Abstract
Background
Serum biomarkers for diagnosis and risk-stratification of childhood solid tumors would improve the accuracy/timeliness of diagnosis and reduce the need for invasive biopsies. We hypothesized that differential expression and/or release of microRNAs by such tumors may be detected as altered serum microRNA profiles.
Methods
We undertook global quantitative-RT-PCR microRNA profiling (n=741) on RNA from 53 serum samples, representing 33 diagnostic cases of common childhood cancers plus 20 controls. Technical confirmation was performed in a subset of 21 cases, plus four independent samples.
Results
We incorporated robust quality-control steps for RNA extraction, qRT-PCR efficiency and hemolysis quantification. We evaluated multiple methods to normalize global profiling data and identified the ‘global-mean’ approach as optimal. We generated a panel of six microRNAs that were most stable in pediatric serum samples and therefore most suitable for normalization of targeted microRNA qRT-PCR data. Tumor-specific serum microRNA profiles were identified for each tumor type and selected microRNAs underwent confirmatory testing. We identified a panel of microRNAs (miR-124-3p/miR-9-3p/miR-218-5p/miR-490-5p/miR-1538) of potential importance in the clinical management of neuroblastoma, as they were consistently highly over-expressed in MYCN-amplified high-risk cases (MYCN-NB). We also derived candidate microRNA panels for non-invasive differential diagnosis of a liver mass (hepatoblastoma vs. combined MYCN-NB/NB), an abdominal mass (Wilms tumor vs. combined MYCN-NB/NB), and sarcoma subtypes.
Conclusions
This study describes a pipeline for robust diagnostic serum microRNA profiling in childhood solid tumors, and has identified candidate microRNA profiles for prospective testing.
Impact
We propose a new non-invasive method with the potential to diagnose childhood solid tumors.
Keywords: biomarker, blood, diagnosis, microRNA, MYCN, neuroblastoma, serum
Introduction
Current challenges in the clinical management of pediatric solid tumors include a requirement for tissue biopsy at initial diagnosis and difficulties in detecting minimal residual disease. The identification of novel body fluid tumor markers that may allow noninvasive diagnosis, risk stratification or follow-up in solid tumors of childhood would be an important advance. One promising approach is measurement of serum microRNA levels.
MicroRNAs are short, non-protein-coding RNAs that post-transcriptionally regulate gene expression (1). Importantly, global microRNA profiles have been shown to classify human cancer tissues, including pediatric solid tumors (1, 2). In pediatric malignant germ cell tumors, for example, the miR-371~373 and miR-302/367 clusters are over-expressed in all cases, regardless of patient age, histologic subtype or anatomic site (1), thus representing universal biomarkers. Packaging of specific microRNAs in membrane-bound exosome particles by tumor cells before release into the bloodstream allows for their detection in the serum (3). Indeed, we showed that microRNAs from the miR-371~373 and miR-302/367 clusters are detectable at high levels in the serum of patients at the time of malignant GCT diagnosis, and that levels fall with treatment (3-5), findings recently replicated by others (6-9). Importantly, these profiles can detect disease at low tumor volumes, demonstrating the sensitivity of this approach (5). MicroRNAs have many qualities that make them suitable tumor markers for translation into clinical practice, including inherent stability and resistance to degradation, even if samples are left at room temperature or subjected to multiple freeze-thaw cycles (10). We therefore hypothesized that other childhood tumors would be characterized by the release of specific microRNAs into the bloodstream, which would be detectable as an altered serum profile compared with control serum samples.
Here, we report the findings of a proof-of-principle study testing this hypothesis at the time of diagnosis of common childhood cancers, using a quantitative reverse transcription polymerase chain reaction (qRT-PCR) approach. We addressed three important questions: whether it was feasible to extract RNA from the serum of children in sufficient amounts to undertake global microRNA profiling; whether serum housekeeping microRNAs in children would be similar to those described for adults, and whether lists of candidate biomarkers could be generated for validation in larger, prospective studies. We identify serum microRNA profiles specific for each of the 11 types of pediatric solid tumor investigated. In particular, we report a panel of serum microRNAs that segregate MYCN-amplified International Neuroblastoma Risk Group (INRG) (11) high-risk neuroblastoma from non-MYCN-amplified INRG low-risk neuroblastoma and other tumors.
Materials and Methods
The study received approval from the Multicenter Research Ethics Committee (reference 02/4/71) and Local Research Ethics Committee (reference 01/128) and was performed with full informed parental consent. All experimental steps were compliant with the Minimum Information for Publication of Quantitative Real-time PCR Experiments (MIQE) (12).
Patient demographics and tumor types analyzed
For the main discovery-phase of the project, we initially recruited 34 patients aged 0-16 years (y) at the time of malignant tumor diagnosis (between April 2010 and June 2013), together with a further 20 age- and gender-matched anonymized samples from a control group of patients without malignant disease. Clinico-pathological details are listed in Table 1. The 34 tumors were from 11 different tumor types. They comprised i) four neuroblastomas [two MYCN-amplified INRG high-risk tumors (MYCN-NB); two non-MYCN-amplified INRG low-risk tumors (NB)]; ii) four hepatoblastomas (HB); iii) seven Wilms tumors (WT); iv) seven lymphomas [five cases of classical nodular-sclerosing Hodgkin’s disease (HD) and two cases of B-cell non-Hodgkin’s lymphoma (B-NHL)]; v) six sarcomas [three rhabdomyosarcoma (RMS), two Ewings sarcoma (ES), one osteosarcoma (OS)]; vi) one DICER1-mutated pleuropulmonary blastoma (PPB) (13) and vii) five central nervous system tumors (gliomas; three WHO 2007 grade I, one grade II and one grade III) (Table 1).
Table 1. Clinico-pathological characteristics of the patients in the tumor and control groups.
| Sample | Cancer Type | Gender | Age (mo) | Additional Comments | Confirmatory Study |
|---|---|---|---|---|---|
| 1 | MYCN-amplified HR neuroblastoma | M | 98 | Undifferentiated | ✓ |
| 2 | MYCN-amplified HR neuroblastoma | M | 6 | Undifferentiated | ✓ |
| 3 | LR neuroblastoma | M | 67 | Ganglioneuroblastoma | ✓ |
| 4 | LR neuroblastoma | F | 21 | Differentiating | ✓ |
| 5 | Hepatoblastoma | M | 14 | Mixed fetal and embryonal type histology | ✓ |
| 6 | Hepatoblastoma | M | 85 | Mixed fetal and embryonal type histology | ✓ |
| 7 | Hepatoblastoma | F | 25 | Fetal type histology | |
| 8 | Hepatoblastoma | F | 14 | Fetal type histology | ✓ |
| 9 | Wilms tumor | F | 54 | Biopsy - anaplastic histology | ✓ |
| 10 | Wilms tumor | M | 22 | Biopsy -stromal type histology; WAGR | ✓ |
| 11 | Wilms tumor | F | 12 | Biopsy - mixed type histology | |
| 12 | Wilms tumor | F | 25 | Biopsy - triphasic histology. Failed QC. | |
| 13 | Wilms tumor | M | 41 | Bilateral tumors | |
| 14 | Wilms tumor | F | 16 | Biopsy - stromal type histology | |
| 15 | Wilms tumor | F | 97 | Biopsy - triphasic histology | |
| 16 | Rhabdomyosarcoma | M | 121 | Abdominal | ✓ |
| 17 | Rhabdomyosarcoma | M | 52 | Pancreatic site; jaundiced; embryonal type | |
| 18 | Rhabdomyosarcoma | F | 104 | Nasopharyngeal site; embryonal type | ✓ |
| 19 | Ewings sarcoma | M | 152 | Bone and soft tissue of cervical vertebrae | |
| 20 | Ewings sarcoma | M | 34 | Soft tissue of lumbo-sacral region | |
| 21 | Osteosarcoma | F | 103 | Previous RMS/Li Fraumeni syndrome | |
| 22 | B-cell non-Hodgkin’s lymphoma | M | 176 | Burkitt’s lymphoma | ✓ |
| 23 | B-cell non-Hodgkin’s lymphoma | M | 110 | Burkitt’s lymphoma | ✓ |
| 24 | Hodgkin’s disease | F | 186 | Classical, nodular sclerosing subtype | |
| 25 | Hodgkin’s disease | M | 156 | Classical, nodular sclerosing subtype | ✓ |
| 26 | Hodgkin’s disease | M | 176 | Classical, nodular sclerosing subtype | ✓ |
| 27 | Hodgkin’s disease | F | 159 | Classical, nodular sclerosing subtype | |
| 28 | Hodgkin’s disease | F | 172 | Classical, nodular sclerosing subtype | |
| 29 | Pleuropulmonary blastoma | F | 133 | Germline DICER1 mutation | |
| 30 | Central nervous system glioma | M | 53 | Pilocytic astrocytoma WHO 2007 grade I | ✓ |
| 31 | Central nervous system glioma | F | 68 | Pilocytic astrocytoma WHO 2007 grade I | |
| 32 | Central nervous system glioma | M | 172 | Gliomatosis cerebri, WHO 2007 grade III | |
| 33 | Central nervous system glioma | F | 17 | Oligodendroglioma WHO 2007 grade II | |
| 34 | Central nervous system glioma | M | 59 | Pilocytic astrocytoma WHO 2007 grade I | ✓ |
| 35 | MYCN-amplified HR neuroblastoma | F | 17 | Undifferentiated; abdominal | ✓ |
| 36 | MYCN-amplified HR neuroblastoma | M | 32 | Undifferentiated; abdominal | ✓ |
| 37 | LR neuroblastoma | M | 5 | Poorly differentiated | ✓ |
| 38 | LR neuroblastoma | M | 3 | Poorly differentiated | ✓ |
| 39 | Control | F | 38 | N/A | |
| 40 | Control | M | 159 | N/A | |
| 41 | Control | F | 34 | N/A | ✓ |
| 42 | Control | F | 142 | N/A | ✓ |
| 43 | Control | F | 129 | N/A | |
| 44 | Control | M | 23 | N/A | |
| 45 | Control | F | 21 | N/A | |
| 46 | Control | M | 57 | N/A | |
| 47 | Control | M | 34 | N/A | |
| 48 | Control | F | 38 | N/A | ✓ |
| 49 | Control | M | 134 | N/A | |
| 50 | Control | M | 48 | N/A | |
| 51 | Control | M | 74 | N/A | |
| 52 | Control | F | 140 | N/A | |
| 53 | Control | M | 181 | N/A | |
| 54 | Control | M | 29 | N/A | |
| 55 | Control | M | 26 | N/A | |
| 56 | Control | F | 50 | N/A | |
| 57 | Control | M | 36 | N/A | ✓ |
| 58 | Control | F | 86 | N/A |
Abbreviations: MYCN-NB = MYCN-amplified high-risk neuroblastoma; NB = non-MYCN-amplified low-risk neuroblastoma; HB = hepatoblastoma; WT = Wilms tumor; RMS = rhabdomyosarcoma; ES = Ewing’s sarcoma; OS = osteosarcoma; B-NHL = B-cell non-Hodgkin’s lymphoma; HD = Hodgkin’s disease; PPB = pleuropulmonary blastoma; GL = Central nervous system glioma; C = control samples. Samples 35-38 were independent samples used for the technical confirmation study only.
For the technical confirmation phase of the study, we used 25 samples. These comprised 17 representative tumor and four control serum samples from the discovery-phase test set (Table 1), plus four independent neuroblastoma serum samples from two MYCN-NB and two NB cases. In total, the confirmatory set comprised four MYCN-NB samples, four NB, three HB, two WT, two RMS, two B-NHL, two HD, two gliomas and four control samples (Table 1). There was no difference in tumor volume between the MYCN-NB and NB cases. While all four of the MYCN-NB cases were undifferentiated, the grade of the NB cases varied from poorly differentiated to differentiating and ganglioneuroblastoma (Table 1).
Data normalization
Samples were obtained and processed as described (3). MicroRNAs were quantified as described (13) using microRNA Ready-to-Use PCR Human Panels I&II (Exiqon, Vedbaek, Denmark). We performed extensive quality control (QC) steps to measure hemolysis and the efficiencies of RNA extraction, reverse transcription and qRT-PCR. A detailed account of this assay pipeline is available in the Supplementary Materials and Methods. Only a single sample (WT-4) failed initial QC; 53 samples (98%) therefore underwent full qRT-PCR profiling. In initial work, we compared a number of different data normalization methods (14) to define the optimal approach to our global serum microRNA profiling study. We assessed:
a) the global mean method, which used the average Ct value of microRNAs expressed in at least one of the 53 samples analyzed, as described (14);
b) the modified global mean method, which did not take into account other samples in the dataset and instead used a sample-specific normalization factor plotted on a linear scale (14);
c) the modified global mean method of common microRNAs (14), i.e. those microRNAs expressed in all 53 samples analyzed;
d) the ten top-ranking microRNAs identified as most stably expressed in the study by geNorm (15);
e) the ten top-ranking microRNAs identified as most stably expressed in the study by NormFinder (16);
f) the six top-ranking overlapping housekeeping microRNAs identified in both d) and e) above;
g) the four housekeeping microRNAs used for initial QC purposes (hkgsQC), namely miR-23a-3p, miR-30c-5p, miR-103a-3p and miR-191-5p;
h) the two small nucleolar RNAs (snoRNAs), RNU38B and RNU49A, present on the Exiqon platform;
i) the single small nuclear RNA (snRNA) RNU6, present on the platform. Both snoRNAs and snRNAs are commonly used for microRNA normalization purposes in tissue samples (14).
Statistical analyses for discovery-phase serum microRNA quantification
Following normalization, we removed miRPlus sequences and microRNAs listed as obsolete according to miRBase (www.mirbase.org/). MicroRNA levels were then quantified using the delta Ct method, with fold-change = 2(Tumor-Ct – mean ‘other tumor’ samples-Ct) and 2(Tumor-Ct – mean control samples-Ct). MicroRNAs that had a ≥2.0 fold-change in expression in the tumor type under consideration compared with the mean expression value from a) the control group and b) the ‘other tumor’ group (comprising all other tumor samples except those under consideration) were called as over-expressed and ranked according to fold-change, as described (13).
Additional analyses were also performed to increase the stringency of our findings further. Firstly, it was necessary for a microRNA to be detected at least 2 Ct values lower in the test group compared with the ‘no template control’ (NTC) sample to be included in the subsequent data analysis. Secondly, a co-efficient of variation (CV) was calculated for each over-expressed microRNA, using the formula CV = standard deviation for that microRNA/n, where n was the mean expression value of all microRNAs within that tumor type. This step identified the level of variation in microRNA expression within the specific tumor type and between the control and ‘other tumor’ group. Thirdly, the Robust Rank Aggregate (RRA) method was employed, as described (17) (Supplementary Materials and Methods). Adjusted RRA scores of p<0.01 were considered significant. Importantly, the RRA method has been used by others in meta-analysis of cancer datasets (18). Differences in serum microRNA expression levels between experimental groups were assessed using a two-tailed t-test (p<0.05 significant).
Serum microRNA qRT-PCR for technical confirmation
To maximise sensitivity in the technical confirmation study, cDNA was diluted 1:7.5, rather than the standard 1:50 dilution. For this targeted work, following QC analysis (Supplementary Materials and Methods), data were normalized using the six top-ranking overlapping housekeeping microRNAs identified in the overlap between the geNorm and NormFinder methods in the full discovery-phase qRT-PCR profiling study. Differences in serum microRNA expression levels between experimental groups were assessed using a two-tailed t-test (p<0.05 significant).
Results
QC and normalization
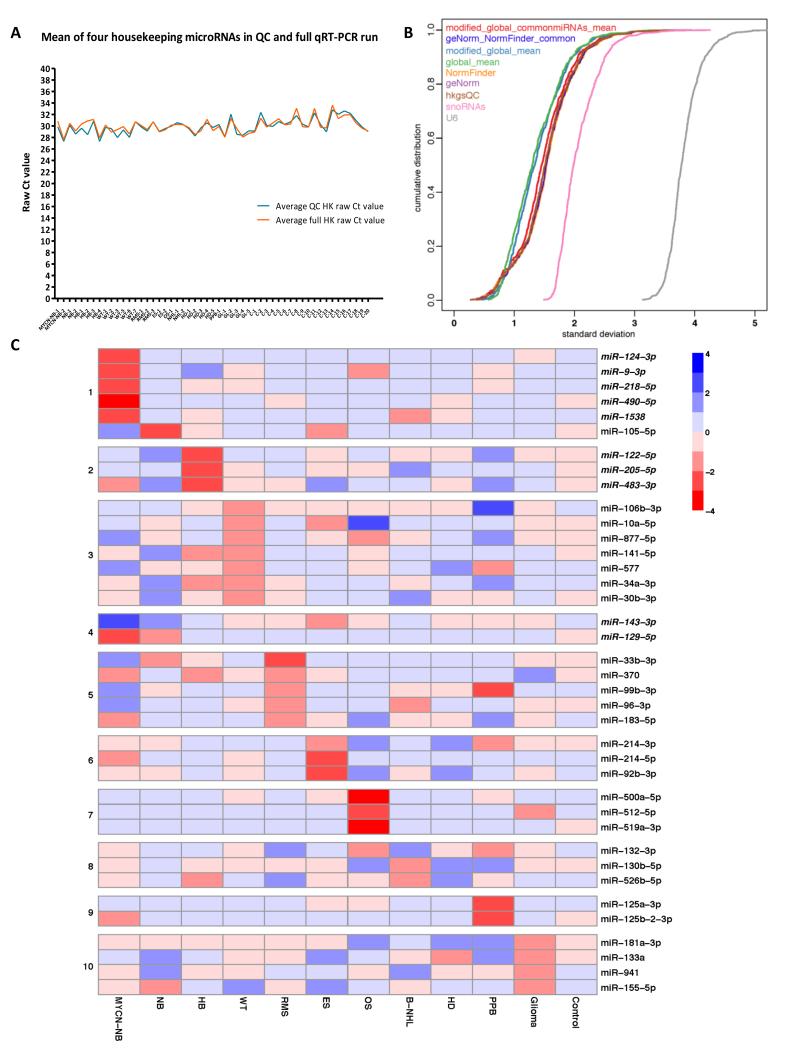
For full details of the QC steps and the assay pipeline developed, see Supplementary Results and Supplementary Figures-S1 to -S5. In the discovery-phase global qRT-PCR profiling study, expression of the four housekeeping microRNAs (19) and their mean raw Ct values for all samples were very similar to the levels obtained in our initial QC qRTPCR (Figure 1A), and were confirmed by linear regression analysis (p<0.0001). The global mean method, using the average Ct value of microRNAs expressed in at least one of the 53 samples analyzed (n=568), was the optimal normalization approach (Figure 1B) and was therefore used in the global profiling study. No additional benefit was identified by using either the modified global mean method or the modified global mean method of common microRNAs (Figure 1B). Use of the single snRNA RNU6 (U6), or the two snoRNAs RNU38B and RNU49A (snoRNAs), whilst often used for normalizing microRNA expression in tissue samples (14), were not appropriate methods for normalizing serum data, as they resulted in increased technical variation of the data compared with other normalization methods (Figure 1B). Assessment of the ten top-ranking microRNAs using the geNorm and NormFinder algorithms showed only marginal inferiority to the global mean method as a normalization approach.
Figure 1. Discovery-phase global qRT-PCR profiling.
A) Comparison of serum housekeeping microRNA expression (miR-23a-3p, miR-30c-5p, miR-103a-3p, and miR-191-5p) in the initial QC and full discovery-phase qRT-PCR analyses. Key: QC = quality control; HK = housekeeping. For tumor abbreviations please refer to Table 1. B) Assessment of normalization approaches for global microRNA profiling in serum samples from pediatric cancer patients and controls using a cumulative distribution graph. C) Heatmap showing overall expression levels of selected serum microRNAs identified by global qRT-PCR profiling. Key: Selected serum microRNA signature for: panel 1=MYCN-NB vs. NB; 2=HB vs. MYCN-NB/NB; 3=WT; 4=MYCN-NB/NB vs. WT; 5/6/7=RMS, ES and OS, respectively, vs. the other two sarcoma subtypes; 8=HD vs. B-NHL; 9=PPB; 10=glioma. For tumor abbreviations see Table 1. Italicized microRNAs were those used for the technical confirmation study. Red=microRNA over-expression; blue=microRNA under-expression.
Normalization using the six top-ranking housekeeping microRNAs common to the top-ten lists generated using NormFinder and geNorm [namely miR-140-3p (chromosomal locus 16q22.1), miR-30b-5p (8q24.22), miR-26a-5p (3p22.2), miR-15b-5p (3q25.33), miR-30c-5p (6q13) and miR-191-5p (3p21.31)] (Supplementary Figure-S6) also performed well in our analysis (Figure 1B). Consequently, these six microRNAs were used for normalization purposes for the technical confirmation phase of the study. Five of these microRNAs are also abundant and stably expressed in serum/plasma in adult patients (19). The four housekeeping microRNAs used in the initial QC checks (hkgsQC) also performed well for normalization purposes in the full discovery-phase profiling study (Figure 1B), indicating their suitability for screening serum samples.
Serum microRNAs in different tumor types
No consistent profile of microRNA over-expression common to all tumor groups was identified when compared with the control group. However, for each of the 11 tumor types studied, we identified microRNAs that were over-expressed compared with both the cohort of other childhood tumors and the control group. The number of such microRNAs ranged from 22-49 (mean 34), depending on tumor type (Supplementary Tables-S1 and -S2). The RRA method refined this to 16-26 (mean 21) serum microRNAs in each tumor group (Supplementary Table-S1). As the MYCN-NB group had the largest coefficient of variance for the delta Ct(miR-23a-3p-miR-451a) hemolysis levels (Supplementary Figure-S1E), we only included microRNAs that were ≥2 fold greater than the ‘other tumor’ group and control group in both of the MYCN-NB samples being interrogated. This reduced the number of microRNAs called as over-expressed in MYCN-NB from 96 to 35 (Supplementary Table-S1), avoiding identification of serum microRNAs that were released from red blood cells rather than being disease-associated.
The lists of over-expressed serum microRNAs for each tumor group are shown in Supplementary Tables-S3 to -S13. Selected findings for the individual tumor groups were used to generate an expression heatmap (Figure 1C). MicroRNAs for the heatmap were chosen based on their overall abundance and potential value in differential diagnosis. For two microRNAs (miR-122 and miR-877), the -5p strands were selected rather than the -3p strands identified in the global profiling study, due to the greater abundance of the -5p strands (20) (Supplementary Figure-S7). Ten microRNAs from the heatmap were chosen for subsequent validation in the technical confirmation study (italics in Figure 1C; Supplementary Table-S2), along with the six top-ranking housekeeping microRNAs that overlapped in the NormFinder and geNorm lists. In the technical confirmation study, the 1:7.5 cDNA dilution increased sensitivity and did not inhibit the PCR reaction. As expected, the 16 microRNAs were detected at approximately 3 Ct values lower in the 1:7.5 dilutions compared with the 1:50 dilutions (Supplementary Figure-S8). The suitability of all 25 samples for the technical confirmation study was verified by assessment of triplicate UniSp6 values (Supplementary Figure-S9A). Expression levels of the six housekeeping microRNAs were assessed in technical triplicate and showed high consistency (Supplementary Figure-S9B).
Taken together, our data strongly suggest important clinical applications based on serum microRNA quantification. The most promising are described in the following sections.
MYCN-amplified high-risk neuroblastoma (MYCN-NB) vs. non-MYCN-amplified low-risk neuroblastoma (NB)
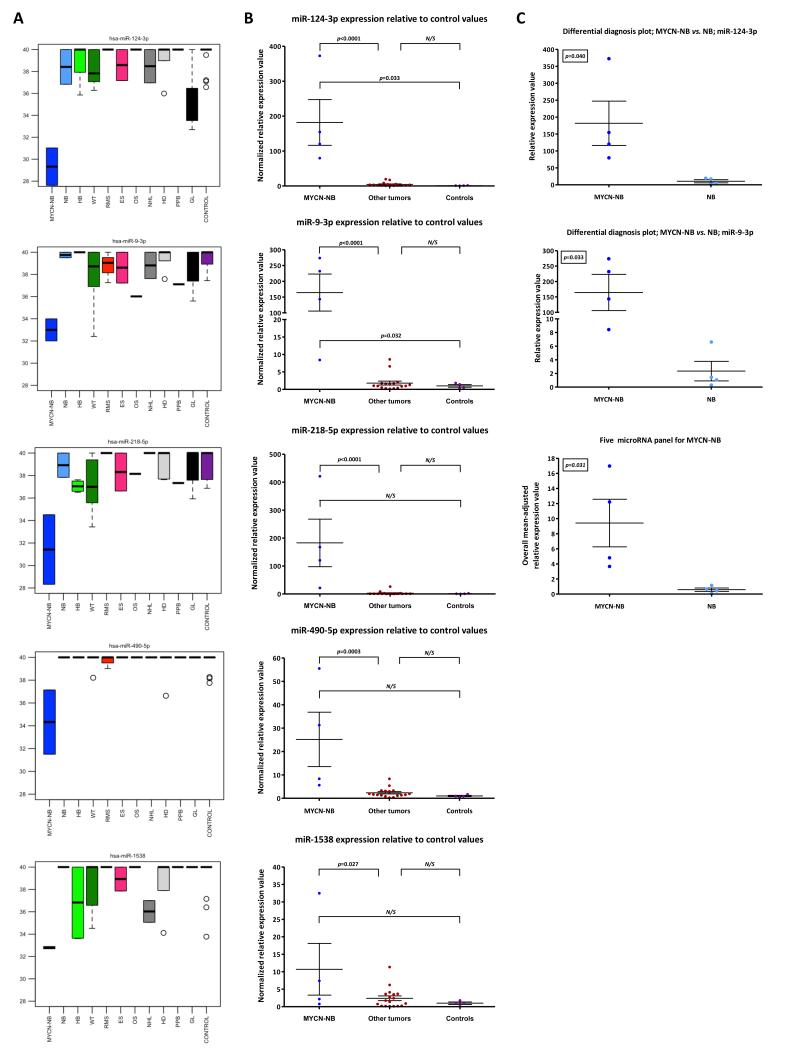
Treatment schedules for neuroblastoma rely on distinguishing tumors by the presence or absence of INRG high-risk molecular abnormalities, such as MYCN-amplification (11). Interestingly, our most striking findings were found in the MYCN-NB group. The five over-expressed serum microRNAs (miR-124-3p/miR-9-3p/miR-218-5p/miR-490-5p/miR-1538) that were top-ranking compared with both the ‘other tumor’ group (including NB samples) and the controls (Supplementary Table-S3) are shown in the heatmap (Figure 1C, panel 1). Levels of these five microRNAs in individual tumor types in the discovery-phase test set are highlighted in boxplot analysis (Figure 2A). Significant over-expression of all five microRNAs in the MYCN-NB samples versus the ‘other tumor’ group was shown in the subsequent confirmatory qRT-PCR experiments, performed in technical triplicate (p<0.05 for all comparisons) (Figure 2B). In a focussed ‘differential diagnosis’ analysis of the MYCN-NB versus the NB group in the confirmatory study, there was a significant difference in expression levels for miR-124-3p and miR-9-3p individually (p<0.05) (Figure 2C). In addition, in the technical confirmation set, the panel of all five microRNAs successfully distinguished MYCN-NB from NB samples (p=0.031) (Figure 2C).
Figure 2. Serum microRNA expression in MYCN-amplified high-risk neuroblastoma (MYCN-NB).
A) Raw Ct values (y-axis) of five highly over-expressed microRNAs in MYCN-NB (miR-124-3p/miR-9-3p/miR-218-5p/miR-490-5p/miR-1538) relative to other tumors and controls in the discovery-phase test set. B) Mean normalized relative expression values for the microRNAs listed in A) above, in MYCN-NB samples versus ‘other tumor’ and control samples, in the technical confirmation qRT-PCR study. C) ‘Mean normalized relative expression values for miR-124-3p and miR-9-3p in MYCN-NB samples versus NB samples, in the technical confirmation qRT-PCR study. In addition, an equally-weighted overall mean-adjusted relative expression value for the specific five microRNA panel was calculated. Key: error bars=standard error of the mean (SEM). For tumor abbreviations see Table 1.
Hepatoblastoma (HB) vs. all neuroblastomas (MYCN-NB/NB)
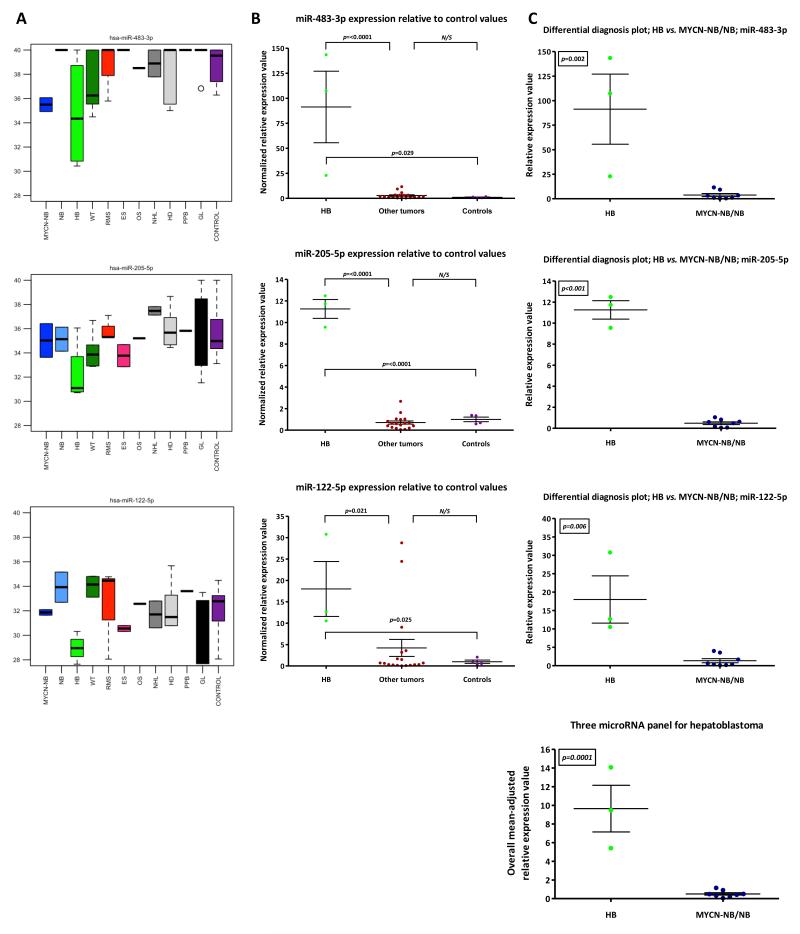
In children presenting with a liver tumor/enlargement, it may be important to distinguish a primary lesion, such as HB, from involvement by a tumor from elsewhere e.g. neuroblastoma. For HB, of the 49 over-expressed serum microRNAs, miR-483-3p, miR-122-3p and miR-205-5p were ranked in the top ten by fold-change (Supplementary Table-S5), with similar fold-changes versus the controls and ‘other tumor’ group. Due to its greater abundance (Supplementary Figure-S7) (20), miR-122-5p, rather than miR-122-3p, was selected for confirmatory testing along with miR-483-3p and miR-205-5p (Figure 1C, panel 2). The findings for these three microRNAs in individual tumor types in the discovery-phase test set are highlighted by boxplot analysis (Figure 3A). Significant over-expression of each of the three microRNAs in the HB group was verified in the technical confirmation study, versus both the ‘other tumor’ group (which included all eight MYCNNB/NB samples, two of which showed liver involvement) and the control group (p<0.05 for all comparisons) (Figure 3B). For the neuroblastomas, there was no association between liver involvement and greater abundance of the HB-associated microRNAs. It was however noted that levels of the liver-specific miR-122-5p (21) were occasionally increased in non-HB samples, for example, in a case of pancreatic RMS presenting with obstructive jaundice (RMS-2) (Figure 3B). Accordingly, miR-122-5p needed to be a part of a larger panel to ensure sufficient specificity for HB. In focussed ‘differential diagnosis’ plots for the HB group versus the combined MYCN-NB/NB group, all three microRNAs were individually significantly elevated in HB (p<0.05 for all comparisons) (Figure 3C). Furthermore, the three microRNA panel also distinguished HB from MYCN-NB/NB (p=0.0001) (Figure 3C).
Figure 3. Serum microRNA expression in hepatoblastoma (HB) vs. all neuroblastomas (MYCN-NB/NB).
A) Raw Ct values (y-axis) of three highly over-expressed microRNAs in HB (miR-483-3p, miR-205-5p and miR-122-5p) relative to other tumors and controls in the discovery-phase test set. B) Mean normalized relative expression values for the microRNAs listed in A) above, in HB samples versus the ‘other tumors’ and control samples, in the confirmatory qRT-PCR study. C) Mean normalized relative expression values for the microRNAs listed in A) above, in HB samples versus MYCN-NB/NB samples, in the technical confirmation qRT-PCR study. In addition, an equally-weighted overall mean-adjusted relative expression value for the specific three microRNA panel was calculated. Key: error bars=SEM. For tumor abbreviations see Table 1.
Wilms tumor (WT) vs. all neuroblastomas (MYCN-NB/NB)
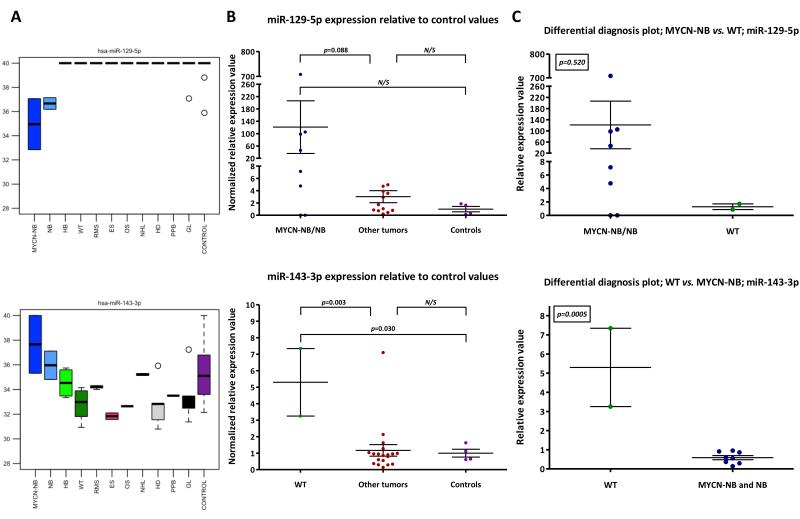
Due to their anatomical proximity, it may be clinically and radiologically difficult to distinguish WT from neuroblastoma; indeed, some neuroblastomas may be intra-renal (22, 23). Due to the different management and prognosis of these two tumor types, it is important to differentiate them diagnostically (23). We screened our profiling data from the discovery-phase test set for microRNAs that might be informative in this differential diagnosis setting. We identified miR-129-5p (over-expressed in both the MYCN-NB and NB lists; Supplementary Tables-S3 and -S4, respectively) and miR-143-3p (10.7 greater fold-change in levels in WT compared with the combined MYCN-NB/NB samples; data not shown) for subsequent testing (Figure 1C, panel 4 and Figure 4A). The technical confirmation study established that miR-143-3p was increased in WT compared with the ‘other tumor’ group (p=0.003), with miR-129-5p being over-expressed in the majority of MYCN-NB/NB cases (Figure 4B). In ‘differential diagnosis’ plots, miR-143-3p distinguished WT from MYCN-NB/NB (p=0.0005), with miR-129-5p being over-expressed in six of eight MYCN-NB/NB cases versus WT (Figure 4C).
Figure 4. Serum microRNA expression in Wilms tumor (WT) vs. all neuroblastomas (MYCN-NB/NB).
A) Raw Ct values (y-axis) of two microRNAs (miR-129-5p and miR-143-3p) relative to other tumors and controls in the discovery-phase test set. B) and C) Mean normalized relative expression values in MYCNNB/NB (miR-129-5p) or WT (miR-143-3p) samples versus B) ‘other tumors’ and control samples or C) each other, in the technical confirmation qRT-PCR study. Key: error bars=SEM. For tumor abbreviations see Table 1.
Sarcoma differential diagnosis
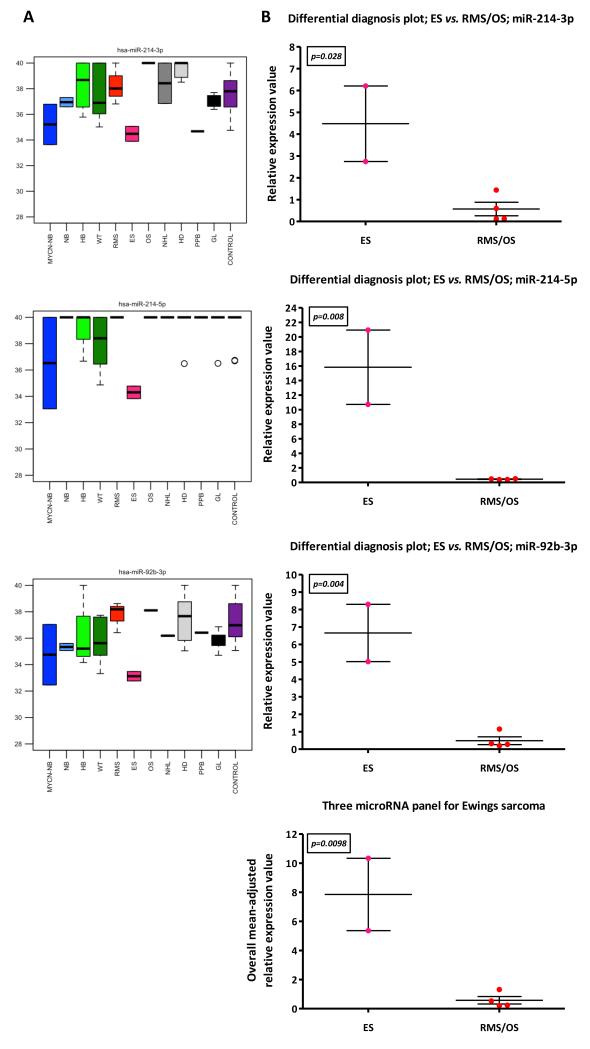
Sarcomas may present with bone lesions (ES/OS), soft tissue masses (RMS/ES), or both (ES). Consequently, discriminating these tumors is important, but may be challenging. Serum microRNAs that were over-expressed for RMS, ES and OS are listed in Supplementary Tables-S7-S9, respectively. Overall levels of representative microRNAs specific for RMS, ES and OS versus the other sarcoma subtypes are illustrated in Figure 1C, (panels 5-7, respectively). In particular, overall analysis (Figure-5A) and ‘differential diagnosis’ plots of the global profiling data (i.e. discovery-phase test set) (Figure-5B) showed that miR-214-3p, miR-214-5p and miR-92b-3p individually segregated ES from RMS/OS (p<0.05 for all comparisons), as did the three microRNA panel (p=0.0098) (Figure-5B). For OS, miR-500a-5p, miR-512-5p and miR-519a-3p showed much higher serum expression than other tumors, including ES (Supplementary Figure-S10).
Figure-5. Serum microRNA expression in sarcoma subtypes.
A) Raw Ct values (y-axis) of three selected microRNAs (miR-214-3p/miR-214-5p/miR-92b-3p) in Ewings sarcoma samples compared with other sarcoma samples (OS/RMS) in the discovery-phase test set. B) ‘Differential diagnosis’ plots for ES vs. RMS/OS in the discovery-phase test set. In addition, an equally-weighted overall mean-adjusted relative expression value for the specific three microRNA panel was calculated. Key: error bars=SEM. For tumor abbreviations see Table 1.
Discussion
We report a robust, quality controlled pipeline suitable for quantifying serum microRNA levels in pediatric patient cohorts (and potentially adult samples), including the assessment of RNA extraction, degree of sample hemolysis and qRT-PCR efficiency. The described approach minimizes technical alterations and maximizes true biological variation between samples, to allow identification of lists of over-expressed serum microRNAs between study groups, as exemplified here for common solid tumors of childhood. We assessed multiple normalization approaches and identified that for high-throughput global serum microRNA qRT-PCR data, the ‘global mean’ method (14) was optimal. In addition, we generated a panel of six housekeeping microRNAs that were most stable in pediatric serum samples and therefore suitable for normalizing qRT-PCR data in more targeted low-throughput studies. Interestingly, the six microRNAs showed substantial overlap with findings from adult samples (19).
The most striking tumor findings were for MYCN-amplified high-risk neuroblastoma (MYCN-NB). The blood-based microRNA panel identified here has potential clinical value, due to the current requirement for surgical biopsy to confirm the diagnosis of neuroblastoma and test for INRG high-risk genomic changes, such as MYCN-amplification (11). Very recently, qRT-PCR detection of neuroblastoma-specific messenger RNAs (mRNAs) in peripheral blood from children at diagnosis of advanced stage neuroblastoma has been reported, with high levels of TH and PHOXB2 representing clinically useful biomarkers of risk (24). However, potential blood-based mRNA biomarkers can be subject to considerable variation in levels, for technical as well as biological reasons (25, 26). In particular, mRNAs are inherently unstable at room temperature and rapidly degrade in blood samples that are not stored correctly (25, 26). In contrast, serum microRNAs offer particular advantages as blood-based biomarkers as they are not prone to such technical variations (10).
The MYCN-NB-specific serum microRNAs identified here are of biological relevance. It has been known for some time that MYCN amplification status of neuroblastoma tissue samples determines global microRNA profiles (27) and our findings are in keeping with this observation. MiR-124-3p is the most abundant neuronal-specific microRNA and silencing of miR-124 in neuroblastoma cells in vitro resulted in their differentiation (28). In addition, expression of miR-9 is activated by MYCN protein, which directly binds to the promoter region of this microRNA (29). In neuroblastoma tissues, high miR-9 levels correlated with MYCN-amplification, tumor grade and metastatic status (29).
Other tumor-specific serum microRNAs are also of biological significance. For example, miR-122 is highly abundant in liver tissues and is considered liver-specific. Recently, reduced miR-122 expression has been shown in HB compared with normal liver tissue (30). Here, we found elevated serum levels of miR-122-5p in HB samples compared with other tumors and controls. Down-regulation of miR-122 in HB (compared with normal liver tissue) (30), but detection at elevated levels in the serum of HB patients is consistent with observations in other tumors (13), and may reflect passive microRNA leakage from tumor cells. It should be noted that miR-122 is a general non-specific marker of liver injury, and is increased in the serum in jaundiced patients, for example (31). Therefore, a wider panel of serum microRNAs, as identified in this study, is likely to offer increased specificity for HB compared with other tumors and disease states. Further discussion of the biological relevance of our findings in individual tumor types is provided in the Supplementary Discussion.
As the number of samples assessed for each of the tumor subtypes is small in the present proof-of-principle study, it will be important to confirm our findings in larger, prospective investigations. The fact that the panel of serum microRNAs that distinguished the initial group (n=4) of MYCN-NB from NB patients was confirmed in a small independent panel of patient samples (n=4) highlights that these changes are promising and worthy of further testing. If confirmed in future studies, we propose that the panels of childhood solid tumor-associated microRNAs identified here represent useful candidate biomarkers for improving the accuracy of pediatric cancer diagnosis. Such markers may reduce or obviate the need for diagnostic histological biopsy and the associated risks of anaesthesia and surgery. Furthermore, compared with the labor-intensive diagnostic techniques in current clinical use, a qRT-PCR approach, based on the analysis pipeline reported here, is likely to be more cost-effective, thereby offering health economic as well as clinical benefits.
Supplementary Material
Acknowledgements
We thank the patients and their families for study participation. We thank Dr David Halsall, Ms Amy Munro and Mr Jonathan Broomfield (Department of Biochemistry, Addenbrooke’s Hospital, Cambridge) for assistance.
Sources of Financial Support: Cancer Research UK (N. Coleman), Medical Research Council (M.J. Murray), SPARKS (N. Coleman, M.J. Murray, J.C. Nicholson) and Children with Cancer UK/Great Ormond Street Hospital Children’s Charity (N. Coleman, M.J. Murray, K.L. Raby, J.C. Nicholson).
Footnotes
Conflict of Interest Statement: There are no conflicts to disclose
References
- 1.Palmer RD, Murray MJ, Saini HK, van Dongen S, Abreu-Goodger C, Muralidhar B, et al. Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010;70:2911–23. doi: 10.1158/0008-5472.CAN-09-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Murray MJ, Halsall DJ, Hook CE, Williams DM, Nicholson JC, Coleman N. Identification of MicroRNAs From the miR-371~373 and miR-302 Clusters as Potential Serum Biomarkers of Malignant Germ Cell Tumors. Am J Clin Pathol. 2011;135:119–25. doi: 10.1309/AJCPOE11KEYZCJHT. [DOI] [PubMed] [Google Scholar]
- 4.Gillis AJ, Rijlaarsdam MA, Eini R, Dorssers LC, Biermann K, Murray MJ, et al. Targeted serum miRNA (TSmiR) test for diagnosis and follow-up of (testicular) germ cell cancer patients: A proof of principle. Mol Oncol. 2013;7:1083–92. doi: 10.1016/j.molonc.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray MJ, Coleman N. Testicular cancer: a new generation of biomarkers for malignant germ cell tumours. Nat Rev Urol. 2012;9:298–300. doi: 10.1038/nrurol.2012.86. [DOI] [PubMed] [Google Scholar]
- 6.Belge G, Dieckmann KP, Spiekermann M, Balks T, Bullerdiek J. Serum levels of microRNAs miR-371-3: a novel class of serum biomarkers for testicular germ cell tumors? Eur Urol. 2012;61:1068–9. doi: 10.1016/j.eururo.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 7.Dieckmann KP, Spiekermann M, Balks T, Flor I, Loning T, Bullerdiek J, et al. MicroRNAs miR-371-3 in serum as diagnostic tools in the management of testicular germ cell tumours. Br J Cancer. 2012;107:1754–60. doi: 10.1038/bjc.2012.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spiekermann M, Belge G, Winter N, Ikogho R, Balks T, Bullerdiek J, et al. MicroRNA miR-371a-3p in serum of patients with germ cell tumours: evaluations for establishing a serum biomarker. Andrology. 2014 doi: 10.1111/j.2047-2927.2014.00269.x. [DOI] [PubMed] [Google Scholar]
- 9.Syring I, Bartels J, Holdenrieder S, Kristiansen G, Muller SC, Ellinger J. Circulating serum microRNA (miR-367-3p, miR-371a-3p, miR-372-3p, miR-373-3p) as biomarkers for patients with testicular germ cell cancers. J Urol. 2014 doi: 10.1016/j.juro.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros PF, Ambros IM, Brodeur GM, Haber M, Khan J, Nakagawara A, et al. International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer. 2009;100:1471–82. doi: 10.1038/sj.bjc.6605014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 13.Murray MJ, Bailey S, Raby KL, Saini HK, de Kock L, Burke GA, et al. Serum levels of mature microRNAs in DICER1-mutated pleuropulmonary blastoma. Oncogenesis. 2014;3:e87. doi: 10.1038/oncsis.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 17.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–80. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132:2884–93. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 19.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59:S1–6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Anglesio MS, Wang Y, Yang W, Senz J, Wan A, Heravi-Moussavi A, et al. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol. 2013;229:400–9. doi: 10.1002/path.4135. [DOI] [PubMed] [Google Scholar]
- 21.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 22.Kessler OJ, Siegel JF, Brock WA. Intrarenal neuroblastoma masquerading as Wilms’ tumor. Urology. 1998;51:313–6. doi: 10.1016/s0090-4295(97)00690-0. [DOI] [PubMed] [Google Scholar]
- 23.Sellaturay SV, Arya M, Banisadr S, Murthi GV, Sebire NJ, Duffy PG. Primary intrarenal neuroblastoma: a rare, aggressive tumour of childhood mimicking Wilms’ tumour. J Pediatr Urol. 2006;2:522–4. doi: 10.1016/j.jpurol.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Viprey VF, Gregory WM, Corrias MV, Tchirkov A, Swerts K, Vicha A, et al. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: a European HR-NBL1/SIOPEN study. J Clin Oncol. 2014;32:1074–83. doi: 10.1200/JCO.2013.53.3604. [DOI] [PubMed] [Google Scholar]
- 25.Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, et al. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–90. [PubMed] [Google Scholar]
- 26.Viprey VF, Corrias MV, Kagedal B, Oltra S, Swerts K, Vicha A, et al. Standardisation of operating procedures for the detection of minimal disease by QRTPCR in children with neuroblastoma: quality assurance on behalf of SIOPEN-R-NET. Eur J Cancer. 2007;43:341–50. doi: 10.1016/j.ejca.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–83. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 28.Huang TC, Chang HY, Chen CY, Wu PY, Lee H, Liao YF, et al. Silencing of miR-124 induces neuroblastoma SK-N-SH cell differentiation, cell cycle arrest and apoptosis through promoting AHR. FEBS Lett. 2011;585:3582–6. doi: 10.1016/j.febslet.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 29.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyugos M, Lendvai G, Kenessey I, Schlachter K, Halasz J, Nagy P, et al. MicroRNA expression might predict prognosis of epithelial hepatoblastoma. Virchows Arch. 2014;464:419–27. doi: 10.1007/s00428-014-1549-y. [DOI] [PubMed] [Google Scholar]
- 31.Shifeng H, Danni W, Pu C, Ping Y, Ju C, Liping Z. Circulating liver-specific miR-122 as a novel potential biomarker for diagnosis of cholestatic liver injury. PLoS One. 2013;8:e73133. doi: 10.1371/journal.pone.0073133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.