with uni-lineage dysplasia (RCUD), in which there is involvement of only one cell line. The 2008 WHO classification scheme also placed emphasis on the key role of cytogenetic analysis in the diagnosis of MDS, particularly in cases where there is otherwise insufficient morphologic evidence to substantiate a diagnosis of MDS. This is reflected in the inclusion of the subtype, myelodysplastic syndrome unclassified (MDS-U), defined by the presence of cytopenia, less than 1% peripheral blasts, less than 10% dysplasia, less than 5% bone marrow blasts with the presence of a cytogenetic abnormality commonly seen in MDS (Table 2). In addition, the WHO 2008 classification now includes “MDS with an isolated del(5q)” as a separate entity. This entity, sometimes referred to as “the 5q- syndrome” 16, had been well known for some time, and is characterized typically by its presentation in middle-aged women with macrocytic anemia, splenomegaly, normal to elevated platelet counts, hypo-lobated megakaryocytes in the bone marrow, and an isolated del(5q).
Table 2.
Recurring Chromosomal Abnormalities in the Myelodysplastic Syndromes
| Disease* | Chromosome abnormality¶ | Frequency | Involved genes+ | Consequence | |
|---|---|---|---|---|---|
|
MDS Unbalanced |
|||||
| +8 | 10% | ||||
| −7/del(7q) ¶ | 10% | ||||
| del(5q)/t(5q) ¶ | 10% | ||||
| del(20q) | 5–8% | ||||
| −Y | 5% | ||||
| i(17q)/t(17p) ¶ | 3–5% | TP53 | Loss of function, DNA damage response |
||
| −13/del(13q) ¶ | 3% | ||||
| del(11q) ¶ | 3% | ||||
| del(12p)/t(12p) ¶ | 3% | ||||
| del(9q) ¶ | 1–2% | ||||
| idic(X)(q13) ¶ | 1–2% | ||||
| Balanced | t(1;3)(p36.3;q21.2) ¶ | 1% | MMEL1 | RPN1 | Deregulation of MMEL1- Transcriptional activation? |
| t(2;11)(p21;q23)/t(11q23) ¶ | 1% | MLL | MLL fusion protein - altered transcriptional regulation |
||
| inv(3)(q21q26.2)/t(3;3)(q21;q26. 2) ¶ |
1% | RPN1 | MDS1/EVI1 | Altered transcriptional regulation by EVI1 |
|
| t(6;9)(p23;q34) ¶ | 1% | DEK | NUP214 | Fusion protein - nuclear pore protein |
|
|
Therapy related MDS |
−7/del(7q) ¶ | 50% | |||
| del(5q)/t(5q) ¶ | 40–45% | ||||
| dic(5;17)(q11.1–13;p11.1–13) ¶ | 5% | TP53 | Loss of function, DNA damage response |
||
| der(1;7)(q10;p10) ¶ | 3% | ||||
| t(3;21)(q26.2;q22.1) ¶ | 3% | RPL22L1 | RUNX1 | RUNX1 fusion protein - altered transcriptional regulation |
|
| t(11;16)(q23;p13.3)/t(11q23) ¶ | 2% | MLL | CREBBP | MLL fusion protein – altered transcriptional regulation |
|
| CMML | t(5;12)(q33.1;p13) | ~2% | PDGFRB | ETV6/TEL | Fusion protein – altered signaling pathway |
MDS, Myelodysplastic Syndrome; CMML, chronic myelomonocytic leukemia.
Cytogenetic abnormalities considered in the WHO 2008 Classification as presumptive evidence of MDS in patients with persistent cytopenias(s), but with no dysplasia or increased blasts.
Genes are listed in order of citation in the karyotype, e.g., for the t(11;16), MLL is at 11q23 and CREBBP at 16p13.3.
Prognosis
The heterogeneity in outcome within the various morphologic categories identified by the FAB and WHO classification systems has led to a proliferation of prognostic tools and scoring systems that attempt to predict outcomes of patients with MDS more accurately. The ability to achieve greater precision in prognostication in MDS is of paramount importance, since therapeutic options vary from supportive care and growth factor use, to more intensive approaches, such as epigenetic modulators, and to approaches associated with significant potential for morbidity and mortality, such as intensive chemotherapeutic strategies and allogeneic stem cell transplantation. At present, the higher intensity approaches are reserved for patients with high risk disease.17 All of the contemporary prognostic systems incorporate the cytogenetic pattern as a key element.18–21
The most widely used, and validated, prognostic scoring system in current use is the International Prognostic Scoring System (IPSS), which incorporates karyotype, bone marrow blasts and number of cytopenias, and identifies these factors as being the most critical in prognostication (Table 3a) 18. Nonetheless, there are a number of limitations that have led to the development of other scoring systems, as well as ongoing attempts to refine the IPSS.
Table 3.
| a: International Prognostic Scoring system | |||||
|---|---|---|---|---|---|
| Prognostic Variable | 0 | 0.5 | 1.0 | 1.5 | 2.0 |
| BM Blasts (%) | <5 | 5–10 | − | 11–20 | 21–30 |
| Karyotype | Good | Intermediate | Poor | ||
| Cytopenias* | 0/1 | 2/3 | |||
| Risk group | IPSS score | Median Survival (yrs) |
Time until 25% of patients developed AML (yrs) |
|---|---|---|---|
| Low | 0 | 5.7 | 9.4 |
| Intermediate-1 | 0.5–1.0 | 3.5 | 3.3 |
| Intermediate-2 | 1.5–2.0 | 1.2 | 1.1 |
| High | ≥ 2.5 | 0.4 | 0.2 |
| b. Cytogenetic Abnormalities in the International Prognosis Scoring System | ||||
|---|---|---|---|---|
| Cytogenetic Abnormalities |
Time until 25% of pts developed AML (mos) |
Median Survival | ||
| Favorable risk | normal karyotype | 5.6 years | 3.8 years | |
| isolated del(5q) | ||||
| isolated del(20q) | ||||
| isolated –Y | ||||
| Intermediate risk | other abnormalities | 1.6 years | 2.4 years | |
| Poor risk | −7/del(7q) | 0.9 years | 0.8 years | |
| complex kayotypes | ||||
Cytopenias were defined as: Hemoglobin < 10 g/dL, ANC < 1500/uL, Platelet count < 100,000/ L.
Limitations of the IPSS include the fact that it was validated in previously untreated patients with de novo MDS, which limits the ability to use this tool to predict outcomes in patients with MDS treated with contemporary treatment approaches. In addition, although the numbers of cytopenias are factored into the IPSS, the severity of cytopenias is not taken into account; in particular, transfusion dependency, which has been identified as a poor prognostic marker in recent scoring systems, is not considered. Furthermore, the IPSS includes a limited number of cytogenetic abnormalities (Table 3b), and there are concerns that high risk cytogenetic aberrations are not accorded sufficient emphasis.
To address some of the concerns associated with the IPSS, newer systems such as the WHO classification based prognostic scoring system (WPSS) have emerged.20 The WPSS is dynamic, and can be applied at diagnosis and during follow up, and is now being validated in patients with MDS undergoing contemporary treatment modalities, such as allogeneic stem cell transplantation.22 Prognostic factors of import included in this model are WHO subtype, transfusion dependency, and karyotype subgroup, as defined by the IPSS. Five risk groups are recognized in the WPSS, with median survivals ranging from 12 months to 103 months.
The MD Anderson Cancer Center (MDACC) prognostic scoring system was also proposed recently to address some of the limitations associated with the IPSS.19 This model is also dynamic, can be applied at any time during the disease course, and encompasses a broader patient population, including patients with t-MDS and CMML. Four risk categories are identified that predict overall survival ranging from a median of 54 months in low risk patients to 6 months in high risk patients.
Despite the development of newer systems, there continues to be significant heterogeneity in outcome, particularly in patients identified by current models as being in the lower risk category, emphasizing the need for continued refinement of these models based on a broader range of anomalies of emerging prognostic significance.23 An updated cytogenetic system has been proposed which involves an international collaborative effort using data sets originating from the German-Austrian Working Group, the International Risk Analysis Workshop, the Spanish Cytogenetics Workshop and the International Cytogenetics Working Group of the MDS Foundation. This 4-tiered cytogenetic risk stratification system (Table 4) involves 20 cytogenetic subgroups; median survival ranged from 5.7 months to 50.6 months, and time until 25% of patients evolved to AML ranged from 3.4 months to 71.9 months.23
Table 4.
Proposed 4-Tier Cytogenetic Risk Stratification System
| Risk group | Karyotype | Median Survival (mos) |
Time until 25% of patients developed AML (mos) |
|---|---|---|---|
| Good | Normal, del(5q), double abnormality including del(5q), der(1;7)(q10;p10)/t(1;7), del(11q), del(12p), +19, del(20q), −Y |
50.6 | 71.9 |
| Intermediate-1 | any other double abnormality (not including del(5q) or −7/del(7q), +8, i(17)(q10), +21, any 7 other single abnormality |
25.7 | 14.7 |
| Intermediate-2 | Double abnormality including −7/del(7q), t(3q26.2), complex with 3 abnormalities |
16 | 9.8 |
| High | Complex (>3 abnormalities) | 5.7 | 3.4 |
There is also ongoing concern that the IPSS accords more prognostic weight to an increase in bone marrow blasts as compared to poor risk karyotypes, e.g., 2 points are assigned for BM blasts in the 21% to 30% range, and 1.5 points for the 11% to 20% range, whereas poor risk karyotypes are assigned 1 point (Table 3a). In a recent retrospective study of 2,351 patients with MDS, the prognostic impact of a poor risk karyotype, was as unfavorable as the presence of a significant increase in blasts >20% blasts.24
In addition to the broad prognostic variables incorporated in the various systems outlined above, it is clear that gene mutations25 and epigenetic aberrations play a critical role in the pathobiology of MDS 26,27 including the determination of prognosis. These genetic and epigenetic determinants, described in more detail in the following sections, deserve consideration for inclusion in a contemporary prognostic model developed for 21st century use.
Cytogenetic Analysis
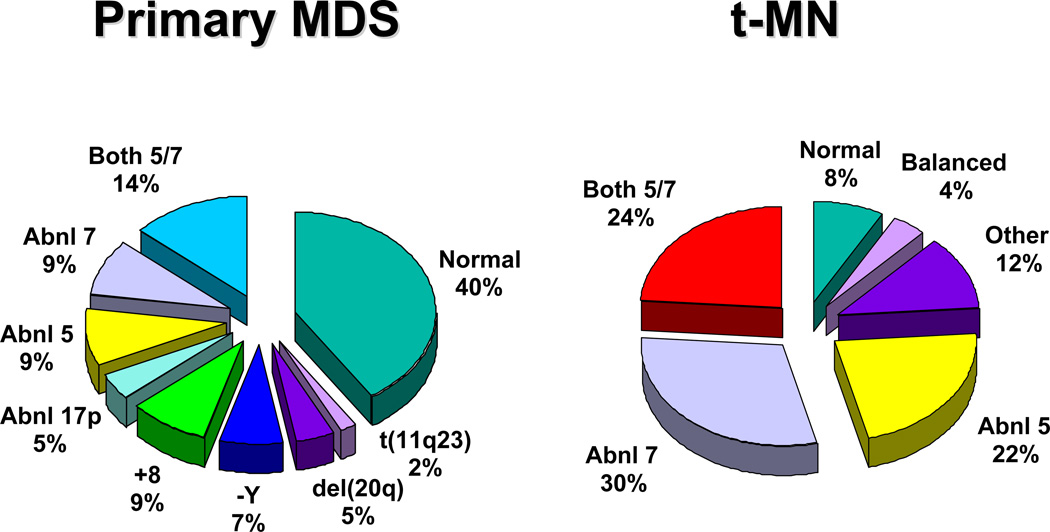
Clonal chromosome abnormalities can be detected in marrow cells of 40%-100% of patients with primary MDS at diagnosis (Tables 1 and 2).5,18 The proportion varies with the risk that a subtype will transform to AML, which is highest for RCMD and RAEB. Most recurring cytogenetic abnormalities found in MDS are unbalanced, most commonly the result of the loss of a whole chromosome, a deletion of part of a chromosome, or an unbalanced translocations (Fig. 2). The most common cytogenetic abnormalities encountered in MDS, del(5q), −7, +8, and del(20q), have been incorporated into the more robust prognostic scoring systems of MDS outlined above. The recurring translocations characteristic of acute leukemia without prior MDS, such as the t(15;17), inv(16) and t(8;21), are almost never seen. With the exception of MDS with isolated del(5q), the chromosome changes show no close association with the morphologic subtypes of MDS. Clones with unrelated abnormalities, one of which typically has +8, are seen at a greater frequency in patients with MDS (5% vs. 1%), than in patients with AML.
Fig. 2.
Recurring chromosomal abnormalities in MDS. Relative frequencies are depicted in the pie chart.
Serial cytogenetic evaluations can be informative, particularly when there is a change in the clinical features of a patient. Cytogenetic evolution is the appearance of an abnormal clone where only normal cells have been seen previously, the acquisition of additional abnormalities within an abnormal clone, or the progression from the presence of a single clone to multiple related, or occasionally unrelated, abnormal clones. Karyotypic evolution in MDS is associated with transformation to acute leukemia in about 60% of cases, and reduced survival, particularly for those patients who evolve within a short period of time (less than 100 days).5,18
Fluorescence In Situ Hybridization Analysis
In the past decade, molecular cytogenetic techniques, such as fluorescence in situ hybridization (FISH) have become a powerful adjunct to classical cytogenetic analysis.28 FISH can be performed on marrow or blood smears, or fixed and sectioned tissue, since it does not require dividing cells. Advantages of FISH include (1) the rapid nature of the method and the ability to analyze large numbers of cells; (2) its high sensitivity and specificity; (3) the ability to obtain cytogenetic data from samples with a low mitotic index or terminally-differentiated cells; and (4) the application to histologically-stained cells allowing a direct correlation of the status of the genetic target within morphologically characterized cells. The major disadvantage is the inability to interrogate more than a few abnormalities, and to identify the full spectrum of abnormalities in each clone, the presence of multiple clones, and clonal evolution.28 In a clinical setting, cytogenetic analysis could be performed at the time of diagnosis to identify the chromosomal abnormalities in bone marrow cells from an individual with MDS. Thereafter, FISH with the appropriate probes could be used to detect residual disease or early relapse, and to assess the efficacy of therapeutic regimens. Commercially-available probes have been developed for the detection of 11q23/MLL translocations, −Y, del(5q)/t(5q), −7/del(7q), +8, del(20q), del(13q), del(11q), and −17/loss of 17p. In MDS, FISH analysis of cases with a normal karyotype by cytogenetic analysis detected recurring abnormalities in 10–15% of cases.29
Cytogenetic Findings in MDS
Normal Karyotype
A normal karyotype is found in ~50% of patients with MDS. This group of patients is almost certainly genetically heterogeneous, where technical factors precluded the detection of chromosomally abnormal cells, or where leukemogenic alterations occur at the molecular level and are not detectable with standard cytogenetic methods. Nonetheless, these cases are a standard reference for comparison of outcomes. The IPSS found that patients with a normal karyotype fall within the favorable risk group (Table 3b).18
−Y
The clinical and biological significance of the loss of the Y chromosome, −Y, is unknown. Loss of the Y chromosome has been observed in a number of malignant diseases, but is also associated with aging in healthy males.30 Patients with a hematological disease have a significantly higher percentage of cells with a –Y (52% vs. 37%, p=0.036), and –Y in >75% of metaphase cells accurately predicted a malignant disease.30 Although loss of a Y chromosome may not be diagnostic of MDS, once the disease is identified by clinical and pathologic means, the IPSS found that −Y as the sole cytogenetic abnormality conferred a favorable outcome.18
+8
A gain of chromosome 8 in MDS is observed in all MDS subgroups in ~10% of patients. 18,31,32 Determining the significance of the gain of chromosome 8 in MDS patients is complicated in that +8 is often associated with other recurring abnormalities known to have prognostic significance, e.g., del(5q)t(5q) or −7/del(7q), and may be seen in isolation as a separate clone unrelated to the primary clone in up to 5% of cases. The IPSS18, as well as the time-dependent WPSS20 ranked this abnormality in the intermediate risk group; however, several subgroups found that +8 as a sole abnormality had a poorer outcome than expected for an intermediate IPSS risk group.31,33 Hematopoietic cells with trisomy 8 express higher levels of many genes that localize to chromosome 8, including the MYC gene, and antiapoptotic genes
del(20q)
A deletion of the long arm of chromosome 20, del(20q), is seen in approximately 5% of MDS cases and 7% of t-MDS cases.5,31 Features characterizing MDS patients with a del(20q) include low risk disease (usually RA), low rate of progression to AML, and prolonged survival (median of 45 months vs. 28 months for other MDS patients), and prominent dysplasia in the erythroid and megakaryocytic lineages.34 The IPSS noted that patients with a del(20q) in the context of a complex karyotype identified a poor-risk group (median survival, 9.6 months), whereas the prognosis for patients with an isolated del(20q) was favorable.18 Although a commonly deleted segment (CDS) has been identified on 20q, containing 19 genes, there is no definitive evidence linking these genes to the pathogenesis of myeloid neoplasms.35,36
Deletions of chromosome 5
A deletion of the long arm of chromosome 5, del(5q), or an unbalanced translocation leading to loss of 5q are observed in 10–20% of MDS patients more commonly, in RAEB 1, 2 in association with a complex karyotype, and 40% of patients with t-MDS (Fig. 3).37 Abnormalities of 5q are associated with a significant occupational exposure to potential carcinogens, as well as previous exposure to standard and high dose alkylating agent therapy, including use in immunosuppressive regimens.38,39 Clinically, the patients with del(5q) coupled with other cytogenetic abnormalities have a high frequency of TP53 mutations, a poor prognosis with early progression to leukemia, resistance to treatment, and short survival.40,41 MDS with an isolated del(5q) represents a distinct clinical syndrome “the 5q minus syndrome”. These patients have a favorable outcome, in fact the best of any MDS subgroup, with low rates of transformation to AML and a relatively long survival of several years duration.16,18 The immunomodulatory agent, lenalidomide, has been associated with significant responses in IPSS low or intermediate-1 risk MDS with the del(5q), including red cell transfusion independency (67% of patients), and complete cytogenetic responses.42
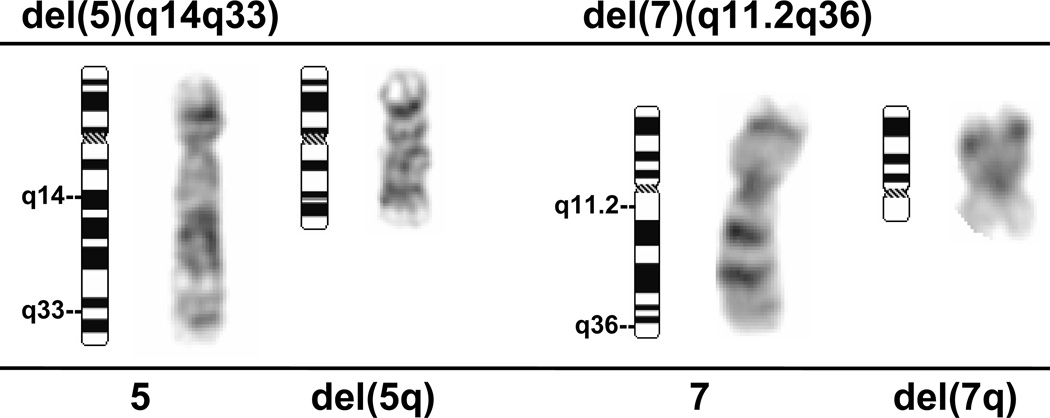
Fig. 3.
Deletions of 5q and 7q in myeloid neoplasms. In this del(5q), breakpoints occur in q14 and q33 resulting in interstitial loss of the intervening chromosomal material. In this del(7q), breakpoints occur in q11.2 and q36. In both cases, the critical commonly deleted segments are lost. Normal chromosome 5 and 7 homologs are shown for comparison.
A recent study showed that minor subclones of TP53-mutant cells were present in some patients prior to lenalidomide therapy, and were associated with lenalidomide-resistance and trend towards a higher risk of evolution to AML.43 The molecular mechanisms underlying the clinical effectiveness of lenalidomide in del(5q) MDS remain obscure, but haploinsufficiency of two dual-specificity cell cycle phosphatases encoded by genes on 5q, CDC25C and PP2A have been implicated in the sensitivity to lenalidomide.44 However, treatment with lenalidomide is unlikely to be curative since del(5q) malignant stem cells persist in remission, and clinical and cytogenetic progression was associated with recurrence or expansion of the del(5q) clone.45
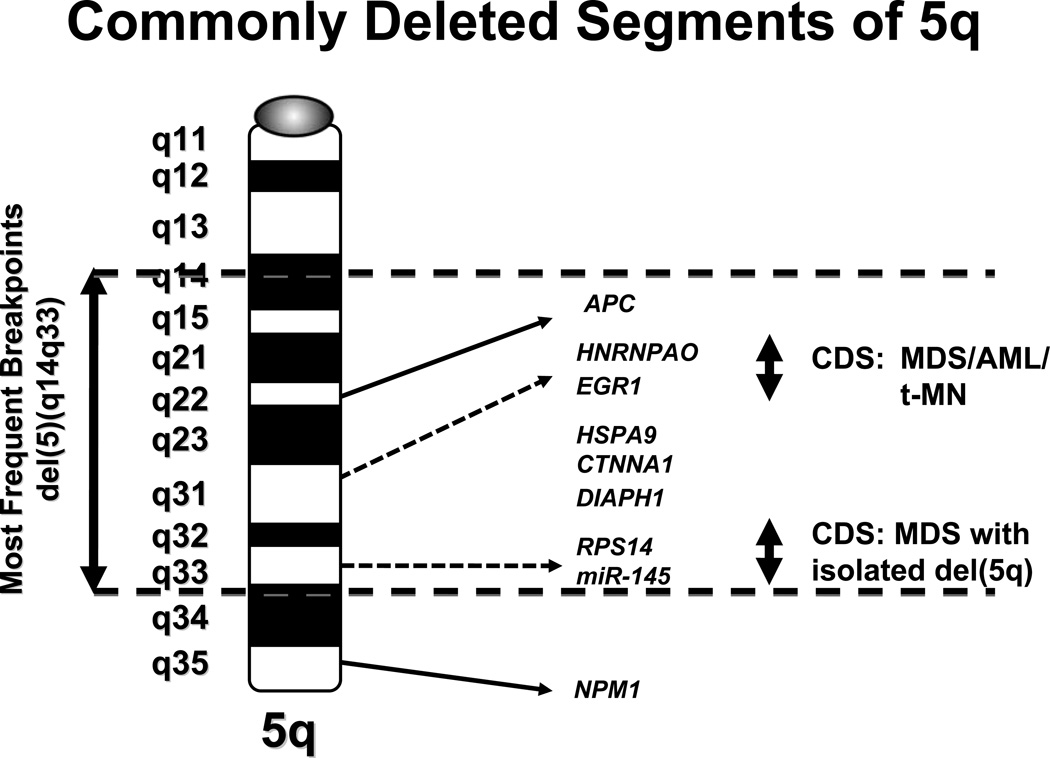
Several groups of investigators have defined a CDS (Fig. 4) on the long arm of chromosome 5, band 5q31.2, predicted to contain a myeloid tumor suppressor gene that is involved in the pathogenesis of the more aggressive forms of MDS and AML.46,47 A second, distal CDS of 1.5 Mb within 5q33.1 has been identified in MDS with an isolated del(5q).48 Despite intense efforts, the identification of tumor suppressor genes (TSGs) on chromosomes 5 has been challenging, as a result of the fact that the deletions of 5q are typically large, and encompass both of these regions. Molecular analysis of the 19 candidate genes within the CDS of 5q31.2 and 44 genes in the 5q33.1 CDS did not reveal inactivating mutations in the remaining alleles, nor was there evidence of transcriptional silencing48–50 [Godley and Le Beau, unpublished data]. Moreover, copy-neutral loss of heterozygosity (also known as acquired uniparental disomy) is not seen on 5q in MDS or AML. These observations are compatible with a haploinsufficiency model in which loss of one allele of the relevant gene(s) on 5q perturbs cell fate, rather than the biallelic inactivation of a tumor suppressor gene.51 A number of genes and several miRNAs located on 5q, including RPS14 52, miRNA-14553,54, EGR1 55, APC56, CTNNA157, HSPA958, and DIAPH159, have been implicated in the development of myeloid disorders due to a gene dosage effect, and several of these are reviewed below. Together, these studies support a haploinsufficiency model, in which loss of a single allele of more than one gene on 5q act in concert to alter hematopoiesis, promote self-renewal of HSPCs, induce apoptosis of hematopoietic cells, and disrupt differentiation.
Fig. 4.
Ideogram of the long arm of chromosome 5 showing candidate genes within the commonly deleted segments (CDSs) as reported by various investigators. The proximal CDS in 5q31.2 was identified in MDS, AML and t-MN, whereas the distal CDS in 5q33.1 was identified in MDS with isolated del(5q).
RPS14
The gene encoding RPS14, which is required for the processing of 18S pre-rRNA, is located at 5q33.1, and is involved in MDS with an isolated del(5q).52 Downregulation of RPS14 in CD34+ bone marrow cells blocks the differentiation and increases apoptosis of erythroid cells via a TP53-dependent mechanism.60 Of interest, the ribosomal processing defect caused by haploinsufficiency of RPS14 in MDS is highly analogous to the functional ribosomal defect seen in Diamond-Blackfan anemia. Other studies have shown that haploinsufficiency of two micro-RNAs, miR-145 and miR-146a, encoded by sequences near the RPS14 gene, cooperate with loss of RPS14.53,54 The Toll-interleukin-1 receptor domain-containing adaptor protein (TIRAP) and FLI1 are targets of these miRNAs. Haploinsufficiency of miR-145 may account for several features of MDS with an isolated del(5q), including megakaryocytic dysplasia; however, neither RPS14 nor miR-145 haploinsufficiency is predicted to confer clonal dominance.
APC
APC is a multifunctional tumor suppressor involved in the pathogenesis of colorectal cancer via regulation of the WNT signaling cascade. The APC gene is located at 5q22.2, and is deleted in >95% of patients with a del(5q).47 Conditional inactivation of a single allele of Apc in mice leads to the development of severe macrocytic anemia, a block in erythropoiesis at the early stages of differentiation, and an expansion of the short-term and long-term HSCs.56 Apc heterozygous myeloid progenitor cells display an increased frequency of apoptosis, and decreased in vitro colony-forming capacity, recapitulating several characteristic features of myeloid neoplasms with a del(5q).
EGR1
The early growth response 1 gene (EGR1) encodes a member of the WT-1 family of zinc finger transcription factors, and mediates the cellular response to growth factors, mitogens, and stress stimuli.61 Recently, Egr1 has been shown to be a direct transcriptional regulator of many known TSGs, e.g., Tp53, Cdkn1a/p21, Tgfb, and Pten, and acts as a TSG in several human tumors, including breast non-small cell lung cancer. 61 Egr1-null mice show spontaneous mobilization of HSPCs into the periphery, identifying Egr1 as a transcriptional regulator of stem cell migration.55,62 Moreover, loss of a single allele of Egr1 cooperates with mutations induced by an alkylating agent in the development of malignant myeloid diseases (MPD with ineffective erythropoiesis) in mice, indicating that Egr1 is a haploinsufficient myeloid suppressor gene.55
Loss of Chromosome 7 or del(7q)
A −7/del(7q) is observed as the sole abnormality in ~5% of adult patients, in ~50% of children with primary MDS, and in ~55% of patients with t-MDS (Fig. 3). 33,37,39,63 As with del(5q), occupational or environmental exposure to mutagens including chemotherapy, radiotherapy, benzene exposure, and smoking, as well as severe aplastic anemia (regularly treated with immunosuppressive agents alone) have been associated with −7/del(7q). 18 The IPSS considers the −7/del(7q) to be a poor prognostic cytogenetic finding.18 “Monosomy 7 Syndrome” has been described in young children, and is characterized by a preponderance of males (~4:1), hepatosplenomegaly, leukocytosis, thrombocytopenia, and a poor prognosis.63 Juvenile myelomonocytic leukemia (JMML) is a MDS/MPD disease in the WHO classification, and shares many features with this entity.63 An emerging paradigm is that −7/del(7q) cooperates with deregulated signaling via the RAS pathway as a result of mutations in the NRAS or KRAS gene, inactivating mutations in the gene encoding NF1, a negative regulator of RAS proteins, or activating mutations in the gene encoding the PTPN11/SHP2 phosphatase, a positive regulator of RAS proteins, as well as RUNX1 mutations, and methylation silencing of the CDKN2B (p15INK4B) gene.41,64,65
To date, three CDSs have been identified on 7q; however, the molecular mutations underlying the development of MDS and AML with del(7q) are poorly-understood66–68 We previously identified two distinct CDSs, a 2.52 Mb CDS within 7q22 spanning the interval containing LRCC17 and SRPK2, and a second, less frequent, region in q32–33.68 Each of the candidate genes within the CDS at 7q22 has been evaluated for mutations69; however, no inactivating mutations have been identified in the remaining allele. Mice with a conditional heterozygous deletion of this region in murine HSPCs had no alterations of hematopoiesis, suggesting that this region does not contain a haploinsufficient myeloid tumor suppressor gene, or that mutations in cooperating genes are required.70 Recently, Dohner et al. reported the analysis of a large series of patients with abnormalities of 7q using FISH. Whereas most patients had large deletions, they identified an ~2 Mb deleted segment in proximal q22, that overlaps with the proximal portion of the CDS defined by Le Beau et al., but extends more proximally, and includes the CUTL1, RASA4, EPO, and FBXL13 genes in 7q22.1.71 The recent recognition of mutations in EZH2, a gene located at 7q36.1, is intriguing; however, myeloid neoplasms with EZH2 mutations typically do not have −7/del(7q), and the del(7q) does not always result in loss of one EZH2 allele.72,73
Rare Recurring Translocations
In MDS, several recurring translocations have been identified, which result in fusion proteins. Translocations involving the MLL gene on 11q23 are noted in 3–5% of MDS and t-MDS cases. The most common translocations are the t(9;11)(p22;q23) and t(11;16)(q23;p13.3). The t(11;16) occurs primarily in t-MDS, but rare cases have presented as t-AML.74 MLL is fused with the CREBBP (CREB binding protein) gene on chromosome 16. The MLL protein is a histone methyltransferase that assembles in protein complexes that regulate gene transcription, e.g., HOX genes during embryonic development, via chromatin remodeling. CREBBP is a histone acetyltransferase involved in transcriptional control via histone acetylation.
The t(5;12)(q33.1;p13) is observed in ~1% of patients with CMML, and results in fusion of the protein encoded by the beta chain of the platelet derived growth factor receptor (PDGFRB) on 5q, and a novel transcriptional repressor gene, ETV6 (also known as TEL) on 12p. Alterations in the PDGFRB kinase activity and function of the ETV6 repressor contribute to the transformed phenotype. The t(5;12) predicts for a response to imatinib mesylate, a selective inhibitor of the PDGFRB kinase activity.75 PDGFRB participates in several other rare translocations in myeloid neoplasms (reviewed in76); a unifying feature is the presence of eosinophilia.
Complex Karyotypes
Complex karyotypes are variably defined, but generally involve the presence of > 3 chromosomal abnormalities. Complex karyotypes are observed in ~20% of patients with primary MDS, and in as many as 90% of patients with t-MDS.37 Abnormalities involving chromosomes 5, 7, or both are identified in most cases with complex karyotypes. There is general agreement that a complex karyotype carries a poor prognosis.18,20,31
Alterations in gene function
A growing body of evidence suggests that mutations of multiple genes mediate the pathogenesis and progression of MDS. The involved genes fall into two main classes, namely, genes encoding hematopoietic transcription factors or proteins that regulate cytokine signaling pathways. There is an increase in the frequency of molecular mutations from low-risk to high- risk MDS, or AML evolving from MDS, emphasizing the role of these mutations in disease progression. A detailed review of these genes is beyond the scope of this chapter, and has been provided elsewhere.25,76 Table 5 provides a partial list and overview of some of the salient features of genes implicated in the pathogenesis of MDS.
Table 5.
Frequency and Significance of Mutated Genes in MDS.
| Mutated Gene | Frequency* | Biological Features and Clinical Significance |
|---|---|---|
| ASXL1 | 10–15% | Polycomb group protein involved in transcriptional regulation Prevalence of frameshift mutations suggest a dominant-negative function Clinical significance unknown |
| ATRX | Rare | Involved in epigenetic modifications of DNA Loss of expression leads to decreased expression of alpha-globin Associated with acquired alpha-thalassemia, often with severe anemia |
| CEBPA | 2–5% | TF involved in hematopoiesis, loss of function impairs granulopoiesis Bi-allelic (N-terminal and C-terminal mutations) No apparent effect on time to progression to AML or overall survival |
| CSF1R | 2–5% | Constitutive activation of the macrophage colony-stimulating factor receptor tyrosine kinase Karyotype predominantly normal Associated with advanced MDS and progression to AML |
| EZH2 | 5% | Encodes a histone 3 lysine 27 methyltransferase regulating transcription Mutations result in loss of function Associated with a poor prognosis |
| FLT3 ITD | 5–10% | Rector tyrosine kinase involved in cytokine signaling, critical in hematopoiesis Associated with progression to AML and a poor prognosis |
| IDH1/2 | Rare | Metabolic enzymes catalyzing oxidative decarboxylation of isocitrate to alpha- ketoglutarate. Missense mutations alter catalytic function converting alpha-KG to 2 hydroxyglutarate, while consuming NAPDH Associated with advanced MDS and progression to AML |
| JAK2 | 5% of RA 60% of RARS-T |
Encodes a tyrosine kinase component of various cytokine signaling pathways Mutations result in constitutive activation of the tyrosine kinase Mutated in 60% of RARS with thrombocytosis, an unclassified MDS/MPD Clinical significance unknown, does not appear to alter prognosis |
| MPL | 5% of RARS-T | Encodes the thrombopoietin receptor Mutations result in constitutive activation of the tyrosine kinase, and are associated with dysmegakaryocytopoiesis Higher expression in advanced MDS is associated with a poor prognosis |
| NPM1 | Rare | Nuclear-cytoplasmic shuttling protein, with pleiotropic functions Terminal frameshift mutations disrupt the nuclear localization signal leading to redistribution to the cytoplasm Unknown clinical significance in MDS |
| NRAS/KRAS | 5% | Encodes a GTPase component of multiple cytokine signaling pathways Activating mutations result in constitutive signal transduction Increased risk of progression to AML |
| PTPN11 | Rare | Encodes the non-receptor SHP2 tyrosine phosphatase, a positive regulator of RAS proteins. Mutations result in protein activation Mutated in 30% of JMML Mutations in NRAS/KRAS, NF1, and PTPN11 are mutually exclusive |
| RUNX1 | 10–15% | Encodes the DNA-binding subunit of the heterodimeric core binding factor transcription factor required for hematopoiesis Point mutations in the RUNT (DNA-binding) domain result in loss of function, and a dominant negative effect Associated with mutations of the RAS pathway and −7/del(7q) Increased risk of progression to AML |
| TET2 | 20% | Epigenetic regulator Clinical significance unknown |
| TP53 | 5–10% (25% in t-MDS) |
Encodes a checkpoint protein which monitors integrity of the genome, arrests the cell cycle in response to DNA damage Associated with chromosomal instability, del(5q), loss of 17p, and complex karyotypes Associated with rapid progression and poor outcome Significantly differentiates worse prognosis within each IPSS subgroup |
Rare mutations occur at a frequency of less than 2%.
Recently, Bejar et al. demonstrated that the integration of mutation analysis into diagnostic classification and prognostic scoring systems in MDS has the potential to stratify a diverse disease into discrete subsets with more consistent clinical phenotypes and prognosis.25,77 For example, mutations in RUNX1, TP53, and NRAS were associated with severe thrombocytopenia and blast percentage. In multivariate analysis, mutations in 5 genes, occurring in one-third of patients, retained independent prognostic significance: TP53, EZH2, ETV6, RUNX1, and ASXL1, and predicted poor overall survival. Mutations of these genes stratified low- and intermediate-1, and intermediate 2 IPSS risk groups into two risk groups each, identifying patients within these subgroups with a poorer prognosis who may require a more intensive therapeutic approach. The genes most commonly mutated in MDS are TET2, ASXL1, EZH2, RUNX1, and TP53, which are described briefly below.
An emerging paradigm in MDS is the high frequency of mutations in genes involved in the regulation of transcription via chromatin modifications (IDH1/2, TET2, EZH2, ASXL1), and the intriguing observation that mutations often occur in more than one gene in the same patient, implying functional cooperation (note that IDH1/2 and TET2 mutations are mutually exclusive). The most frequently mutated gene in MDS is TET2 (20%); point mutations are observed in all cytogenetic subsets.25,77 TET2 converts 5-methylcytosine to 5-hydroxymethylcytosine, thereby altering the epigenetic mark created by DNA methyltransferases.78 Recent studies suggest no impact of TET2 mutations on overall survival in MDS.77,79
ASXL1 mutations are observed in 10–15% of MDS.25,80 ASXL1 is a member of the polycomb family of chromatin-binding proteins, and is involved in the epigenetic regulation of gene expression (typically repression). Mutated proteins are predicted to function as dominant negative proteins inhibiting the function of the wild type proteins as well as other members of the polycomb complex. The prognostic significance of ASXL1 mutations in MDS is not yet known.
EZH2 (enhancer of zeste homolog 2) mutations occur in 5% of MDS. 25,72,73 EZH2 encodes a histone methyltransferase that trimethylates histone 3 lysine 27, an epigenetic mark that confers gene silencing. In MDS, the mutations lead to loss of the catalytic activity, and are predicted to increase HSC expansion.81 Although EZH2 is located at 7q36.1, loss or mutation of EZH2 does not appear to be the sole driver of myeloid neoplasms associated with −7/del(7q).
Point mutations in the Runt-related transcription factor 1 (RUNX1) have been reported in AML and MDS (12%), particularly in MDS secondary to treatment with cytotoxic therapy, and increase with the severity of the disease.82 RUNX1, also known as CBFA2 or AML1, encodes the DNA-binding subunit of the heterodimeric core-binding factor (CBF) complex, which is essential for definitive hematopoiesis.83 RUNX1 mutations impair DNA binding and act as dominant negative proteins, and are associated with activating mutations of the RAS pathway, −7/del(7q), and a shorter overall survival.82 Germline mutations of RUNX1 cause a rare human disease called familial platelet disorder; affected individuals have an MDS-like phenotype with thrombocytopenia, and/or dysfunctional platelets, and a predisposition to progress to AML.84
The TP53 tumor suppressor gene encodes an essential checkpoint protein that monitors the integrity of the genome, and arrests cell cycle progression in response to DNA damage. Mutations of TP53 (exons 4–8) or loss of an allele, typically as a result of a cytogenetic abnormality of 17p, are observed in MDS (5–10%) and t-MDS (25–30%), particularly in patients who have received alkylating agent therapy.40 TP53 mutations may occur as either an early or late event in the course of the disease, and are associated with rapid progression, and a poor outcome. In t-MDS, TP53 mutations are associated with −5/del(5q) and a complex karyotype.
JAK2V617F is a constitutively active cytoplasmic tyrosine kinase that is able to activate JAK-STAT signalling and mediate transformation to cytokine independent growth in MPN, and has been identified in rare cases of MDS (2–5%) and CMML (3%).85 An exception is RARS-T, in which 60% of patients have the JAK2V617F mutation.86 RARS-T patients with JAK2V617F mutations present with higher WBCs and platelet counts.
The role of epigenetic changes in the pathogenesis and treatment of MDS is becoming increasingly important. Transcriptional silencing via DNA methylation of the CDKN2B (p15INK4B) gene increases with progression from RA to RAEB-T, and is observed in a high percentage of patients with t-MDS, and is associated with −7/del(7q), and a poor prognosis.64 Recent genome-wide studies have demonstrated that increases in promoter hypermethylation are predictive of survival in MDS, even when age, sex, and IPSS risk groups are considered. Moreover, increases in promoter methylation are seen during progression to AML.27 These observations form the rationale for use of demethylating agents in MDS. Similarly, inhibition of histone-modifying enzymes represents another rational target for MDS therapy.
Emerging Technologies
Recent advances in microarray technology have enabled high-resolution genome-wide genotyping using single nucleotide polymorphisms (SNPs) for the identification of disease susceptibility loci, as well as the identification of acquired genetic imbalances, and loss of heterozygosity that occurs without concurrent changes in the gene copy number, which can be attributed to somatic mitotic recombination (referred to as copy-neutral LOH or acquired uniparental disomy). Gondek et al. used high-density 250K arrays to examine 174 patients (94, MDS; 33, AML following MDS; 47, MDS/MPD).87 Acquired, copy-neutral LOH was identified in 20% of MDS, 23% of MDS->AML, and 35% of MDS/MPD (particularly CMML). Collectively, abnormalities were detected in a higher proportion of cases as compared to conventional cytogenetic analysis (78% vs. 59% for MDS). New lesions detected by microarray analysis included copy-neutral LOH of 6p21.2-pter, 11q13.5-qter, 4q23-qter, 7q11.23-qter, and 7q22.1. When the presence of newly-identified SNP array lesions were factored into the IPSS classification, the survival curves diverged for patients originally classified as IPSS Intermediate- 1, suggesting that SNP arrays provide additional information allowing for better prognostic resolution (median survival 28 vs. 9 mos., p=0.03). Thus, the results of these studies suggest that SNP array analysis may have future diagnostic application, and may complement conventional cytogenetic analysis in risk stratification and the selection of therapy.
SUMMARY
Cytogenetic analysis in MDS remains a critical genetic test for establishing the diagnosis and prognosis, and for therapeutic decision-making. The IPSS and the WHO classification systems incorporate only the most common chromosomal abnormalities. An international effort is underway to develop a comprehensive cytogenetic scoring system for MDS that incorporates rare cytogenetic subsets, which will inform an ongoing revision of the IPSS.23 With the advent of more sensitive techniques already available in the research setting, including next generation genome and transcriptome sequencing and SNP arrays, the rate of discovery will accelerate, and the compendium of genetic alterations in MDS will undoubtedly expand. Defining the genetic complexity of MDS holds tremendous promise for elucidating the pathogenesis of these diseases, refining the prognostic scoring systems, and identifying novel therapeutic targets.88
Acknowledgments
This work was supported by Grant No. CA40046 from the National Cancer Institute.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors have nothing to disclose
REFERENCES
- 1.Brunning RD, Orazi A, Germing U, et al. Myelodysplastic Syndromes/Neoplasms, Overview. In: Swerdlow SH, Campo E, Harris NL, et al., editors. Classification WHO of Tumors of Haematopoietic, Tissues Lymphoid. Lyons: IARC; 2008. 2008. [Google Scholar]
- 2.Cazzola M, Malcovati L. Myelodysplastic syndromes--coping with ineffective hematopoiesis. N Engl J Med. 2005 Feb 10;352(6):536–538. doi: 10.1056/NEJMp048266. [DOI] [PubMed] [Google Scholar]
- 3.Sekeres MA, Schoonen WM, Kantarjian H, et al. Characteristics of US patients with myelodysplastic syndromes: results of six cross-sectional physician surveys. J Natl Cancer Inst. 2008 Nov 5;100(21):1542–1551. doi: 10.1093/jnci/djn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007 Dec 15;110(13):4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 5.Olney HJ, Le Beau MM. Evaluation of recurring cytogenetic abnormalities in the treatment of myelodysplastic syndromes. Leuk Res. 2007 Apr;31(4):427–434. doi: 10.1016/j.leukres.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Lichtman MA. Myelodysplasia or myeloneoplasia: thoughts on the nosology of clonal myeloid diseases. Blood Cells Mol Dis. 2000 Dec;26(6):572–581. doi: 10.1006/bcmd.2000.0335. [DOI] [PubMed] [Google Scholar]
- 7.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982 Jun;51(2):189–199. [PubMed] [Google Scholar]
- 8.Germing U, Gattermann N, Minning H, Heyll A, Aul C. Problems in the classification of CMML--dysplastic versus proliferative type. Leuk Res. 1998 Oct;22(10):871–878. doi: 10.1016/s0145-2126(97)00192-6. [DOI] [PubMed] [Google Scholar]
- 9.Voglova J, Chrobak L, Neuwirtova R, Malaskova V, Straka L. Myelodysplastic and myeloproliferative type of chronic myelomonocytic leukemia--distinct subgroups or two stages of the same disease? Leuk Res. 2001 Jun;25(6):493–499. doi: 10.1016/s0145-2126(00)00159-4. [DOI] [PubMed] [Google Scholar]
- 10.Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Third Edition: Lyon: International Agency for Research on Cancer (IARC) 2001 [Google Scholar]
- 11.Rosati S, Mick R, Xu F, et al. Refractory cytopenia with multilineage dysplasia: further characterization of an ‘unclassifiable’ myelodysplastic syndrome. Leukemia. 1996 Jan;10(1):20–26. [PubMed] [Google Scholar]
- 12.Germing U, Strupp C, Kuendgen A, et al. Prospective validation of the WHO proposals for the classification of myelodysplastic syndromes. Haematologica. 2006 Dec;91(12):1596–1604. [PubMed] [Google Scholar]
- 13.Foucar K. Myelodysplastic/myeloproliferative neoplasms. Am J Clin Pathol. 2009 Aug;132(2):281–289. doi: 10.1309/AJCPJ71PTVIKGEVT. [DOI] [PubMed] [Google Scholar]
- 14.Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Fourth Edition. Lyon: IARC Press; 2008. [Google Scholar]
- 15.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009 Jul 30;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 16.Boultwood J, Lewis S, Wainscoat JS. The 5q- syndrome. Blood. 1994 Nov 15;84(10):3253–3260. [PubMed] [Google Scholar]
- 17.Garcia-Manero G, Fenaux P. Hypomethylating agents and other novel strategies in myelodysplastic syndromes. J Clin Oncol. 2011 Feb 10;29(5):516–523. doi: 10.1200/JCO.2010.31.0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997 Mar 15;89(6):2079–2088. [PubMed] [Google Scholar]
- 19.Kantarjian H, O’Brien S, Ravandi F, et al. Proposal for a new risk model in myelodysplastic syndrome that accounts for events not considered in the original International Prognostic Scoring System. Cancer. 2008 Sep 15;113(6):1351–1361. doi: 10.1002/cncr.23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malcovati L, Germing U, Kuendgen A, et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J Clin Oncol. 2007 Aug 10;25(23):3503–3510. doi: 10.1200/JCO.2006.08.5696. [DOI] [PubMed] [Google Scholar]
- 21.Morel P, Declercq C, Hebbar M, Bauters F, Fenaux P. Prognostic factors in myelodysplastic syndromes: critical analysis of the impact of age and gender and failure to identify a very-low-risk group using standard mortality ratio techniques. Br J Haematol. 1996 Jul;94(1):116–119. doi: 10.1046/j.1365-2141.1996.6122056.x. [DOI] [PubMed] [Google Scholar]
- 22.Alessandrino EP, Della Porta MG, Bacigalupo A, et al. WHO classification and WPSS predict posttransplantation outcome in patients with myelodysplastic syndrome: a study from the Gruppo Italiano Trapianto di Midollo Osseo (GITMO) Blood. 2008 Aug 1;112(3):895–902. doi: 10.1182/blood-2008-03-143735. [DOI] [PubMed] [Google Scholar]
- 23.Schanz J, Tuechler H, Sole F, et al. Cytogenetic risk features in MDS-Update and present state. Blood (ASH annual meeting abstracts) 2009:114. [Google Scholar]
- 24.Schanz J, Steidl C, Fonatsch C, et al. Coalesced multicentric analysis of 2,351 patients with myelodysplastic syndromes indicates an underestimation of poor-risk cytogenetics of myelodysplastic syndromes in the international prognostic scoring system. J Clin Oncol. 2011 May 20;29(15):1963–1970. doi: 10.1200/JCO.2010.28.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011 Feb 10;29(5):504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueroa ME, Skrabanek L, Li Y, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009 Oct 15;114(16):3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen L, Kantarjian H, Guo Y, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28(18):3098. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gozzetti A, Le Beau MM. Fluorescence in situ hybridization: uses and limitations. Semin Hematol. 2000 Oct;37(4):320–333. doi: 10.1016/s0037-1963(00)90013-1. [DOI] [PubMed] [Google Scholar]
- 29.Cherry AM, Brockman SR, Paternoster SF, et al. Comparison of interphase FISH and metaphase cytogenetics to study myelodysplastic syndrome: an Eastern Cooperative Oncology Group (ECOG) study. Leuk Res. 2003 Dec;27(12):1085–1090. doi: 10.1016/s0145-2126(03)00104-8. [DOI] [PubMed] [Google Scholar]
- 30.Wiktor A, Rybicki BA, Piao ZS, et al. Clinical significance of Y chromosome loss in hematologic disease. Genes Chromosomes Cancer. 2000 Jan;27(1):11–16. [PubMed] [Google Scholar]
- 31.Hasle H. Myelodysplastic and myeloproliferative disorders in children. Curr Opin Pediatr. 2007 Feb;19(1):1–8. doi: 10.1097/MOP.0b013e3280128ce8. [DOI] [PubMed] [Google Scholar]
- 32.Paulsson K, Sall T, Fioretos T, Mitelman F, Johansson B. The incidence of trisomy 8 as a sole chromosomal aberration in myeloid malignancies varies in relation to gender, age, prior iatrogenic genotoxic exposure, and morphology. Cancer Genet Cytogenet. 2001 Oct 15;130(2):160–165. doi: 10.1016/s0165-4608(01)00486-1. [DOI] [PubMed] [Google Scholar]
- 33.Sole F, Espinet B, Sanz GF, et al. Incidence, characterization prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Grupo Cooperativo Espanol de Citogenetica Hematologica. Br J Haematol. 2000 Feb;108(2):346–356. doi: 10.1046/j.1365-2141.2000.01868.x. [DOI] [PubMed] [Google Scholar]
- 34.Kurtin PJ, Dewald GW, Shields DJ, Hanson CA. Hematologic disorders associated with deletions of chromosome 20q: a clinicopathologic study of 107 patients. Am J Clin Pathol. 1996 Nov;106(5):680–688. doi: 10.1093/ajcp/106.5.680. [DOI] [PubMed] [Google Scholar]
- 35.Bench AJ, Nacheva EP, Hood TL, et al. Chromosome 20 deletions in myeloid malignancies: reduction of the common deleted region, generation of a PAC/BAC contig and identification of candidate genes. UK Cancer Cytogenetics Group (UKCCG) Oncogene. 2000 Aug 10;19(34):3902–3913. doi: 10.1038/sj.onc.1203728. [DOI] [PubMed] [Google Scholar]
- 36.Wang PW, Eisenbart JD, Espinosa R, 3rd, Davis EM, Larson RA, Le Beau MM. Refinement of the smallest commonly deleted segment of chromosome 20 in malignant myeloid diseases and development of a PAC-based physical and transcription map. Genomics. 2000 Jul 1;67(1):28–39. doi: 10.1006/geno.2000.6215. [DOI] [PubMed] [Google Scholar]
- 37.Godley LA, Larson R. J.M. Bennett. New York: Marcel Dekker Inc.; 2002. The syndrome of therapy-related myelodysplasia and myeloid leukemia. The Myelodysplastic Syndromes: Pathobiology and Clinical Management; pp. 136–176. [Google Scholar]
- 38.Allan JM, Travis LB. Mechanisms of therapy-related carcinogenesis. Nat Rev Cancer. 2005 Dec;5(12):943–955. doi: 10.1038/nrc1749. [DOI] [PubMed] [Google Scholar]
- 39.Smith SM, Le Beau MM, Huo D, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003 Jul 1;102(1):43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 40.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001 Mar 1;19(5):1405–1413. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- 41.Side LE, Curtiss NP, Teel K, et al. RAS, FLT3, and TP53 mutations in therapy-related myeloid malignancies with abnormalities of chromosomes 5 and 7. Genes Chromosomes Cancer. 2004 Mar;39(3):217–223. doi: 10.1002/gcc.10320. [DOI] [PubMed] [Google Scholar]
- 42.List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006 Oct 5;355(14):1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 43.Jadersten M, Saft L, Smith A, et al. TP53 Mutations in Low-Risk Myelodysplastic Syndromes With del(5q) Predict Disease Progression. J Clin Oncol. 2011 May 20;29(15):1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 44.Wei S, Chen X, Rocha K, et al. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12974–12979. doi: 10.1073/pnas.0811267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tehranchi R, Woll PS, Anderson K, et al. Persistent malignant stem cells in del(5q) myelodysplasia in remission. N Engl J Med. 2010 Sep 9;363(11):1025–1037. doi: 10.1056/NEJMoa0912228. [DOI] [PubMed] [Google Scholar]
- 46.Fairman J, Chumakov I, Chinault AC, Nowell PC, Nagarajan L. Physical mapping of the minimal region of loss in 5q- chromosome. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7406–7410. doi: 10.1073/pnas.92.16.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao N, Stoffel A, Wang PW, et al. Molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases to 1–1.5 Mb and preparation of a PAC-based physical map. Proc Natl Acad Sci U S A. 1997 Jun 24;94(13):6948–6953. doi: 10.1073/pnas.94.13.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boultwood J, Fidler C, Strickson AJ, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002 Jun 15;99(12):4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 49.Graubert TA, Payton MA, Shao J, et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS One. 2009;4(2):e4583. doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai F, Godley LA, Joslin J, et al. Transcript map and comparative analysis of the 1.5-Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q) Genomics. 2001 Jan 15;71(2):235–245. doi: 10.1006/geno.2000.6414. [DOI] [PubMed] [Google Scholar]
- 51.Shannon KM, Le Beau MM. cancer: hay in a haystack. Nature. 2008 Jan 17;451(7176):252–253. doi: 10.1038/451252a. [DOI] [PubMed] [Google Scholar]
- 52.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008 Jan 17;451(7176):335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar M, Narla A, Nomami A, et al. Coordinate loss of a microRNA mir145 and a protein-coding gene RPS14 cooperate in the pathogenesis of 5q–Syndrome. Blood. 2009;114:947. doi: 10.1182/blood-2010-12-324715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010 Jan;16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 55.Joslin JM, Fernald AA, Tennant TR, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007 Jul 15;110(2):719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Fernald AA, Anastasi J, Le Beau MM, Qian Z. Haploinsufficiency of Apc leads to ineffective hemopoiesis. Blood. 2010 Apr 29;115(17):3481–3488. doi: 10.1182/blood-2009-11-251835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu TX, Becker MW, Jelinek J, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007 Jan;13(1):78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 58.Chen TH, Kambal A, Krysiak K, et al. Knockdown of Hspa9, a del(5q31.2) gene, results in a decrease in hematopoietic progenitors in mice. Blood. 2011 Feb 3;117(5):1530–1539. doi: 10.1182/blood-2010-06-293167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisenmann KM, Dykema KJ, Matheson SF, et al. 5q- myelodysplastic syndromes: chromosome 5q genes direct a tumor-suppression network sensing actin dynamics. Oncogene. 2009 Oct 1;28(39):3429–3441. doi: 10.1038/onc.2009.207. [DOI] [PubMed] [Google Scholar]
- 60.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010 Jan;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006 Feb;13(2):115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min IM, Pietramaggiori G, Kim FF, Passegue E, Stevenson KE, Wagers AJ. The transciption factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;10:380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 63.Luna-Fineman S, Shannon KM, Lange BJ. Childhood monosomy 7: epidemiology, biology, and mechanistic implications. Blood. 1995 Apr 15;85(8):1985–1999. [PubMed] [Google Scholar]
- 64.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2003 Sep;17(9):1813–1819. doi: 10.1038/sj.leu.2403054. [DOI] [PubMed] [Google Scholar]
- 65.Loh ML, Vattikuti S, Schubbert S, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004 Mar 15;103(6):2325–2331. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 66.Fischer K, Frohling S, Scherer SW, et al. Molecular cytogenetic delineation of deletions and translocations involving chromosome band 7q22 in myeloid leukemias. Blood. 1997 Mar 15;89(6):2036–2041. [PubMed] [Google Scholar]
- 67.Johnson EJ, Scherer SW, Osborne L, et al. Molecular definition of a narrow interval at 7q22.1 associated with myelodysplasia. Blood. 1996 May 1;87(9):3579–3586. [PubMed] [Google Scholar]
- 68.Le Beau MM, Espinosa R, 3rd, Davis EM, Eisenbart JD, Larson RA, Green ED. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996 Sep 15;88(6):1930–1935. [PubMed] [Google Scholar]
- 69.Curtiss NP, Bonifas JM, Lauchle JO, et al. Isolation and analysis of candidate myeloid tumor suppressor genes from a commonly deleted segment of 7q22. Genomics. 2005 May;85(5):600–607. doi: 10.1016/j.ygeno.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 70.Wong JC, Zhang Y, Lieuw KH, et al. Use of chromosome engineering to model a segmental deletion of chromosome band 7q22 found in myeloid malignancies. Blood. 2010 Jun 3;115(22):4524–4532. doi: 10.1182/blood-2009-07-232504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dohner K, Habdank M, Rucker FG, et al. Molecular characterization of distinct hot spot regins on chromosome 7q in myeloid leukemias. Blood. 2006:108. [Google Scholar]
- 72.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010 Aug;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 73.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010 Aug;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 74.Rowley JD, Reshmi S, Sobulo O, et al. All patients with the t(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997 Jul 15;90(2):535–541. [PubMed] [Google Scholar]
- 75.Apperley JF, Gardembas M, Melo JV, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. 2002 Aug 15;347(7):481–487. doi: 10.1056/NEJMoa020150. [DOI] [PubMed] [Google Scholar]
- 76.Olney HJ, Le Beau MM. Myelodysplastic Syndromes. In: Heim S, Mitelman F, editors. Cancer Cytogenetics. 3rd Ed. Hoboken, NJ: John Wiley & Sons; 2008. in press. [Google Scholar]
- 77.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009 Jul;41(7):838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 78.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010 Dec 9;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kosmider O, Gelsi-Boyer V, Cheok M, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009 Oct 8;114(15):3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 80.Gelsi-Boyer V, Trouplin V, Adelaide J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009 Jun;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 81.Majewski IJ, Ritchie ME, Phipson B, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010 Aug 5;116(5):731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]
- 82.Chen CY, Lin LI, Tang JL, et al. RUNX1 gene mutation in primary myelodysplastic syndrome--the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br J Haematol. 2007 Nov;139(3):405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 83.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002 Jul;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 84.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999 Oct;23(2):166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 85.Steensma DP, Dewald GW, Lasho TL, et al. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005 Aug 15;106(4):1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zipperer E, Wulfert M, Germing U, Haas R, Gattermann N. MPL 515 and JAK2 mutation analysis in MDS presenting with a platelet count of more than 500 × 10(9)/l. Ann Hematol. 2008 May;87(5):413–415. doi: 10.1007/s00277-007-0409-0. [DOI] [PubMed] [Google Scholar]
- 87.Gondek LP, Tiu R, O’Keefe CL, Sekeres MA, Theil KS, Maciejewski JP. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008 Feb 1;111(3):1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Odenike O, LeBeau MM. The Dawn of the Molecular Era of the Myelodysplastic Syndromes. New Engl J Med. doi: 10.1056/NEJMe1102921. in press. [DOI] [PubMed] [Google Scholar]