Abstract
At hippocampal synapses, activation of group I metabotropic glutamate receptors (mGluRs) induces long-term depression (LTD), which requires new protein synthesis. However, the underlying mechanism remains elusive. Here we describe the translational program that underlies mGluR-LTD and identify the translation factor eIF2α as its master effector. Genetically reducing eIF2α phosphorylation, or specifically blocking the translation controlled by eIF2α phosphorylation, prevented mGluR-LTD and the internalization of surface AMPA receptors (AMPARs). Conversely, direct phosphorylation of eIF2α, bypassing mGluR activation, triggered a sustained LTD and removal of surface AMPARs. Combining polysome profiling and RNA sequencing, we identified the mRNAs translationally upregulated during mGluR-LTD. Translation of one of these mRNAs, oligophrenin-1, mediates the LTD induced by eIF2α phosphorylation. Mice deficient in phospho-eIF2α–mediated translation are impaired in object-place learning, a behavioral task that induces hippocampal mGluR-LTD in vivo. Our findings identify a new model of mGluR-LTD, which promises to be of value in the treatment of mGluR-LTD-linked cognitive disorders.
Information storage is widely believed to have a physical basis in long-lasting synaptic changes in selective circuits in the brain1,2. Repeated activity in a given neuronal pathway changes the efficacy of synaptic connections. For instance, high activity strengthens the efficacy of synaptic transmission, yielding long-term potentiation (LTP), whereas low activity weakens it, yielding LTD. An intriguing aspect of long-term memory is that different types of learning are associated with either LTP or LTD. In contrast to LTP, which remains the best characterized model of synaptic plasticity2, much less is known about the mechanism(s) underlying LTD. At hippocampal CA1 synapses, activation of group I mGluRs induces LTD by lowering the surface density of AMPA-type glutamate receptors (AMPARs)3–5. Unlike LTD induced by stimulation of NMDA-type receptors (NMDARs), LTD induced by activation of mGluRs (mGluR-LTD) requires protein syn-thesis6. Several signaling pathways impinging on translation, including the phosphoinositide 3-kinase (PI3K)–Akt–mechanistic target of rapamycin (mTOR) pathway and the extracellular signal–regulated kinase (ERK) pathway, have been implicated in mGluR-LTD3,5, but little is known about the detailed translational events and translational program underlying mGluR-LTD. Furthermore, while the expression of some proteins is known to be increased in response to mGluR activation3,5, there has been no large-scale and unbiased attempt to specifically identify the mRNAs that are translationally upregulated during mGluR-LTD. Given that alterations in mGluR-LTD have been linked to drug addiction, Alzheimer’s disease and intellectual disability3, unraveling the basic molecular mechanism(s) of mGluR-LTD will not only provide insight into our understanding of the basic aspects of synaptic plasticity and memory storage but could also lead to the development of mechanism-based treatments of mGluR-LTD-linked cognitive disorders.
Protein synthesis occurs in three steps: initiation, elongation and termination. Translation initiation, the rate-limiting step, is a major target for translational control7 and can be regulated in two independent ways. The first is by regulation of the eIF4F complex via mTOR complex 1 (mTORC1)8,9. Though experiments originally suggested that mGluR activation is coupled to protein synthesis via the PI3K–Akt–mTORC1 signaling pathway10, recent findings have challenged this view11,12. The second mechanism is by regulation of ternary complex formation via phosphorylation of the translation initiation factor eIF2α7. Phosphorylation of eIF2α blocks general translation but, paradoxically, results in translational upregulation of a subset of mRNAs that contain upstream open reading frames (uORFs) in their 5′ untranslated region (UTR)13,14. Here we report that activation of mGluRs, through phosphorylation of eIF2α, induces LTD by downregulating surface AMPAR density at synapses. mGluR activation selectively engages the type of translational program that is regulated by eIF2α, thus silencing general mRNA translation but also initiating the translation of specific mRNAs (synthesis of ‘LTD proteins’). One of these mRNAs, Ophn1, contains ORFs in its 5′ UTR and explains the protein synthesis–dependent LTD induced by eIF2α phosphorylation. Hence, our results demonstrate a novel translational program of mGluR-LTD. In addition, object-place learning, a task that triggers hippocampal LTD in vivo15,16, is impaired in mice with altered eIF2α phosphorylation-mediated translational control, or those with reduced OPHN1 levels in the hippocampus, thus highlighting the wider significance of eIF2α phosphorylation for mnemonic processes.
RESULTS
Deficient eIF2α phosphorylation prevents mGluR-LTD
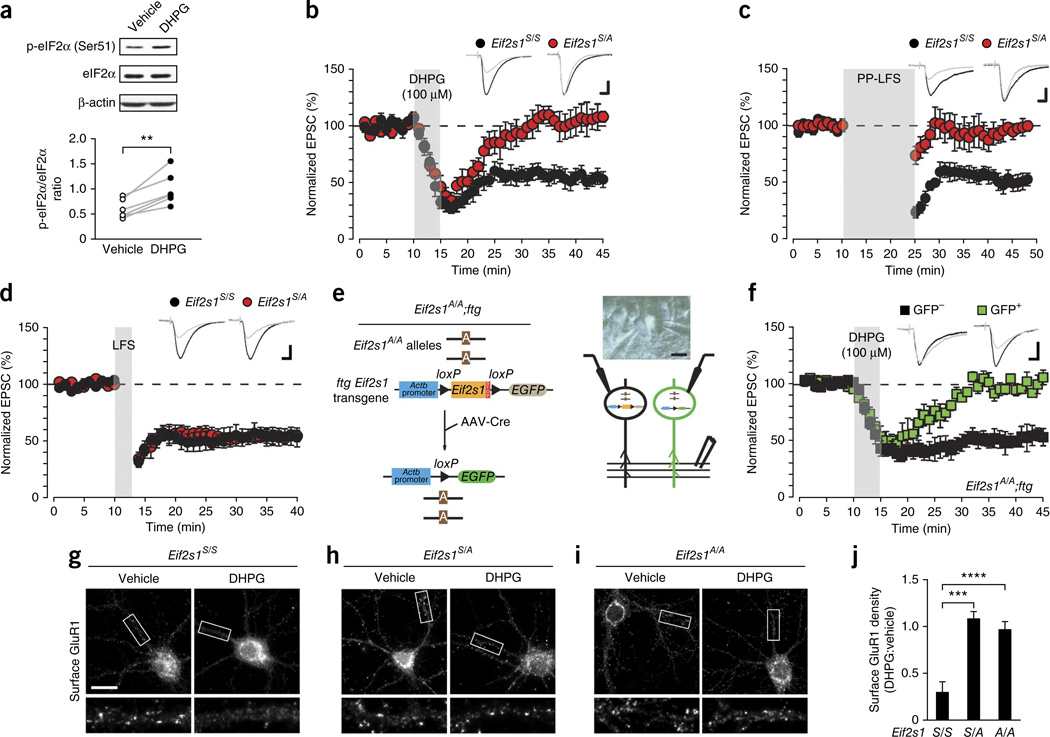
To investigate the role of eIF2α-mediated translational control in LTD, we treated hippocampal slices with (RS)-dihydroxyphenylglycine (DHPG), a selective mGluR1/5 agonist, and examined the phosphorylation state of eIF2α. DHPG (100 µM, 5 min) consistently increased eIF2α phosphorylation (Fig. 1a). To test whether eIF2α phosphorylation is required for mGluR-LTD at CA1 synapses, we first studied eIF2α+/S51A (Eif2s1S/A) heterozygous knock-in mice, containing one allele encoding eIF2α in which the single phosphorylation site at Ser51 is replaced by alanine17. In these mice, eIF2α phosphorylation is reduced in the hippocampus (Supplementary Fig. 1a). As expected, DHPG induced a typical depression of AMPAR-mediated excitatory postsynaptic currents (EPSCs) in wild-type (WT) control (Eif2s1S/S) slices (Fig. 1b). However, in Eif2s1S/A slices the same stimulation protocol failed to generate an mGluR-LTD (Fig. 1b). In agreement with previous reports18,19, paired-pulse low frequency stimulation (LFS) elicited a normal mGluR-LTD in control slices, whereas in Eif2s1S/A slices the magnitude of EPSCs evoked by Schaffer collateral stimulation returned to baseline by 5 min after the end of stimulation (Fig. 1c). NMDAR-LTD elicited by LFS20, which is not protein synthesis dependent6, occurred normally in Eif2s1S/A slices (Fig. 1d), indicating that eIF2α phosphorylation is necessary only for protein synthesis– dependent mGluR-LTD.
Figure 1.
Deficient eIF2α phosphorylation selectively prevents protein synthesis-dependent mGluR-LTD. (a,b) In WT hippocampal slices DHPG (100 µM, 5 min) increases eIF2α phosphorylation (a; n = 6 independent experiments, t = 5.067, P = 0.004, paired t-test) and induces LTD in CA1 neurons of WT Eif2s1S/S mice (b; 45.2 ± 6.4%, n = 7 cells from 4 mice, t = 7.6, P = 0.00030, paired two-sided t-test), but not in Eif2s1S/A slices (n = 7 cells from 4 mice, 5.2 ± 11.5%, t = 0.59, P = 0.57, paired two-sided t-test). Insets are examples of EPSCs before and after DHPG application. (c) LTD induced by paired-pulse low-frequency stimulation (PP-LFS; 900 pairs of stimuli at 50-ms intervals) at 1 Hz in the presence of the NMDAR antagonist D-2-amino-5-phosphonovalerate (AP5) is suppressed in Eif2s1S/A slices (n = 6 cells from 2 mice, 7.6 ± 7.7%, t = 0.94, P = 0.39, paired two-sided t-test), but not Eif2s1S/S slices (n = 7 cells from 4 mice, 45.4 ± 5.9%, t = 6.5, P = 0.00065, paired two-sided t-test). (d) Protein synthesis-independent LTD evoked by LFS (5 Hz, 3 min) was similar (t = 0.03, P = 0.9, unpaired two-sided t-test) in Eif2s1S/A (49.2 ± 6.1%, t = 5.59, P = 0.00082, n = 8 cells from 6 mice, paired two-sided t-test) and Eif2s1S/S slices (47.1 ± 7.0%, t = 7.87, P = 0.00053, n = 6 cells from 6 mice, paired two-sided t-test). (e) Left, in Eif2s1A/A;ftg mice both Eif2s1 alleles contain the S51A mutation; the ftg transgene contains WT Eif2s1 and a stop signal flanked by two loxP sites followed by EGFP. After infection with Cre-expressing AAV virus, WT Eif2s1 and the stop signal are cleaved by Cre recombinase. Right, paired recordings from GFP+ and GFP CA1 neurons from Eif2s1A/A;ftg mice. (f) DHPG induces LTD in GFP- (n = 6 cells from 6 mice, 47.7 ± 6.3%, t = 11.1, P = 0.0001, paired t-test), but not in GFP+ CA1 neurons (n = 6 cells from 6 mice, 0.4 ± 2.5%, t = 1.29, P = 0.25, paired t-test). (g-i) Surface staining of GluR1 in neurons from Eif2s1S/S (g), Eif2s1S/A (h) and Eif2s1A/A (i) mice. Cultures fixed 60 min after treatment with DHPG (100 µM, 5 min) or vehicle and labeled with antibody against N terminus of GluR1 without permeabilization. (j) Grouped data reveal significantly reduced surface GluR1 after DHPG treatment in Eif2s1S/S but not in Eif2s1S/A or Eif2s1A/A neurons (Eif2s1S/S, n = 51 cells; Eif2s1S/A, n = 51 cells; Eif2s1A/A, n = 34 cells; cells cultured from 3 mice per genotype; Eif2s1S/S versus Eif2s1S/A, ***P = 0.0007; eIF2α S/S versus Eif2s1A/A, ****P = 5 × 10−8; genotype × treatment interaction, F(2,12) = 19.25). Calibration bars in b,d,f: 10 ms, 40 pA. Scale bar in g-i, 20 µm. t-test in a-d,f; two-way ANOVA in j. Error bars represent ± s.e.m. Full-length blots are shown in Supplementary Figure 10.
To further support these findings, we engineered an Eif2s1 transgenic mouse line (Eif2s1A/A;ftg), in which the homozygous Eif2s1S51A/S51A genotype (Eif2s1A/A) is complemented by a WT Eif2s1 floxed transgene (ftg) flanked by loxP sites. (The breeding strategy used to generate Eif2s1A/A;ftg is depicted in Supplementary Fig. 1b.) We next excised the complementing WT transgene in a sparse population of CA1 pyramidal neurons by local infection with a virus carrying the Cre recombinase. Cre-mediated deletion coordinately induced the expression of green fluorescent protein (GFP), thereby enabling the identification of mutant neurons under the microscope (Fig. 1e). We performed simultaneous paired recordings from GFP+ neurons (in which eIF2α cannot be phosphorylated) and GFP− control neurons. DHPG evoked a sustained LTD in control neurons, but not in GFP+ neurons (Fig. 1f). Interestingly, LTD was blocked both in Eif2s1S/A and Eif2s1A/A slices, presumably because phosphorylation is already sufficiently impaired in Eif2s1S/A slices. A nonspecific change in synaptic transmission due to GFP expression cannot account for the impaired mGluR-LTD in GFP+ neurons from Eif2s1A/A;ftg mice because DHPG elicited a normal LTD in GFP+ neurons in WT mice (Supplementary Fig. 1c). We conclude that eIF2α phosphorylation is necessary for the induction of mGluR-LTD.
Because activation of mGluRs induces mGluR-LTD by persistently decreasing the AMPARs surface expression3,21, we examined whether eIF2α phosphorylation is important for this event. To this end, we measured changes in the surface expression of the AMPAR GluR1 in cultured hippocampal pyramidal neurons. DHPG-mediated activation of mGluRs reduced surface GluR1 density in WT control neurons (Fig. 1g,j and Supplementary Fig. 2) but not in Eif2s1S/A or Eif2s1A/A neurons (Fig. 1h–j and Supplementary Fig. 2). These data provide direct evidence that eIF2α phosphorylation is necessary for mGluRs to elicit a persistent decrease in surface expression of AMPARs.
Increased eIF2α phosphorylation induces mGluR-LTD
We hypothesized that increasing eIF2α phosphorylation by an alternative approach should also induce LTD. To test this idea, we incubated control slices with a low concentration of DHPG (10 µM, 5 min) or Sal003 (5 µM, 10 min), a selective inhibitor that blocks eIF2α phosphatases22,23. While either treatment alone failed to induce LTD (Fig. 2a,b) and to increase eIF2α phosphorylation (Supplementary Fig. 3), the combined application of low concentrations of DHPG and Sal003 increased eIF2α phosphorylation (Supplementary Fig. 3) and generated a sustained LTD in control neurons (Fig. 2c,d), but not in GFP+ neurons from Eif2s1A/A;ftg mice, in which eIF2α cannot be phosphorylated (Fig. 2d). These data demonstrate that the synergistic activation of mGluR-LTD by DHPG and Sal003 depends on eIF2α phosphorylation. Notably, the LTD generated by combining low concentrations of DHPG and Sal003 was of similar magnitude to that generated by 100 µM DHPG (compare Fig. 1b to Fig. 2c).
Figure 2.
Direct stimulation of eIF2α phosphorylation induces LTD. (a,b) In WT slices, no LTD is induced by low concentrations of DHPG (a; 10 µM, 5 min, n = 10 cells from 4 mice, 0.2 ± 1.8%, t = 0.93, P = 0.36) or Sal003 (b; 5 µM, 10 min), a selective inhibitor of eIF2α phosphatases (n = 8 cells from 4 mice, 1.4 ± 1.6%, t = 0.45, P = 0.66). (c) At the same sub-threshold doses, Sal003 followed by DHPG triggers LTD (n = 5 cells from 3 mice, 49.7 ± 4.0%, t = 9.44, P = 0.0007), mimicking the LTD induced by 100 µM DHPG. (d) The same combination of Sal003 and DHPG induces LTD in control GFP- neurons from Eif2s1A/A;ftg mice (n = 6 cells from 3 mice, 46.9 ± 7.3%, t = 5.3, P = 0.0031), but not GFP+ neurons (n = 6 cells from 3 mice, 4.2 ± 7.2%, t = 0.51, P = 0.62). (e) A higher concentration of Sal003 (20 µM, 10 min) alone induces sustained LTD in GFP neurons (n = 5 cells from 3 mice, 60.7 ± 6.3, t = 10.87, P = 0.00011), but not in GFP+ neurons (n = 5 cells from 3 mice, 5.5 ± 7.8%, t = 2.15, P = 0.098) from Eif2s1A/A;ftg mice. (f) Experiments with fPKR transgenic mice. Left: fPKR mice carry transgene under control of Camk2a promoter, containing lacZ and stop signal flanked by loxP sites and followed by the FKBP-PKR fusion gene. When neurons are infected with Cre-expressing AAV virus and EGFP, the stop signal is cleaved and FKBP-PKR fusion protein expressed. The small compound AP20187 induces dimerization and activation of FKBP-PKR only in GFP+ neurons, leading to eIF2α phosphorylation. Right: paired recordings from GFP+ and GFP neurons in slices from AAV-Cre-GFP-injected fPKR mice. (g) AP20187 (1 µM, 10 min) induces a sustained LTD in GFP+(n = 10 cells from 5 mice, 41.5 ± 4.6%, t = 6.73, P = 9 × 10−5) but not in GFP neurons (n = 10 cells from 5 mice, 3.3 ± 1.5%, t = 0.42, P = 0.68). (h,i) Immunostaining of surface GluR1 in neurons from Eif2s1S/S (h) and Eif2s1A/A (i) mice. Cultures were fixed 60 min after applying Sal003 (20 µM, 10 min) or vehicle, and labeled with GluR1 antibody without permeabilization. Sal003 reduced surface GluR1 density in control Eif2s1S/S, but not Eif2s1A/A neurons (framed areas are shown expanded below). (j) Normalized GluR1 surface density data (Eif2s1S/S: n = 51 cells, 3 mice; Eif2s1A/A: n = 34 cells, 3 mice; genotype × treatment interaction, F(2,12) = 5.1; *P = 0.012). Calibration bars in a-e,g: 10 ms, 40 pA. Scale bar in h,i, 20 µm. Paired t-test in a-e,g; two-way ANOVA in j. Error bars represent ± s.e.m.
We next tested whether direct stimulation of eIF2α phosphorylation is sufficient to generate a sustained LTD. To this end, we applied a concentration of Sal003 (20 µM, 10 min) that robustly increases phosphorylation of eIF2α in control slices (see below) and showed that it resulted in a sustained depression of EPSC amplitude (Supplementary Fig. 4a). In paired recordings, Sal003 induced a sustained LTD only in GFP− control neurons (Fig. 2e). The absence of LTD in GFP+ cells (in which eIF2α cannot be phosphorylated) supports the notion that Sal003 induces LTD by promoting phosphorylation of eIF2α.
Unlike the LTD induced by DHPG, Sal003-induced LTD was insensitive to mGluR1 and mGluR5 antagonists (2-methyl-6-(phenylethynyl)pyridine (MPEP) and LY367385; Supplementary Fig. 4b,c). Thus, the Sal003-mediated increase in eIF2α phosphorylation bypasses the activation of mGluR1/5 and is sufficient to generate LTD. Furthermore, like DHPG, Sal003 reduced surface GluR1 expression in control neurons, but not in neurons deficient in eIF2α phosphorylation (Fig. 2h–j and Supplementary Fig. 5).
As an independent, complementary test, we employed a chemical genetic strategy to selectively activate eIF2α phosphorylation in CA1 neurons. We used a transgenic mouse, fPKR, in which the eIF2α kinase PKR, fused to FK506 binding protein (FKBP), is conditionally expressed upon recombination mediated by a virus expressing a Cre-EGFP fusion protein (Fig. 2f). FKBP-PKR autoactivation was then induced by a chemical dimerizer, AP20187, leading to eIF2α phosphorylation24. In paired recordings, AP20187 generated a sustained LTD only in GFP+ neurons, in which FKBP-PKR is expressed (Fig. 2g). Taken together, these data support the idea that eIF2α phosphorylation weakens synaptic strength.
Translational control by eIF2α is necessary for mGluR-LTD
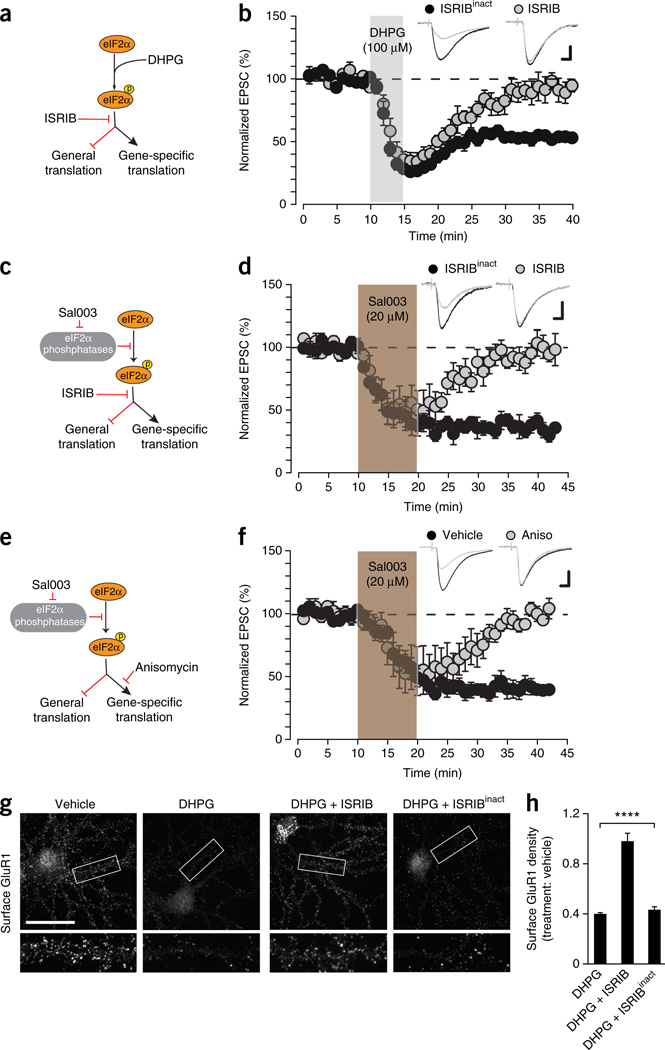
Because eIF2α phosphorylation decreases general mRNA translation but also increases the translation of specific mRNAs13,14, we asked whether mGluR-LTD requires eIF2α-mediated translational control. To answer this question, we used a recently discovered small molecule inhibitor, ISRIB25, which prevents the translational effects that are mediated by eIF2α phosphorylation (Fig. 3a). Preincubation with ISRIB (but not its inactive analog ISRIBinact; Supplementary Fig. 6) abolished both DHPG-induced LTD (Fig. 3b) and the reduction in surface expression of GluR1 induced by mGluR activation (Fig. 3g,h). Hence, the translational program governed by eIF2α phosphorylation is necessary for mGluR-LTD and the underlying reduction in surface AMPARs.
Figure 3.
mGluR-LTD requires eIF2α-mediated translational control. (a) Schematic of effect of ISRIB on eIF2α signaling. By promoting eIF2α phosphorylation DHPG depresses general translation but facilitates translation of specific mRNAs. ISRIB selectively blocks both specific and general translational effects mediated by eIF2α phosphorylation. (b) ISRIB (50 nM), but not its inactive analog ISRIBinact (50 nM, n = 5 cells from 5 mice, 8.9 ± 6.1%, t = 14.85, P = 0.00012), prevented LTD induction by DHPG in WT slices (n = 5 cells from 5 mice, 45.9 ± 2.2%, t = 1.55, P = 0.2). (c) Sal003 promotes the eIF2α phosphorylation by inhibiting eIF2α phosphatases. (d) ISRIB but not ISRIBinact (both 50 nM) blocked LTD induction by Sal003 (20 µM, 10 min) (n = 6 cells from 5 mice each, ISRIBinact: 1.2 ± 6.1%, t = 0.19, P = 0.86; ISRIB: 58.1 ± 4.4%, t = 13.1, P = 5 × 10−5; group difference t = 8.4, P = 3.7 × 10−6). (e) The mRNA translation inhibitor anisomycin blocks increase in gene-specific translation caused by eIF2α phosphorylation. (f) Anisomycin (Aniso; 1 h, 25 µM) blocked Sal003-induced (20 µM, 10 min) LTD in WT slices (n = 5 cells from 3 mice, 2.5 ± 1.9%, t = 0.82, P = 0.46) versus vehicle-treated slices (n = 5 cells from 4 mice, 58.4 ± 5.6%, t = 28.1, P = 1 × 10−5; group difference t = 13.8, P = 7.4 × 10−6). (g) Surface staining of GluR1 in WT neurons (19 d in vitro) treated under different conditions; framed areas in g are shown expanded below. Cultures were fixed 60 min after treatment and labeled with GluR1 antibody without permeabilization. ISRIB prevented reduction in surface GluR1 induced by DHPG, while control ISRIBinact had no effect. (h) Surface GluR1 density normalized to vehicle (n = 20 cells per condition; F(4,95) = 36.78, ****P = 1.4 × 10−7). Experiments were in triplicate. Calibration bars in b,d,f: 10 ms, 40 pA. Scale bar in g, 40 µm. Paired two-sided t-test in b,d,f; one-way ANOVA in h. Error bars represent ± s.e.m.
As expected, ISRIB also blocked Sal003-induced LTD (Fig. 3c,d). Next, to investigate whether Sal003-induced LTD is caused by a decrease in general translation or by increased translation of specific mRNAs, we blocked the putative increase in translation induced by Sal003-mediated eIF2α phosphorylation with the protein synthesis inhibitor anisomycin (Fig. 3e). Preincubation with anisomycin prevented the long-lasting depression of EPSCs induced by Sal003 (Fig. 3f), suggesting that the Sal003-mediated increase in eIF2α phosphorylation induces LTD by activating the translation of specific mRNAs rather than by blocking general translation.
Translation of Ophn1 by eIF2α is required for mGluR-LTD
To determine which specific mRNAs need to be translated to generate mGluR-LTD, we employed an unbiased approach that combines polysomal RNA profiling with whole-transcriptome shotgun sequencing (RNA-seq). First, we found that DHPG caused a reduction in general translation, indicated by the decrease in the polysome population and concomitant increase in monosomes (notably the 80S peak; Fig. 4a), as expected from the increase in eIF2α phosphorylation induced by mGluR activation. Accordingly, DHPG consistently decreased global protein synthesis, as determined by a nonradioactive surface sensing of translation (SUnSET) assay (Fig. 4b).
Figure 4.
Activation of mGluR selectively promotes translation of Ophn1 mRNA by eIF2α phosphorylation. (a) Polysome profiles from cultured mouse neurons show that DHPG (100 µM, 5 min) decreases general translation. (b) DHPG reduces protein synthesis in cultured mouse neurons, as determined by SUnSET. Western blotting (left) reveals decrease in puromycin incorporation in DHPG-treated (100 µM, 5 min) versus vehicle-treated neurons. Line scans plot relative intensity along vertical axis of vehicle (black) and DHPG (red) lanes. Quantification of puromycinabeled peptides (n = 4 independent experiments, 16 cultures (4 per condition per experiment), t = 6.051, **P = 0.0091, paired t-test). β-actin was measured as loading control. (c) RNA-seq analysis of RNA isolated from total extracts and polysomal fractions from vehicle- and DHPG-treated neurons. Heat map shows corrected translational profile. Genes were ranked in decreasing order according to polysomal fraction/free ribosomes ratio between vehicle- and DHPG-treated cells. Neuronal genes are in bold. (d,e) qRT-PCR analysis reveals increased amount of Ophn1 mRNA in heavy polysome fractions (d) but no major change in β-actin (Actb) mRNA in DHPG-treated neurons (e), versus control neurons. (f) Diagram of the 5′ UTR-OPHN1-Fluc reporter (top), consisting of the 5′ UTR of Ophn1 mRNA fused to the coding region of firefly luciferase (Fluc) and control Fluc reporter. Ratio represents translation of 5′UTR-OPHN1-Fluc mRNA versus control Fluc mRNA relative to a control Renilla luciferase (Rluc) reporter vector. Reporters were transfected into HEK293T cells. Fluc values were normalized against Rluc values. Fluc/Rluc ratio of expression shows that the 5′ UTR of Ophn1 represses translation (bottom; n = 3 independent experiments, ***P = 0.00054, t = 10.12, unpaired t-test). (g) 5′ UTR-OPHN1-Fluc and Rluc reporters were co-transfected in HEK293T cells, subsequently treated with vehicle or 200 nM thapsigargin (Tg, for 4 h), which increased expression of 5′ UTR-OPHN1-Fluc (n = 6 independent experiments; ***P = 0.00013, t = 6.03, unpaired t-test). t-test in b,f,g. Error bars represent ± s.e.m. Full-length blots are shown in Supplementary Figure 10.
To identify the mRNAs that are translationally upregulated by mGluR activation, we used RNA-seq to analyze total mRNA abundance, mRNAs that are poorly or not translated (monosome fraction) and the actively translating mRNAs (polysome fraction) in control and DHPG-treated cultured neurons. Specifically, we focused on mRNAs with (i) low or no change in total mRNA level (<1.5 fold), (ii) an induction of translation (>2.0 fold) and (iii) the presence of uORFs in their 5′ UTRs (a key feature for eIF2α phosphorylation-mediated translational control of specific mRNAs13,14). For a complete list of mRNAs with or without 5′ ORFs that are translationally upregulated upon mGluR activation, see Supplementary Table 1. These analyses revealed that oligophrenin-1 (Ophn1) mRNA was one of the highest ranked neuronal mRNAs (Fig. 4c). OPHN1 was recently implicated in mGluR-LTD, where it was shown to be required for AMPAR endocytosis26. To validate these data, we studied the recruitment of ribosomes to Ophn1 mRNA using quantitative reverse transcription–PCR (qRT-PCR). In this assay, the individual mRNAs are tracked throughout the polysome distribution and their position is determined by the number of associated ribosomes. Under baseline conditions, Ophn1 mRNA was primarily associated with monosomes and light polysomes, consistent with a moderate translation rate (Fig. 4d). By contrast, when neurons were treated with DHPG, a large fraction of Ophn1 mRNA shifted toward the heavier polysome fractions as a result of increased translation (Fig. 4d), whereas the distribution of the abundant β-actin (Actb) mRNA remained unaltered (Fig. 4e).
eIF2α phosphorylation triggers the translation of mRNAs containing uORFs in the 5′ UTR, such as GCN4 in yeast and Atf4 in mammalian cells14,27. Notably, the Ophn1 mRNA harbors two uORFs in its 5′ UTR (Fig. 4f). To directly examine the role of 5′ UTR uORFs in the regulation of Ophn1 mRNA translation, we constructed a reporter in which the 5′ UTR of Ophn1 was fused to the coding region of the firefly luciferase (Fluc) vector (5′UTR-OPHN1-Fluc; Fig. 4f, top). The expression of 5′UTR-OPHN1-Luc in HEK293T (human embryonic kidney) cells was reduced when compared to control vector (Fluc; Fig. 4f, bottom), as would be expected from an mRNA containing uORFs in its 5′ UTR. To test whether the 5′ UTR of Ophn1 enables translation of this mRNA in response to eIF2α phosphorylation, HEK293T cells were transfected with 5′UTR-OPHN1-Luc and subsequently treated with thapsigargin, which is known to induce endoplasmic reticulum stress and the phosphorylation of eIF2α25,28. Similarly to its action on a luciferase reporter containing the 5′ UTR of Atf4 mRNA25,28, the treatment with thapsigargin increased the expression of 5′UTR-OPHN1-Luc (Fig. 4g), suggesting that the 5′ ORFs control the translation of Ophn1 mRNA.
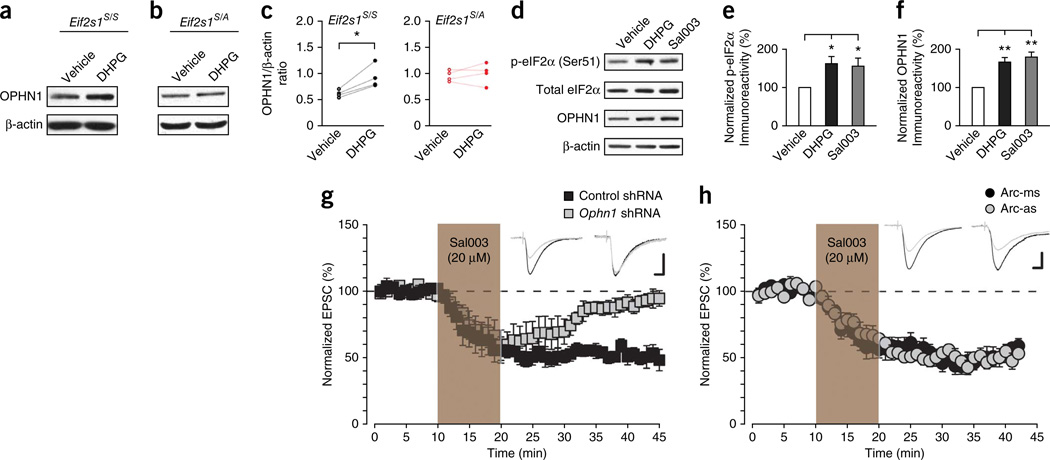
Consistent with the enrichment of Ophn1 mRNA in the polysome fractions, DHPG increased OPHN1 protein levels in control neurons (Fig. 5a,c), but not in eIF2α phosphorylation–deficient neurons (Fig. 5b,c). As expected, like DPHG, Sal003 increased eIF2α phos-phorylation (Fig. 5d,e) and OPHN1 levels (Fig. 5d,f). Furthermore, at individual synapses there was a positive correlation between the levels of OPHN1 and phosphorylated eIF2α in control neurons and those treated with DHPG and Sal003 (Supplementary Fig. 7).
Figure 5.
eIF2α phosphorylation selectively mediates the translation of OPHN1 required for mGluR-LTD. (a,b) Compared to vehicle-treated slices, OPHN1 protein level was in creased by DHPG treatment in WT Eif2s1S/S slices (a; n = 4 independent experiments, t = 4.13, P = 0.026), but not in Eif2s1S/A slices (b; n = 4 independent experiments, t = 0.64, P = 0.57). (c) Quantification of OPHN1 levels using β-actin as a loading control (n = 4 independent experiments). (d) Western blotting revealed that both DHPG (100 µM, 5 min) and Sal003 (20 µM, 10 min) increased the phosphorylation of eIF2α and the expression level of OPHN1 in WT slices. (e,f) Quantification of p-eIF2α versus total eIF2α (e) and OPHN1 versus β-actin (f), n = 4 independent experiments (e: F(2,11) = 4.503, *P = 0.037 as compared to vehicle, f: F(2,11) = 18.527, **P = 0.0003 as compared to vehicle). (g) Sal003 (20 µM, 10 min) induced LTD in CA1 neurons expressing control scrambled shRNA (n = 5 cells from 3 mice, 49.6 ± 4.7%, t = 8.27, P = 7 × 10−5), but not in neurons expressing an shRNA against Ophn1 (n = 5 cells from 3 mice, 7.4 ± 3.3%, t = 1.40, P = 0.20). (h) Sal003-induced LTD does not require Arc mRN A translation. Sal 003 (20 µM, 10 min) induced a similar LTD in neurons infused with either an Arc antisense oligonucleotide (Arc-as) (n = 8 cells from 3 mice, 43.5 ± 3.8%, t = 7.91, P = 9.8 × 10−5) or a mismatched oligonucleotide (Arc-ms) (n = 5 cells from 4 mice, 49.5 ± 2.5%, t = 8.57, P = 0.001) from the whole-cell recording pipette. Calibration bars in g,h: 10 ms, 40 pA. Two-sided t-test in c,g,h; one-way ANOVA in e,f. Error bars represent ± s.e.m. Full-length blots are shown in Supplementary Figure 10.
To establish a causal relationship between OPHN1 translational upregulation and the Sal003-induced LTD, we used a specific short hairpin RNA to lower OPHN1 levels. We first confirmed that Ophn1 shRNA blocked DHPG-induced LTD (Supplementary Fig. 8a), as previously reported26. Ophn1 shRNA also prevented Sal003-induced LTD (Fig. 5g), confirming that OPHN1 is essential for both processes that elicit eIF2α phosphorylation-mediated LTD. Because the synthesis of the activity-regulated cytoskeleton-associated protein Arc has also been implicated in mGluR-LTD and underlying changes in surface AMPAR density29,30, we asked whether Arc translation is important for Sal003-induced LTD. Unlike DHPG-induced LTD (Supplementary Fig. 8b), Sal003-induced LTD was not suppressed by an antisense oligonucleotide that blocks Arc synthesis (Fig. 5h). In addition, while Sal003 boosted eIF2α phosphorylation, it did not affect Arc levels (Supplementary Fig. 8c). Taken together, these data demonstrate that translational control of Ophn1 mRNA underlies, at least in part, the LTD induced by eIF2α phosphorylation.
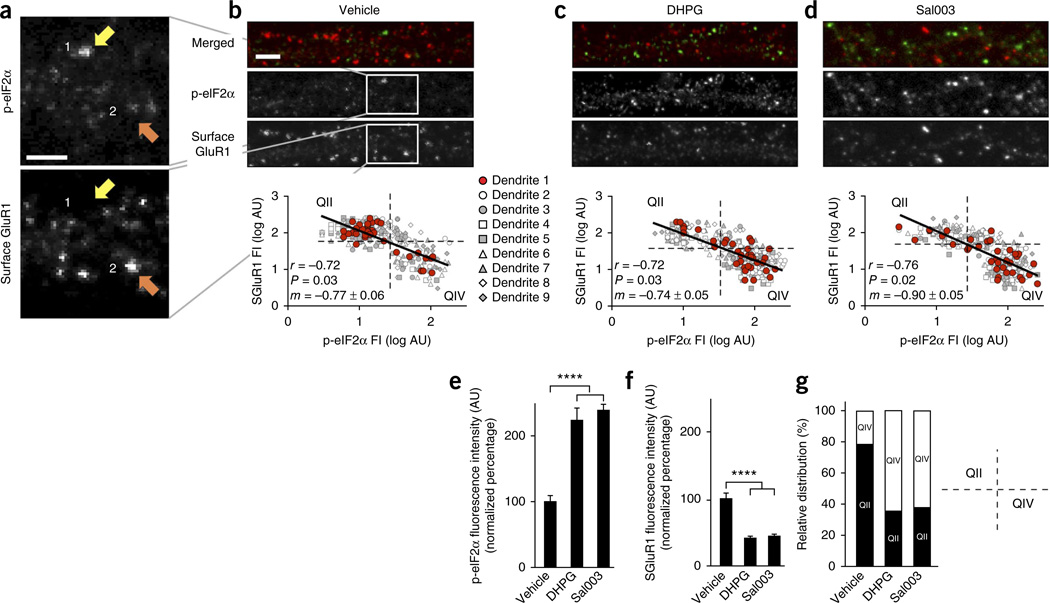
To examine whether the phosphorylation state of eIF2α could be a predictor of whether an individual synapse undergoes LTD in response to mGluR activation, we monitored eIF2α phosphorylation and surface GluR1 expression at individual synapses. Strikingly, under baseline conditions, surface GluR1 and phospho-eIF2α (p-eIF2α) immunoreactivities at single synapses were negatively correlated (Fig. 6a,b): when p-eIF2α levels were high at individual synapses, surface GluR1 density was low, and vice versa. We next tested whether these observations were relevant to changes in synaptic activity triggered by either DHPG or Sal003. After blind quantification of p-eIF2α immunoreactivity and surface GluR1 expression, we found that application of either Sal003 or DHPG, at concentrations sufficient to induce LTD, progressively shifted the intensity distribution toward a state of increased p-eIF2α and reduced surface GluR1 (Fig. 6c–g). Taken together, our findings support the notion that GluR1 surface reduction and the consequent mGluR-LTD are triggered by phosphorylation of eIF2α (and increased translation of Ophn1) at individual synapses.
Figure 6.
At hippocampal synapses phosphorylation of eIF2α and surface GluR1 immunoreactivity are negatively correlated. (a,b) Representative two-channel immunostaining for p-eIF2α and surface GluR1 of a dendritic segment from control, DHPG-, or Sal003-treated hippocampal neuron cultures. Framed areas (white squares) in b are expanded in a. Some synapses had low surface GluR1 signals but high p-eIF2α signals (yellow arrows), whereas others showed opposite pattern (orange arrows). Both DHPG and Sal003 increased proportion of synapses with high p-eIF2α signals and low surface GluR1. (b,c) Regression lines of log-transformed fluorescence intensity (FI) data, in arbitrary units (AU), from 315 synapses, representing 9 cells analyzed per condition, with corresponding slopes (m) and correlation coefficients (r). Scale bars in a-d, 2 µm. bP = 0.03; cP = 0.03; dP = 0.02. (e,f) Average p-eIF2α (e) and surface GluR1 (f) fluorescence intensity values ± s.e.m. (arbitrary units) normalized to vehicle-treated values (en = 9 cells, 35 synapses per cell; F(2,24) = 29.9, ****P = 3.0 × 10−7; fn = 9 cells, 35 synapses per cell; F(2,24) = 32.8, ****P = 1.4 × 10−7). (g) Quadrant analysis of asymmetrical distribution of data in plots (b-d) contrasts the low p-eIF2α/high SGluR1 ratios in upper left quadrant (QII) and high-p-eIF2α/lowSGluR1 in lower right quadrant (QIV). Normalized values: control, 78.2% versus 21.8%; DHPG, 35.4% versus 64.6%; Sal003, 37.8% versus 62.2%. Chi-squared tests comparing frequency distribution of vehicle versus DHPG and vehicle versus Sal003 were highly significant (n = 9 cells, 35 synapses per cell; P = 1.7 × 10−5, 1.3 × 10−5, respectively), whereas comparison of DHPG versus Sal003 was not (n = 9 cells, 35 synapses per cell; P = 0.62). Statistical significance assessed by Pearson product-moment correlation coefficient (r) in b-d, by one-way ANOVA in e,f and by chi-squared test in g.
Translation by eIF2α is required for spatial recognition
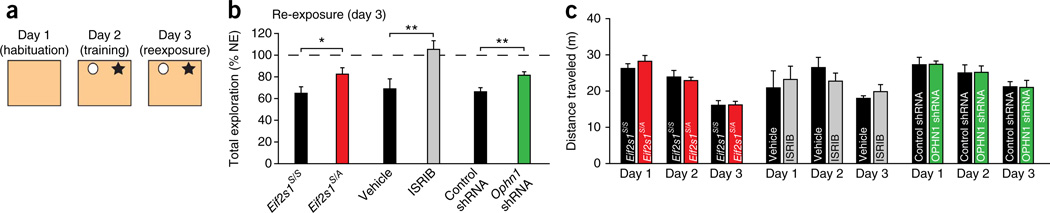
Hippocampal LTD is crucial for spatial learning of object-place recognition15,16. Spatial recognition of objects triggers LTD at Schaffer collateral–CA1 synapses in freely moving animals15,16. Briefly, in this task, mice are exposed on day 1 to an empty box for habituation (Fig. 7a). Exposure to two novel objects in the same box on day 2 elicits a sustained LTD in the hippocampus. However, when reexposed to the same objects on the following day (day 3), animals spend less time exploring the now familiar objects and no LTD is induced in CA1 (refs. 15,16). Notably, such LTD depends on mGluR activation, since its blockage shortly before novel object exploration (on day 2) inhibits the LTD in vivo and completely blocks the reduction in reexploration time on day 3 (ref. 16), suggesting that this behavioral task is a direct consequence of hippocampal LTD.
Figure 7.
Increased eIF2α-mediated translational control is needed for successful learning of novel object-space configuration. (a) Experimental design for the spatial recognition task. On day 1, mice were placed in an empty training box. On day 2, in the same box, mice were exposed to two novel objects (novelty exploration). On day 3 (reexposure), the mice were reexposed to the same objects, in the same spatial configuration. (b) Compared to WT mice (n = 14 mice), Eif2s1S/A mice (n = 15) spent more time exploring the objects during reexposure (t = 2.5, *P = 0.019). Mice injected with a single dose of ISRIB (2.5 mg/kg intraperitoneally) 2 h before exposure to the objects (on day 2) (n = 9) spent more time exploring during reexposure (t = 3.4, **P = 0.004) versus vehicle-injected mice (n = 6). Compared to control mice injected intrahippocampally with scrambled shRNA (n = 11), Ophn1 shRNA–injected mice spent significantly more time exploring the objects during reexposure (n = 9, t = 3.08, **P = 0.006). NE, novelty exploration. (c) WT and Eif2s1S/A mice traveled similar distances on days 1 and 2 (day 1: t = 1.36, P = 0.18; day 2: t = 0.70, P = 0.49; day 3: t = 0.15, P = 0.88). Vehicle- and ISRIB-injected mice traveled similar distances on days 1 and 2 (n = 6 for vehicle and 9 for ISRIB; day 1: t = 0.38, P = 0.71; day 2: t = 1.17, P = 0.27; day 3: t = 1.06, P = 0.31). Mice injected with control shRNA (n = 11) and Ophn1 shRNA (n = 9) traveled similar distances on days 1 and 2 (day 1: t = 0.093, P = 0.93; day 2: t = 0.035, P = 0.97; day 3: t = 0.12, P = 0.91). Unpaired t-test in b,c. Error bars represent ± s.e.m.
To investigate the role of eIF2α phosphorylation–mediated translational control in object-place learning, we first examined whether eIF2α phosphorylation and OPHN1 levels are increased in the hippocampus as a result of behavioral learning. Briefly, we found that exposure to the two objects on day 2 led to an increase in eIF2α phosphorylation at both 10 min and 90 min after training (Supplementary Fig. 9a,b). Accordingly, OPHN1 levels were also higher at 90 min, but not at 10 min, suggesting that at this early time the synthesis of OPHN1 was not completed in all hippocampal synapses that are engaged during spatial recognition (Supplementary Fig. 9c,d). Hence, like mGluR-LTD, LTD-dependent behavior increases eIF2α phosphorylation and OPHN1 levels.
Given that mGluR-LTD is blocked in hippocampal slices from eIF2α-deficient mice, we then predicted that, when reexposed to the same objects, these mice should show little or no reduction in exploration time. Indeed, compared to WT Eif2s1S/S mice, Eif2s1S/A mice exhibited much less reduction in exploratory activity during reexposure (Fig. 7b and Supplementary Fig. 9e). A nonspecific change in locomotor behavior in Eif2s1S/A mice is unlikely since the distance traveled was similar for WT and mutant mice on all three days (Fig. 7c). Hence, eIF2α phosphorylation is required for object-place recognition.
We next investigated whether ISRIB, an agent that prevents the translational effects of eIF2α phosphorylation, also blocks object-place learning in WT mice. While administration of ISRIB to WT mice before novelty exploration had no effect on distances traveled or exploration times during training (Fig. 7c and Supplementary Fig. 9f), it prevented the decrease in object exploration during reexposure (Fig. 7b). Finally, we found that spatial object recognition also required OPHN1. Compared to mice injected in the hippocampus with control shRNA, those injected with Ophn1 shRNA show less reduction in exploration when reexposed to the same objects (Fig. 7b and Supplementary Fig. 9g). We therefore conclude that, like eIF2α phosphorylation–mediated translational control, OPHN1 is necessary for object-place learning mediated by hippocampal mGluR-LTD.
DISCUSSION
A crucial feature of mGluR-LTD is its dependence on new protein synthesis3,6, which is subjected to fine-tuned regulation of translation. Our results identify the translational program that underlies protein synthesis–mediated mGluR-LTD. This program is governed in a highly selective manner by the translation factor eIF2α at the translation initiation level. We found that mGluR-LTD and the underlying reduction of surface AMPARs were blocked or facilitated by single-cell genetic and pharmacogenetic strategies that either suppress or promote eIF2α phosphorylation, respectively. Furthermore, ISRIB, which prevents eIF2α phosphorylation-mediated translation25, also blocked mGluR-LTD and the removal of surface AMPARs. Unexpectedly, mGluR activation, which leads to a protein synthesis–mediated LTD, triggered the phosphorylation of eIF2α and blocked general translation, as determined by the shift in mRNA sedimentation toward monosomal fractions and the reduction of general protein synthesis seen by SUnSET. However, the same stimulation protocol led to the synthesis of specific ‘LTD proteins’. In this process, the silencing of general translation reduces the competition of mRNAs for the translational machinery, thus favoring a selective enhancement of translation of the specific mRNAs needed for mGluR-LTD. Using polysome profiling combined with RNA-seq, we determined the mRNAs that are newly translated during LTD and identified uORFs in these mRNAs that enable their translation when eIF2α is phosphorylated. One of these mRNAs, Ophn1, which contains two uORFs in its 5′ UTR, is translationally upregulated in an eIF2α phosphorylation–dependent manner. In addition to the 5′ uORFs, we cannot exclude a possible involvement of other elements in the 5′ UTR or 3′ UTR of the mRNA in the expression OPHN1.
We also provide direct evidence that OPHN1 is required for both Sal003-induced LTD and DHPG-induced LTD. An important point is that OPHN1, by binding to the endocytic machinery, promotes the removal of AMPARs from the surface membrane and thus induces mGluR-LTD26. We therefore propose that the increase in Ophn1 mRNA translation mediated by eIF2α phosphorylation reduces AMPAR surface density and thereby induces a protein synthesis– dependent LTD.
Arc is another protein implicated in mGluR-LTD29,30. Interestingly, we found that blocking Arc translation prevented DHPG-induced LTD, but had no effect on Sal003-induced LTD. A possible explanation for this apparently paradoxical observation is that DHPG-mediated activation of mGluRs leads to both Arc expression and an eIF2α phosphorylation-mediated increase in OPHN1 levels, whereas during Sal003-induced LTD only OPHN1 is translationally upregulated. Thus, since Sal003 failed to increase Arc levels, its absence did not affect this type of LTD. We postulate that mGluR stimulation can activate two translational programs: one that triggers Ophn1 mRNA translation and is regulated at the initiation level by p-eIF2α and a second one that increases Arc protein and appears to be independently regulated at the elongation level by the eukaryotic elongation factor 2 (eEF2)30. Because mGluRs consist of two subtypes, mGluR1 and mGluR5, the selective activation of mGluR subtypes (mGluR1 or mGluR5) could favor the specific induction of one or the other of these translational programs. Finally, although the microtubule-associated protein 1b (MAP1b) and fragile × mental retardation protein (FMRP) have been reported to be synthesized in response to mGluRs31,32, we did not see a shift of these mRNAs from monosomes to polysomes following mGluR activation. Possible explanations for this are that the translation of these mRNAs is regulated at the elongation level33,34, rather than at the initiation level, or differences in the experimental procedures.
Could the degree of eIF2α phosphorylation predict whether a given synapse undergoes LTD or LTP? Our results indicate that this could be the case. First, a direct increase in eIF2α phosphorylation led to a protein synthesis–mediated LTD. Second, eIF2α phosphorylation varied inversely with surface AMPAR density. Third, because LTP requires the insertion of AMPARs into the surface membrane, we can expect that synapses undergoing local reductions in eIF2α phosphorylation would be potentiated. Indeed, in Eif2s1S/A slices or slices lacking the eIF2α kinase GCN2 or PKR, in which p-eIF2α is reduced, LTP is facilitated35–37. Fourth, increased eIF2α phosphorylation blocks LTP24,36. Fifth, behavioral studies show that hippocampal LTP is suppressed by a spatial recognition task known to induce hippocampal LTD in vivo15. This LTD-inducing task, which depends on mGluRs and NMDARs16, raised eIF2α phosphorylation in the hippocampus on day 2 and was impaired in mice with a deficient p-eIF2α-mediated translation program (Eif2s1S/A mice or WT mice treated with ISRIB), in which hippocampal mGluR-LTD is impaired. By contrast, an LTP-associated learning paradigm (for example, contextual fear conditioning), which reduces eIF2α phosphorylation in the hippocampus, leads to enhanced long-term memory in Eif2s1S/A mice or WT mice treated with ISRIB25,36.
Given that different types of hippocampus-dependent learning selectively trigger LTP or LTD, we propose that this bidirectional plasticity (and the resulting behaviors) depends on the type of learning. For instance, if the behavioral task triggers eIF2α phosphorylation (for example, object-place learning), which induces LTD, mice in which the p-eIF2α-mediated translation program is deficient will show impaired learning in this task. However, if the behavioral tasks (for example, contextual fear conditioning) suppress eIF2α phosphorylation (and induce LTP), the performance of the same mice will be enhanced25,36. Hence, as a key regulator of protein synthesis–dependent increases (LTP) or decreases (LTD) in synaptic strength, gene-specific eIF2α-mediated translational control emerges as a central mechanism of hippocampal synaptic plasticity and related behaviors.
In conclusion, we introduce a model that explicitly links two key mechanisms required for hippocampal long-lasting changes in synaptic strength in the hippocampus: protein synthesis and AMPA receptor trafficking. Our results show that the translation driven by eIF2α phosphorylation is both necessary and sufficient for mGluR-LTD and a correlated learning behavior. In addition, our findings hold promise that targeting phosphorylation of eIF2α could result in therapies for mGluR-LTD–linked cognitive disorders, including Alzheimer’s disease, Parkinson’s disease, intellectual disability and drug addiction3.
ONLINE METHODS
mouse husbandry
Eif2s1S/A mice were maintained on C57Bl/6 background as previously described36. Eif2s1A/A;ftg were generated as depicted in Supplementary Figure 1b. First, Eif2s1S/A;ftg mice were obtained by crossing ftg/0 mice with Eif2s1S/A mice (backcrossed to C57Bl/6J background for more than 10 generations as described by Back et al.38). Second, Eif2s1S/A;ftg mice were then bred with Eif2s1S/A mice to generate Eif2s1A/A;ftg strain. fPKR mice were generously provided by K. Nakazawa24. All experiments were performed on male mice 12–20 weeks old. Mice were kept on a 12 h light/dark cycle and had access to food and water ad libitum. Animal care and experimental procedures were approved by the animal care committee of Baylor College of Medicine, according to NIH guidelines.
Slice electrophysiology
Electrical recordings were performed as previously described37. The investigator was blind to the genotypes. Briefly, mice were anesthetized with a mixture of ketamine (100 mg/kg), xylazine (10 mg/kg) and acepromazine (3 mg/kg). The animals were transcardially perfused with ice-cold, oxygenated low-Na+ and low-Ca2+ medium containing (in mM) 87 NaCl, 2.5 KCl, 1.25 NaH2PO4 1.25mM, 26 NaHCO3, 25 dextrose, 75 sucrose, 0.5 CaCl2, 7 MgCl2. The brain was removed, trimmed and glued (with its ventral surface up) to a block of agar (4%) attached to the vibratome tray. Horizontal slices (300 µm) were cut with a vibratome (Leica VT 1000S, Leica Microsystems, Buffalo Grove, IL) and then transferred to a holding chamber filled with standard artificial cerebrospinal fluid (ACSF) containing (in mM) 120 NaCl, 3.3 KCl, 1.25 NaH2PO4, 25 NaHCO3, 10 dextrose, 1 MgCl2 and 2 CaCl2 saturated with 95% O2/5% CO2, pH 7.4, at 34 °C for 40 min and then at room temperature for at least 30 min before recording. The slices were submerged in a chamber perfused continuously with oxygenated ACSF (2–3 ml/min), containing picrotoxin (0.1 mM), at about 32 °C. Pyramidal neurons in the stratum pyramidale of CA1 were visualized under a Zeiss microscope (Axio Examiner D1) set for differential interference contrast optics and equipped with a 40× water-immersion lens. A bipolar stimulating electrode was placed in the stratum radiatum. To evoke excitatory postsynaptic currents (EPSCs), stimuli (100 µs) were delivered at 0.05 Hz. The stimulation strength was set to yield EPSCs with peak amplitudes of 100–200 pA.
Whole-cell patch-clamp recordings were performed with glass micropipettes (3–5 MΩ), pulled from standard borosilicate glass. Cells were voltage clamped at −70 mV. The liquid-junction potential was not compensated. The pipette solution contained (in mM) 117 CsMeSO3, 0.4 EGTA, 20 HEPES, 2.8 NaCl, 2.5 Mg-ATP, 0.25 Na-GTP, 5 TEA chloride, adjusted to pH 7.3 with CsOH and 290 mOsm. Data were obtained with a MultiClamp 700B amplifier, digitized at 20 kHz with a Digidata 1440A, recorded using Clampex 10 and analyzed with Clampfit 10 software (Molecular Devices). Recordings were filtered online at 4 kHz with a Bessel low-pass filter. A 2-mV hyperpolarizing pulse was applied before each EPSC to evaluate the access resistance (Ra). Data were discarded when Ra was either unstable or greater than 25 MΩ, holding current was above 200 pA, input resistance dropped more than 20% during the recording or EPSCs baseline changed by more than 10%. Only one cell was recorded from one slice in each case except for pair recordings, where two cells were recorded in a single slice. Traces illustrated in figures are averages of ten consecutive traces. Paired two-sided t-tests were performed to compare EPSCs at 5 min and 40 min.
Slice treatment for western blotting
Hippocampal slices were cut (300 µm) with a McIlwain tissue chopper (Mickle, UK) and incubated for at least 1 h at room temperature in oxygenated (95% O2, 5% CO2) ACSF as described39,40. They were kept at 32 °C for another hour before treatment with DHPG (100 µM) for 5 min or with Sal003 (20 µM) for 10 min, before snap-freezing over dry ice. In all instances, similar slices were treated with vehicle as control. Frozen slices were lysed in homogenizing buffer (200 mM HEPES, 50 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 50 mM NaF, 2 mM Na3VO4, 25 mM β-glycerophosphate and EDTA-free complete ULTRA tablets (Roche, Indianapolis, IN)). Western blotting was performed as described by Huang et al.40.
Primary antibodies for western blotting were rabbit anti–p-eIF2α (Ser51) (1: 500, 3398, Cell Signaling and Technology Laboratories, Danvers, MA), mouse anti-total eIF2α (1: 1,000, 2103, Cell Signaling), mouse anti-β-actin (1:10,000, ab8227, Abcam, Cambridge, MA), mouse anti-OPHN1 (1:1,000, sc-374330, Santa Cruz Biotechnology, Santa Cruz, CA) and mouse anti-Arc (1: 200, sc-17839, Santa Cruz Biotechnology). Secondary antibodies for western blotting were peroxidase-conjugated goat anti-rabbit (1:10,000, 111–035–144, Jackson ImmunoResearch) and goat anti-mouse (1:10,000, 115–035–003, Jackson ImmunoResearch).
Polysome profiling and RNA isolation
We prepared fresh 12-ml 10–50% sucrose density gradients (10 mM HEPES-KOH (pH 7.4), 5 mM MgCl2, 150 mM KCl, 5 U/ml RNasin RNase inhibitor (Promega, Madison, WI)), as previously described35,41. Gradients were kept in a cold room for 2 h before use. Primary mouse cortical neurons at 14 DIV were treated with DHPG (100 µM) or vehicle control for 10 min and then washed twice with ice-cold PBS plus 100 µg/ml cycloheximide (Sigma-Aldrich, St. Louis, MO). Cells were then collected in polysome lysis buffer (10 mM HEPES-KOH (pH 7.4), 5 mM MgCl2, 150 mM KCl, 0.5 mMDTT, 100 U/mL RNasin RNase inhibitor (Promega, Madison, WI), 100 µg/ml cycloheximide and EDTA-free protease inhibitors (Roche Indianapolis, IN)), supplemented with 1% NP-40 and kept on ice for at least 10 min. Samples were centrifuged at 10,000g for 5 min at 4 °C. The supernatant was either layered onto a sucrose gradient or reserved for total RNA isolation. Gradients were centrifuged in an SW-41Ti rotor (Beckman Coulter) at 32,000 r.p.m. at 4 °C for 2 h and then analyzed by piercing the tube with a Brandel tube piercer, passing 70% sucrose through the bottom of the tube, and monitoring the absorbance of the material eluting from the tube using an ISCO UA-6 UV detector. Fractions were collected throughout and RNA extracted with TRIzol following the manufacturer’s instructions (Life Technologies, Carlsbad, CA). 5 ng of poly(A)+ luciferase mRNA (Promega) was added to each fraction for normalization.
RNA-seq and quantitative PcR
All experiments were performed twice (biological replicates). Extracted RNA samples were divided. One half was pooled into three categories according to the ribosomal association: free ribonucleoprotein complex, light polysome and heavy polysome. RNA-seq template was generated from each of these categories using the TruSeq RNA sample preparation kit v2 (Illumina) and was sequenced on an Illumina HiSeq. From the remaining half, we prepared cDNA with the Superscript III reverse transcriptase (Life Technologies) and oligo(dT) primers, according to the manufacturer’s instructions. Quantitative PCR was performed to determine the abundance of transcripts using SYBR Green PCR mix (Life Technologies) and respective primers. Measurements were then normalized to luciferase abundance.
Oligonucleotides used for quantitative PCR of mouse mRNAs are as follows: Ophn1: forward 5′-CAAACCCCTGGAAACTTTTCG-3′, reverse 5′-ATGACAGATGTAAGTGGCGG-3′; Actb: forward 5′-TTCTTTGCA GCTCCTTCGTTGC-3′, reverse 5′-TGGATGGCTACGTACATGGCTG-3′; luciferase: forward 5′-ATCCGGAAGCGACCAACG-3′, reverse 5′- GTCGGGAAGACCTGCCAC-3′.
Bioinformatic analyses
RNA sequencing reads were aligned to the mouse reference genome version mm10 using TopHat 2.0.7 aligner42 followed by reference-guided transcript assembly using Cufflinks 2.1.1 (ref. 43). Gene expression values were obtained for all samples by summing the fragments per kilobase of exon per million (FPKM) values for different transcripts of each gene as assembled by cufflinks. Isoforms with FPKM <0.3 were excluded.
To identify genes with ORFs (uORFs) in the 5′ UTR, a systematic analysis of all gene sequences, as annotated by NCBI RefSeq mm10, was conducted using in-house Perl scripts.
The gene ontology terms associated with genes that were translationally upregulated and contained more than one upstream ORF were determined using GOTERMFINDER (http://go.princeton.edu/). A total of 25,496 genes from Mouse Genome Informatics Mus musculus annotation set were used to calculate the background distribution of gene ontology terms. A P-value cutoff of 0.01 was employed and the false discovery rate for the predictions was projected to be as low as 0.05%.
SUnSET
Protein synthesis was measured using SUnSET, a nonradioactive labeling method to monitor protein synthesis, as previously described44. Briefly, cortical neurons were isolated and maintained 18 d in culture, as described above. Neurons were treated with media containing either vehicle or 100 µM DHPG for 5 min at 37 °C. Treated media were then washed out and replaced with fresh media containing 10 µg/ml puromycin. After puromycin labeling for 10 min, cells were immediately washed with ice-cold PBS and lysed in homogenization buffer (in mM: 40 Tris HCl, pH 8.0, 150 NaCl, 25 β-glycerophosphate, 50 NaF, 2 Na3VO3, protease inhibitor cocktail, 10% glycerol, 1% Triton X-100). Puromycin incorporation was detected by western blot using the 12D10 antibody to puromycin (1:10,000, Millipore). The density of the resulting bands was quantified using ImageJ and statistical significance assessed by Student’s t-test.
Hippocampal and cortical neuron cultures
Primary embryonic hippocampal neurons were cultured essentially as in Kaech and Banker45, in the absence of the glial feeder layer. Briefly, hippocampi were isolated from E17 mouse or E18 rat pups and digested in 0.25% trypsin for 15 min at 37 °C. Cells were then centrifuged 5 min at 1,200g to remove enzymes and lysed cells, and the resulting pellet was resuspended in neuronal growth medium (Neurobasal medium without l-glutamine, with 1% Glutamax, 2% B-27 supplement (Life Technologies)). Hippocampal neurons were cultured at a density of 250 cells/mm2 on glass coverslips coated with poly-d-lysine and laminin. Half of the neuronal growth medium was replaced with fresh medium 24 h after plating and then again every 4 d in culture. Neurons were treated with 0.4 µM cytosine arabinoside overnight at 10 DIV to kill any contaminating glia. Hippocampal neurons were maintained at least 2–3 weeks in culture.
For the polysome profiling, cortical neurons, cultured like hippocampal neurons, were plated at a density of approximately 2,600 cells/mm2. Neuron extraction and polysome profiling was performed at 14 DIV.
Drug treatments of cultured neurons
Hippocampal neurons at 14–19 DIV were treated in one of the following five ways (as specified in the text and figure legends): (1) 5 min 100 µM DHPG, (2) 10 min 20 µM Sal003, (3) 50 nM ISRIBinact for 1 h before 5 min 100 µM DHPG + 50 nM ISRIBinact, (4) 50 nM ISRIB for 1 h before 5 min 100 µM DHPG + 50 nM ISRIB, or (5) vehicle. Cells were fixed for either 15 or 60 min after treatment onset.
Immunostaining
Neurons were allowed to mature to 14–19 DIV. After treatment with DHPG or Sal003, neurons were immediately fixed in ice-cold pH 7.2 4% PFA, 4% sucrose for 20 min at 4 °C. The PFA/sucrose solution was aspirated and the cells were washed three times with ice-cold PBS.
Surface GluR1 staining
Nonspecific binding was blocked for at least 30 min in 0.1 M phosphate buffer (PB) containing 10% goat-serum (or 10% BSA) without TX-100. Cells were then incubated overnight with a primary antibody targeting the extracellular N terminus of GluR1 and then washed with 10% goat serum (or 10% BSA) in PB for 2 h to remove excess primary antibody. The cells were then permeabilized in 0.3% TX-100, 10% goat serum (or 10% BSA) PB for 30 min, at which point additional primary antibodies targeting intracellular epitopes of indicated proteins were added and incubated overnight. Neurons were kept at 4 °C throughout the entire primary antibody stage. Excess primary antibodies were thoroughly removed and species-specific Alexa dye–conjugated secondary antibodies were added for 1 h in the dark at room temperature. The coverslips were then washed, air-dried, and mounted with mounting medium (KPL).
OPHN1 staining
Nonspecific binding was blocked for at least 30 min in buffer containing 10% BSA with 0.3% TX-100. Goat anti-OPHN1 and other primary antibodies were added and incubated at 4 °C overnight. Excess primary antibodies were removed by three 5-min washes with 10% BSA, 0.3% TX-100 PB. Cells were first incubated in Alexa dye–conjugated donkey anti-goat secondary antibodies for 1 h at room temperature. The anti-goat secondaries were removed by three 5-min washes with 10% BSA, 0.3% TX-100 PB and the cells were then incubated in 0.3% TX-100 PB with 10% goat serum to block nonspecific binding of secondary antibodies. Alexa dye–conjugated secondaries targeting mouse, rabbit and/or chicken primary antibodies used in conjunction with the goat anti-OPHN1 antibodies for multi-channel labeling were added for 1 h at room temperature. Coverslips were washed, air-dried and mounted. Cultures, treatments and analyses on neurons isolated from Eif2s1S/S, Eif2s1S/A and Eif2s1A/A mouse embryos were performed blind to genotype.
Fluorescent imaging was performed on an AxioImager.Z2m microscope (Carl Zeiss MicroImaging) mounted with an AxioCam digital camera (Carl Zeiss MicroImaging). Images were collected using AxioVision acquisition software (Carl Zeiss MicroImaging). All images in a given data set were collected at equivalent exposure times to allow comparison of signal intensity. Fluorescence intensity was measured with ImageJ (NIH) with the PunctaAnalyzer plugin. In some images, contrast and brightness were linearly adjusted using Photoshop (Adobe). Image processing was applied uniformly across all images within a given data set.
Antibodies for immunostaining
The following primary antibodies were used: mouse anti–GluR1 N terminus (surface GluR1) (1:200, clone RH95, Millipore), mouse anti–PSD-95 (1:400, K28/43, NeuroMab), rabbit anti–p-eIF2α (Ser51) (1:50, 9721, Cell Signaling Technology), rabbit anti-synapsin-1 (1:400, 5297, Cell Signaling Technology), rabbit anti-VGluT1 (1:200, 135 303, Synaptic Systems), chicken anti–MAP2 (1:1,000, CPCA-MAP2, EnCor Biotechnology Inc.) and goat anti-OPHN1 (1:100, sc8374, Santa Cruz Biotechnology). The following secondary antibodies were used for immunostaining: goat anti-mouse Alexa 594 (A-1 1032, Invitrogen), goat anti-rabbit Alexa 488 (A-11034, Invitrogen), goat anti-rabbit Alexa 594 (A-11037, Invitrogen), goat anti-chicken AMCA (103–155–155, Jackson ImmunoResearch), donkey anti-goat Alexa 488 (705–456–147, Jackson ImmunoResearch). All secondary antibodies were diluted 1:1,000.
Virus injection
AAV5-Cre (titer: 1.0 × 1013 GC/ml) was obtained from Vector Biolabs (cat. no. 7012, Philadelphia, PA); AAV1.hSynap.HI.eGFP-Cre.WPRE.SV40 (titer: 1.83 × 1013 GC/ml) was from Penn Vector Core (University of Pennsylvania, Philadelphia, PA). Lentiviral constructs expressing Ophn1 shRNA and scrambled shRNA were a generous gift from L. van Aelst26, and viruses were produced by the Gene Vector Core Laboratory (Baylor College of Medicine). For all injections, mice were anesthetized with isoflurane (2–3%) and mounted in a stereotaxic frame (Leica Angle Two system, Leica Microsystems, Buffalo Grove, IL). Viruses were injected bilaterally from a glass pipette (tip diameter about 20 µm) connected to a 10-µl Hamilton syringe propelled by a syringe pump (KD Scientific, Holliston, MA). The glass needle was inserted targeting two sites of the hippocampus in the left and right hemispheres (coordinates: site 1 −3.40 AP, ± 3.53 ML, −3.69 DV; site 2 −3.28 AP, ± 3.40 ML, −3.02 DV from bregma); 1–2 µl of purified concentrated viruses per site were delivered at the rate of 0.1 µl/min followed by additional 10 min to allow diffusion of viral particles. For behavioral experiments, viruses were injected at high titer. After removal of the injection needle, the open skin was sutured. Mice were then returned to home cages and body weight and signs of illness were monitored until full recovery from surgery (∼1 week). Electrophysiological and behavioral experiments were performed 3 weeks after lentiviral injection.
Plasmid construction, transfection and luciferase assay
The 5′ UTR of mouse Ophn1 mRNA was amplified from total RNA extracted from adult mouse brain. Primer sequences based on NCBI Reference Sequence (NM_05 2976.3) were as follows: sense 5′-CTAGCTAGCAGTTTCCGTAGGGAAGCGC-3′ and antisense 5′-TAGGCTAGCATACTTG AGCCTCTGGCGG-3′. The PCR product was then cloned into the pCIneo-Luciferase (Fluc) reporter vector. The resulting 5′UTR-OPHN1-Fluc construct was confirmed by DNA sequencing. HEK293T cells were grown in 12-well plates in Gibco DMEM+Glutamax (Life Technologies) supplemented with 10% FBS, 100 units of penicillin/streptomycin per ml. 5′UTR-OPHN1-Fluc or control Fluc vector were transfected with Renilla luciferase plasmid pRL-TK (Promega, Madison, WI) into HEK293T cells at 50–80% confluency using Lipofectamine LTX plus (Life Technologies). HEK293T cells were treated 24 h after transfection with 150 nM thapsigargin (Sigma) or vehicle (0.1% DMSO) for 6 h. Cell extracts were prepared in passive buffer and assayed for Rluc (internal control) and Fluc activity using a dual-luciferase reporter assay system (Promega). The ratio of Fluc to Rluc activity was reported as relative light units (RLU).
Spatial recognition
Spatial recognition was performed as previously described15,16. Briefly, behavior was recorded by cameras positioned above the training chamber. Exploration of the objects was defined as sniffing (with nose contact or head directed to the object) within a 2-cm radius of the object. Sitting or standing on the objects was not scored as exploration. The investigators performing and scoring the behavior were blind to the genotype. Data are expressed as a percentage of reexploration (on day 3) relative to the initial exploration time (on day 2).
Statistical analyses
Data are presented as means ± s.e.m. The statistics were based on the two-sided Student’s t-test or one- or two-way ANOVA with Bonferroni post hoc analysis to correct for multiple comparisons, unless otherwise indicated. P < 0.05 was considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Normality was tested by the Shapiro-Wilk or the Jarque-Bera test. Equal variances were assessed by F-test. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications6,18,19,21,26,29,30,37,40. For the linear regression, both the correlation coefficient (r) and P value were calculated to evaluate the strength and significance of the linear correlation. r values were indicated for individual linear regressions. For behavior and culture studies, animals or cultures were randomly assigned to control and treatment groups whenever appropriate. These experiments were performed and analyzed blind to treatment conditions and/or genotype. No animals or data points were excluded from analyses.
A Supplementary methods checklist is available.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Nakazawa and L. Van Aelst for the fPKR frozen embryos and Ophn1 shRNA, respectively. This work was supported by grants from the US National Institutes of Health to M.C.-M. (NIMH 096816, NINDS 076708) and R.J.K. (DK042394, DK088227, HL052173), the Intellectual Disability Research Center (P30HD024064) and Dan L. Duncan Cancer Center (P30CA125123) Genomic and RNA Profiling Cores, and the Cancer Prevention and Research Institute of Texas (CPRIT) RP100861.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.C.-M., G.V.D.P., W.H. and S.A.B. designed the experiments and wrote the manuscript; G.V.D.P. conducted electrophysiology and behavioral experiments and analyzed data; W.H. conducted behavioral, polysome profiling, qRT-PCR and immunoblotting experiments and analyzed data; S.A.B. conducted neuron culture, immunostaining, SUnSET and immunoblotting experiments and analyzed data; C.-C.H. performed firefly luciferase reporter experiments; P.B. analyzed RNA-seq data; A.P. contributed to the discussion of the electrophysiological experiments; C.S. and R.K. contributed to the characterization of ISRIB and the generation of Eif2s1A/A;ftg mice, respectively; K.K. and P. W. contributed to in-depth discussion of the project and editing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat. Rev. Neurosci. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 3.Luscher C, Huber KM. Group 1 mGluR-dependent synaptic long-term depression: mechanisms and implications for circuitry and disease. Neuron. 2010;65:445–459. doi: 10.1016/j.neuron.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collingridge GL, Peineau S, Howland JG, Wang YT. Long-term depression in the CNS. Nat. Rev. Neurosci. 2010;11:459–473. doi: 10.1038/nrn2867. [DOI] [PubMed] [Google Scholar]
- 5.Gladding CM, Fitzjohn SM, Molnar E. Metabotropic glutamate receptor-mediated long-term depression: molecular mechanisms. Pharmacol. Rev. 2009;61:395–412. doi: 10.1124/pr.109.001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 7.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational Control of Long-Lasting Synaptic Plasticity and Memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J. Neurosci. 2004;24:6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile × syndrome. J. Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism defne an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dever TE. Gene-specifc regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 15.Goh JJ, Manahan-Vaughan D. Spatial object recognition enables endogenous LTD that curtails LTP in the mouse hippocampus. Cereb. Cortex. 2013;23:1118–1125. doi: 10.1093/cercor/bhs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh JJ, Manahan-Vaughan D. Endogenous hippocampal LTD that is enabled by spatial object recognition requires activation of NMDA receptors and the metabotropic glutamate receptor, mGlu5. Hippocampus. 2013;23:129–138. doi: 10.1002/hipo.22072. [DOI] [PubMed] [Google Scholar]
- 17.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 18.Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J. Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis-dependent long-term depression in hippocampal area CA1. J. Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- 20.Morishita W, Marie H, Malenka RC. Distinct triggering and expression mechanisms underlie LTD of AMPA and NMDA synaptic responses. Nat. Neurosci. 2005;8:1043–1050. doi: 10.1038/nn1506. [DOI] [PubMed] [Google Scholar]
- 21.Snyder EM, et al. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat. Neurosci. 2001;4:1079–1085. doi: 10.1038/nn746. [DOI] [PubMed] [Google Scholar]
- 22.Boyce M, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 23.Robert F, et al. Initiation of Protein Synthesis by Hepatitis C Virus Is Refractory to Reduced eIF2·GTP·Met-tRNAiMet Ternary Complex Availability. Mol. Biol. Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z, et al. eIF2alpha phosphorylation-dependent translation in CA1 pyramidal cells impairs hippocampal memory consolidation without affecting general translation. J. Neurosci. 2010;30:2582–2594. doi: 10.1523/JNEUROSCI.3971-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadif Kasri N, Nakano-Kobayashi A, Van Aelst L. Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron. 2011;72:300–315. doi: 10.1016/j.neuron.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ron D, Harding HP. In: Translational Control in Biology and Medicine Cold Spring Harbor. Mathews MB, Sonenberg N, Hershey JWB, editors. Vol. 39. Cold Spring Harbor Laboratory Press; 2007. pp. 345–368. [Google Scholar]
- 28.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 29.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park S, et al. Elongation factor 2 and fragile × mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiler IJ, et al. Fragile × mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J. Neurosci. 2007;27:13273–13278. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graber TE, et al. Reactivation of stalled polyribosomes in synaptic plasticity. Proc. Natl. Acad. Sci. USA. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung YJ, et al. The fragile × mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. J. Biol. Chem. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- 35.Costa-Mattioli M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costa-Mattioli M, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu PJ, et al. Suppression of PKR promotes network excitability and enhanced cognition by interferon-gamma-mediated disinhibition. Cell. 2011;147:1384–1396. doi: 10.1016/j.cell.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Back SH, et al. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoica L, et al. Selective pharmacogenetic inhibition of mammalian target of rapamycin complex I (mTORC1) blocks long-term synaptic plasticity and memory storage. Proc. Natl. Acad. Sci. USA. 2011;108:3791–3796. doi: 10.1073/pnas.1014715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, et al. mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat. Neurosci. 2013;16:441–448. doi: 10.1038/nn.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan J, Khan SN, Harvey I, Merrick W, Pelletier J. Eukaryotic protein synthesis inhibitors identifed by comparison of cytotoxicity profles. RNA. 2004;10:528–543. doi: 10.1261/rna.5200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 45.Kaech S, Banker G. Culturing hippocampal neurons. Nat. Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.