Abstract
SOSIP gp140 trimers represent a soluble, stabilized, proteolytically cleaved form of the HIV-1 envelope (Env) glycoproteins. SOSIP gp140 derived from a subtype A HIV-1 isolate, KNH1144, forms exceptionally stable trimers that resemble virion-associated Env in antigenicity and topology. Here, we used electron microscopy to demonstrate that KNH1144 SOSIP gp140 trimers bound three soluble CD4 molecules in a symmetrical orientation similar to that seen for native Env. We compared the immunogenicities of KNH1144 SOSIP gp140 trimers and gp120 monomers in rabbits and found that the trimers were superior at eliciting neutralizing antibodies (NAbs) to homologous virus as well as neutralization-sensitive subtype B and C viruses. The NAb specificities for SOSIP antisera mapped in part to the CD4 binding site on gp120. We also observed adjuvant-dependent induction of antibodies to the residual levels of host cell proteins (HCPs) contained in the purified Env preparations. When present, HCP antibodies enhanced pseudovirus infection. Our findings are relevant for the further development of Env-based vaccines for HIV-1.
Keywords: SOSIP gp140, KNH1144, neutralizing antibody, HIV-1
INTRODUCTION
Human immunodeficiency virus type-1 (HIV-1) vaccine candidates have been designed to stimulate neutralizing antibodies (NAbs), vigorous cell-mediated immunity (CMI) or both [1]. Broadly neutralizing antibodies (bNAbs) have the potential to provide protection or blunt the early stages of HIV-1 viremia [2–6], and elicitation of bNAbs remains a major goal of HIV-1 vaccine research [7–9]. The HIV-1 envelope (Env) spike proteins responsible for receptor binding and viral entry are also the targets of NAbs [10;11]. The most widely studied subunit immunogen, monomeric gp120 (called “gp120” herein), has not been shown to induce bNAbs at significant titers [12–22]. Similar results have been reported for gp41-based subunit immunogens [23].
We previously described the design of SOSIP HIV-1 gp140 trimers (called “SOSIP” herein) [24;25], which are stabilized by a gp120-gp41 intersubunit disulfide bond (termed “SOS”) and an isoleucine to proline mutation (“IP”) at gp41 residue 559. These SOSIP mutations have been effectively applied to several HIV-1 strains [25–28], including subtype A KNH1144 [26;29], subtype B JR-FL [25;27;28] and subtype C Du151 (unpublished data). Of these, KNH1144 SOSIP gp140 formed particularly stable and homogeneous trimers and retained favorable antigenic properties [26;29].
We previously compared the immunogenicity of JR-FL SOSIP gp140 trimers, uncleaved gp140 trimers and gp120 monomers when administered to rabbits via protein-only and DNA-prime/protein-boost immunization regimens [27;30]. SOSIP gp140 trimers more frequently elicited NAbs effective against the homologous, neutralization-resistant strain JR-FL, than did the other immunogens. All three proteins induced NAbs against more sensitive strains, but the breadth of activity against heterologous primary isolates was limited.
Here, electron micrograph structures of KNH1144 SOSIP gp140 trimers in complex with soluble CD4 (sCD4) are described and compared with models for native gp120 trimers in the CD4-bound state. We also describe a comparative immunogenicity study of KNH1144 SOSIP gp140 trimers and gp120 monomers in rabbits. SOSIP antisera were superior to gp120 antisera at neutralizing homologous virus and neutralization-sensitive heterologous viruses. We investigated the specificity of the neutralizing activity elicited here and that previously elicited by our clade B prototype SOSIP gp140 [30]. In addition, methods are described for assessing the potential presence of Abs to host cell proteins and their effects on neutralization assays.
MATERIALS AND METHODS
Production of trimeric SOSIP and monomeric gp120 immunogens
The Env expression constructs, KNH1144 SOSIP gp140 with the hexaarginine (R6) mutation and KNH1144 monomeric gp120, have been described previously [26;29]. SOSIP gp140 and gp120 proteins were expressed in human embryonic kidney (HEK) 293T cells as previously described [29]. Briefly, 293T cells were seeded into triple flasks (Corning Life Sciences, Lowell, MA) in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum and supplements 24 hr prior to transfection. Cells were co-transfected with SOSIP-PPI4 and furin-pcDNA3.1 using polyethylenimine (Polysciences, Inc., Warrington, PA). Following transfection, cells were cultured for 48 hr in DMEM containing 0.05% bovine serum albumin. Supernatants were harvested, clarified and stabilized with protease inhibitors (Sigma-Aldrich, St. Louis, MO).
SOSIP gp140 trimers were purified as described previously [29]. Briefly, clarified 293T supernatant was concentrated ~50-fold by tangential flow filtration over a 100 kDa molecular weight cut-off (MWCO) membrane (Pall Corporation, East Hills, NY). Concentrated cell culture fluid was precipitated by ammonium sulfate, and the supernatant containing SOSIP gp140 was captured on a Galanthus nivalis agglutinin (GNA) lectin affinity column (Vector Laboratory, Burlingame, CA). Impurities, such as monomers, dimers and α2-macroglobulin, were removed by two sequential DEAE anion exchange columns. Tween 20 was added prior to the second DEAE column to convert non-covalent aggregates into gp140 trimers [29]. KNH1144 gp120 was purified by GNA lectin, Q Sepharose, and Superdex 200 size-exclusion chromatographies (GE Healthcare, Piscataway, NJ) [31].
MAbs and soluble CD4
Monoclonal antibodies (mAbs) directed to HIV-1 gp120 included mAbs b12 and 15e, both directed to epitopes that overlap the CD4bs [32;33]; mAb 2G12 directed to a carbohydrate cluster on the heavily glycosylated silent face of gp120 [34;35]; E51 directed to a CD4-inducible (CD4i) epitope [36]; and 447-52D, F425, and LE311 directed to the V3 loop [31;37–39]. Anti-gp41 mAbs included 2F5, Z13e1 and 4E10 directed to the membrane-proximal external region (MPER) [40–42]; and 7B2 directed to the cluster I epitope. MAbs b12, 2G12, 2F5, Z13e1 and 4E10 are broadly neutralizing [43]; mAb E51 can neutralize some primary isolates in the presence of sCD4 [36], and V3 loop-specific mAbs neutralize some primary isolates [44;45]. MAbs 2G12 and 2F5 were purchased from Polymun Scientific Inc. (Vienna, Austria). MAbs b12 and 4E10 and two-domain soluble CD4 (2D-sCD4) were provided by Drs. Ralph Pantophlet and Robert Pejchal (The Scripps Research Institutes, La Jolla, CA). Four-domain soluble sCD4 and CD4-IgG2 were produced as described [46;47].
Analysis of purified trimeric SOSIP gp140 and monomeric gp120 immunogens
Blue Native polyacrylamide gel electrophoresis (BN-PAGE) and sodium dodecyl sulfate (SDS)-PAGE analyses were performed as previously described [31]. Thyroglobulin (669 kDa) and ferritin (440 kDa) (GE Healthcare, Piscataway, NJ) were used as molecular weight standards in BN-PAGE. Gels were stained with colloidal Coomassie blue (Invitrogen, Carlsbad, CA). Protein purity was determined by densitometric analysis of the stained gels by the use of ImageQuant software (Molecular Devices, Sunnyvale, CA).
Trimer-sCD4 complexes were prepared by co-incubation overnight at 4°C. Free ligands were removed using a 100 kDa MWCO spin filter (Sartorius, Edgewood, NY). The formation of trimer-ligand complex was detected by trimer molecular weight shift in BN-PAGE analysis [37]. The molecular weight and binding stoichiometry of trimer-sCD4 complexes were estimated with reference to the standards, using ImageQuant software.
Env protein concentration was determined using the bicinchoninic acid (BCA) assay (Bio-Rad Laboratories Inc., Hercules, CA). Purified JR-FL gp120 was used as a concentration standard [48]. The level of residual HCP in SOSIP trimers or gp120 monomers was determined using a HEK 293T ELISA kit (Cygnus, Southport, NC), according to the manufacturer’s protocol. Endotoxin levels were determined using the Limulus Amebocyte Lysate (LAL) Pyrotell assay kit (Associates of Cape Cod Inc., Falmouth, MA).
Electron microscopy
Detergent can interfere with negative stain electron microscopy (EM). Consequently, detergent was removed from KNH1144 SOSIP with the mini Detergent-OUT™ Detergent Removal Kit (Calbiochem, Gibbstown, NJ), according to the manufacturer’s protocol. Detergent-depleted KNH1144 SOSIP was incubated with four-domain sCD4 at room temperature for 30 minutes and then analyzed by negative-stain EM as previously described [29]. Briefly, two micrograms of complex were diluted in 200 μl BSB (0.1 M H3BO3, 0.025 M Na2B4O7, 0.075 M NaCl, pH 8.3), and affixed to a carbon support membrane. The membrane was stained with 1% uranyl formate, and mounted on 600-mesh copper grids for analysis. Electron micrographs were recorded at 100,000x magnification at 100 kV on a JOEL JEM 1200 electron microscope.
i) Immune complex image processing and averaging
Electron micrographs were digitalized on an AGFA DUOSCAN T2500 Negative Scanner (Ridgefield Park, NJ) at 1000 pixels per inch scanning resolution. Putative SOSIP complexes with three sCD4 molecules were selected and windowed as 180 × 180 pixel images. A total of 86 randomly oriented complexes were windowed and then brought into approximate alignment utilizing SPIDER software [49]. For further averaging and analysis, the windowed images of complexes were manually assigned to two categories (clockwise and counterclockwise) based on the chiral appearance of the liganded sCD4 molecules. The clockwise-oriented images were mirrored and subjected to averaging with counterclockwise-oriented images. The images were centered, normalized by scaling to adjust the image pixel values to a mean of 1.0, and masked to exclude the surrounding area. The complexes were then subjected to an alignment through classification. Particles were sorted into three classes using k-means clustering [49]. The images in each class were averaged and the three class averages were then used in a multi-reference alignment scheme in which each complex was matched and aligned to the most similar of the three reference images. The newly aligned complexes were then reclassified and class averages were calculated to generate new references for the next round of multi-reference alignment. The classification and multi-reference alignment steps were repeated until no further improvement in the image averages was observed. Angular measurements of the sCD4-gp120 interaction were made using Image-Pro Plus software (Media Cybernetics, Inc., Bethesda, MD).
ii) Molecular modeling of sCD4-SOSIP complexes
The final sCD4-SOSIP image class averages were compared and found to be similar in appearance. The class average subjectively judged to display the most inherent symmetry was selected and rotationally averaged (trimerized). To facilitate atomic model fitting, 12 identical trimerized images were stacked and imported into the Chimera software package (http://www.cgl.ucsf.edu/chimera/). The X-ray structure of the gp120 core with V3 loop in complex with 2D-sCD4 (pdb code: 2B4C) [50] was manually fitted into one subunit of the stacked sCD4-SOSIP images. The X-ray structure of 4-domain sCD4 (pdb code: 1wio) [51] was then superimposed onto this image. Domains D3 and D4 of 4-domain sCD4 were adjusted slightly to fit into the corresponding density in the sCD4-SOSIP image. The sCD4-gp120 atomic model was then rotationally trimerized based on the fitted position to generate the atomic model of sCD4-gp120 trimers.
Rabbit immunizations
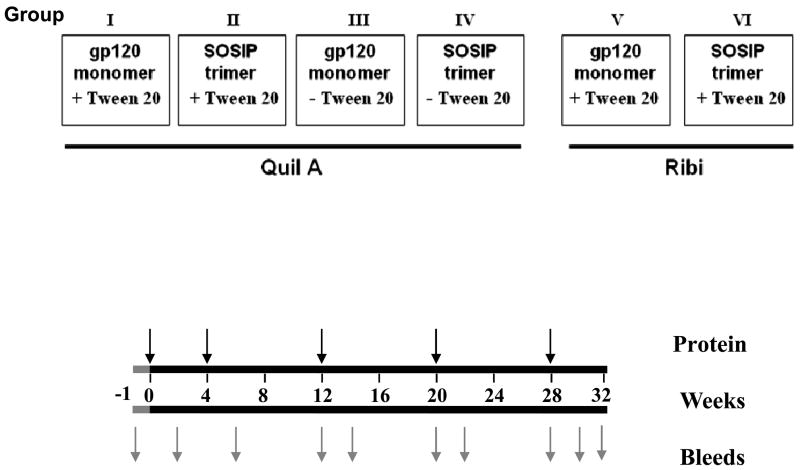
Fig. 1 outlines the rabbit immunogenicity study. Female New Zealand White rabbits (6 to 10 weeks old; Millbrook Farm, Concord, MA) were immunized (n = 4 per group) with KNH1144 SOSIP gp140 or gp120 in the presence of Quil A (Accurate Chemical and Scientific Corp., Westbury, NY) or Ribi (Sigma-Aldrich, St Louis, MO) adjuvant, as indicated. Doses of 100 μg/animal of Quil A were used in Groups I–IV. In groups V and VI, each animal received 25 μg Ribi adjuvant (Sigma Cat # M6536) containing monophosphoryl Lipid A (MPL) and trehalose dicorynomycolate (TMD) for the first dose, followed by Ribi adjuvant (Sigma Cat # M6661) containing MPL, TDM and cell wall skeleton from a tubercle bacillus for the subsequent doses. Vaccines were administered at multiple anatomical sites (300 μl intramuscularly into each hind leg, 50 μl intradermally at six sites, and 100 μl subcutaneously into the neck region, for a total of 1 ml) [27] at 0, 4, 12, 20 and 28 weeks.
Fig. 1. Design of the immunogenicity study comparing KNH144 SOSIP trimer with gp120 monomer.
Animals in Groups I to IV received Quil A as adjuvant in the presence or absence of 0.05% Tween 20, as indicated. Animals in Groups V and VI received Ribi adjuvant. Animals in Groups V and VI received an initial prime with 100 μg Env, followed by 30 μg boosts. A 30 μg dose of Env was used for all immunizations of animals in Groups I to IV.
Animals in groups I–IV received 30 μg doses of gp120 monomer and SOSIP trimer that were formulated with or without 0.05 % Tween 20; Quil A was used as adjuvant. Animals in groups V and VI received Tween-treated Env proteins, Ribi adjuvant, and a 100 μg priming dose of proteins followed by 30 μg booster injections. Sera were collected at weeks -1 (pre-immunization), 2, 6, 12, 14, 20, 22, 28, 30, and 32.
ELISA for Ab binding to monomeric gp120
Enzyme-linked immunosorbent assay (ELISA) was used to measure binding of serum Abs to monomeric KNH1144 gp120 immobilized on plates via a sheep Ab to the gp120 C-terminus (Cat # 6205; Cliniqa Corp., Fallbrook, CA), as previously described [52–54]. Goat anti-sheep IgG (H + L) alkaline phosphatase conjugate (Jackson ImmunoResearch, West Grove, PA) was used as the secondary Ab. Midpoint binding titers were estimated by interpolation using Prism (GraphPad Software, Inc., La Jolla, CA).
In additional experiments, we measured the binding titers of SOSIP and gp120 antisera to gp120s derived from HIV-1 strains MN and HXB2. Both gp120 proteins were expressed in HEK 293 cells using vaccinia virus vectors and purified through GNA lectin column chromatography as previously described [55]. The purified gp120 proteins from these two isolates were coated on ELISA plates directly and midpoint serum titers were measured as above.
Virus neutralization assays
The neutralizing activities of pre- and post-immune sera were evaluated using the PhenoSense™ HIV Entry assay (Monogram Biosciences, South San Francisco, CA). This assay measures the infectivity of Env-complemented, luciferase-encoding pseudoviruses in a single-cycle assay using CCR5- or CXCR4-expressing U87.CD4 cells [43;56–58]. Rabbit sera were heat-inactivated at 56°C for 30 min before use. Neutralizing activity is reported as the dilution of each serum required to confer 50% inhibition of infection. Specific neutralization of HIV-1 required that the titer was > 1:10 and was ≥ 2.5-fold higher than that observed against the control amphotropic murine leukemia virus (MLV). Virus-serum combinations were tested in triplicate. NL4-3 was run as a control virus in all assays.
Flow cytometry analysis of serum binding to U87 and HEK 293T cells
U87-CD4-CCR5 or HEK 293T cells at 5×105 cells per sample were incubated with pre-immune (pre-bleed) and post-immune rabbit sera at serial dilutions for 30 min on ice. After washing, secondary Alexa fluor® 488 conjugated goat anti-rabbit IgG (H+L) (Invitrogen, Carlsbad, CA) was added for 15 min on ice. Flow cytometry analysis was performed on a BD FACSCalibur instrument (BD Biosciences, NJ) after washing the cells. Data were analyzed using FloJo software (TreeStar, Ashland, OR).
Mapping neutralization specificities
i) Plasmids and pseudovirus production
KNH1144 gp160 Env sequence [59] was synthesized using optimized codons and expressed from the pCDNA3.1 plasmid (Invitrogen, Carlsbad, CA). The wild-type KNH1144 Env sequence [60] was cloned into pCAGGS truncated at amino acid 709 (HXB2 numbering system), leaving 3 amino acids of the gp41 cytoplasmic tail. Mutations to introduce a gp120-gp41 disulfide bridge (SOS mutant) have been described previously [42;61]. SOS and wild-type (WT) versions of HIV-1 subtype B JR-FL Env with a similar C-terminal truncation have been described earlier [37;61]. A plasmid expressing vesicular stomatitis virus G protein (pVSV-G) has also been previously described [62]. Pseudoviruses were produced by transient cotransfection of HEK 293T cells with an Env-expressing plasmid and plasmid pNL-LucR-E-, which encodes the luciferase gene and an HIV-1 genome truncated in the Env and Nef genes, with a frameshift in Vpr [61]. For virus capture experiments, cells were triply transfected with the above two plasmids and pVSV-G [62].
ii) Virus capture competition
Virus capture competition measures the ability of serum to inhibit capture of pseudovirions by various mAbs [62–64]. The mAbs were coated on microwells overnight at 5 μg/ml. The wells were then washed and blocked with 3% bovine serum albumin in PBS. The sera in graded dilutions were incubated with KNH1144 pseudoviruses for 30 min. As the HIV-1 pseudoviruses may be neutralized by the competing Ab, pseudoviruses expressing VSV-G were used to provide an independent readout of captured virus irrespective of neutralization. For E51 mAb capture, a fixed concentration of 5μg/ml sCD4 was added. The virus-serum mixtures were then added to mAb-coated wells for 3 hrs followed by washing with PBS, and CF2.CD4.CCR5 cells were overlaid [65]. Two days later, infection was measured by luciferase. The reciprocal serum dilution that inhibited mAb-mediated virus capture by 50% was then recorded.
iii) Neutralization assays using CF2 cell targets
Neutralization was assessed in various formats, as described previously [37;61]. All assays involved a 15 minute ‘spinoculation’ step at room temperature, followed by a 2 hr infection at 37°C [61], after which the medium was changed. The cells were cultured for three more days, and then luciferase activity was measured. Assays were performed at least four times and representative data are shown.
a) Standard format
The standard protocol provides a reference for neutralization in other formats. Essentially, Ab or serum was mixed with virus for 1 hr before adding to target cells.
b) Post-CD4 format
In post-CD4 neutralization assays, virus was incubated with a fixed concentration of 5 μg/ml sCD4 and graded concentrations of mAb or sera. Subsequently, infection was assayed on CF2.CCR5 cells.
c) Post-CD4/CCR5 format
Post CD4/CCR5 binding neutralization was measured using SOS pseudovirus [61]. These particles can engage CD4 and CCR5, but require a reducing agent to break the gp120-gp41 disulfide, allowing fusion to proceed. SOS pseudovirions were allowed to attach to CF2.CD4.CCR5 cells for 2 hr. Unbound particles were washed away and graded concentrations of mAbs were added. Following a 1 hr incubation, infection was activated using 5 mM dithiothreitol for 10 minutes to reduce the gp120-gp41 disulfide bond [61].
iv) Blue Native PAGE trimer shifts
We investigated Ab binding to native, virion-derived trimers by BN-PAGE [37;62;63;66]. Briefly, concentrated pseudoviruses were mixed with rabbit sera for 5 minutes. The viruses were disrupted in 1% Triton X-100/1% NP40 in 1 mM EDTA in 1.5 M aminocaproic acid to liberate envelope protein. In the case of 2G12, 40 μl of protein A Dynabeads (Invitrogen, Carlsbad, CA) were added. After 5 minute incubation at room temperature, tubes were applied to a magnet to separate the beads. An equal volume of 2x sample buffer containing 100 mM morpholinepropanesulfonic acid (MOPS), 100 mM Tris-HCl, pH 7.7, 40% glycerol, and 0.1% Coomassie blue was then added. Samples were loaded onto a 4–12% Bis-Tris NuPAGE gel (Invitrogen, Carlsbad, CA). Samples were electrophoresed at 4°C for 3 hr at 100 V with 50 mM MOPS/50 mM Tris, pH 7.7, containing 0.002% Coomassie blue as the cathode buffer and the same buffer without Coomassie blue as the anode buffer. The gel was then blotted onto polyvinylidene difluoride, destained, and probed by mAbs b12, 2G12, E51, 39F, 2F5 and 4E10 [63]. Goat anti-human Fc alkaline phosphatase conjugate was then used for detection (Jackson ImmunoResearch, West Grove, PA). Band densities were determined using UN-SCAN-IT software, as were approximate molecular weights of trimer complexes [66].
Statistical analyses
Inter-group comparisons of midpoint ELISA titers or of neutralization titers at a specified time point were performed by two-tailed student t-tests using JMP software v6.0.3 (Cary, NC).
RESULTS
Production and characterization of Env immunogens
Purified KNH1144 SOSIP migrated in BN-PAGE analysis as a gp140 trimer (Fig. 2), consistent with our prior results [29]. The trimers were free of gp140 monomers, dimers, or high molecular weight aggregates. No gp140 aggregates or uncleaved gp140 were observed in SDS-PAGE analysis (data not shown). Monomeric KNH1144 gp120 was prepared to a similar degree of purity (not shown). SOSIP gp140 trimers were recognized by mAbs 2G12, b12, b6, 15e, 17b (±sCD4) and CD4-IgG2 in immunoprecipitation assays [26].
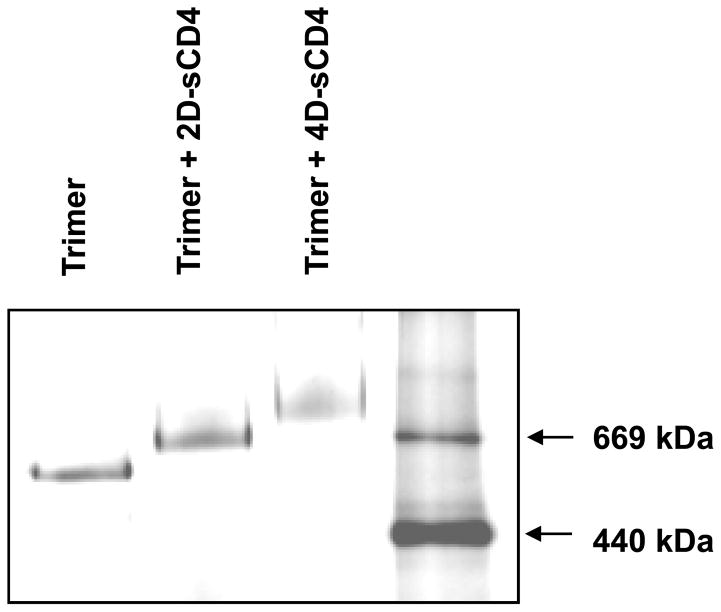
Fig. 2. BN-PAGE analysis of KNH1144 SOSIP in complex with soluble CD4 proteins.
5 μg of two-domain sCD4 (2D-sCD4) or 4-domain sCD4 (4D-sCD4) was incubated with 1 μg of KNH1144 trimer overnight at 4°C. BN-PAGE analysis was performed as previously described [29;42], except that Invitrogen 4–16% Bis-Tris Native PAGE and buffers were used. Thyroglobulin (669 kDa) and ferritin (440 kDa) were used as molecular weight standards.
We investigated the binding of 2D-sCD4 and four-domain sCD4 to KNH1144 SOSIP trimers by BN-PAGE gel-shift analysis (Fig. 2). Each CD4-SOSIP complex migrated as a single homogeneous band, with molecular weight shifts of ~75 kDa for 2D-sCD4 and ~165 kDa for sCD4. These results are consistent with the binding of 3 molecules of each ligand (molecular weights of ~25 kDa for 2D-sCD4 and ~55 kDa for sCD4). These shifts were associated with a depletion of unliganded trimer.
HCP levels were 10.8 parts per million (ppm, defined as ng HCP per mg product) for KNH1144 SOSIP trimer and less than 1 ppm for gp120. The differences in HCP levels in the SOSIP and gp120 preparations may reflect differences in the methods used to produce and purify the proteins. In addition, HCP may interact differently with SOSIP and gp120 proteins. Endotoxin levels were less than 0.6 endotoxin units per mg for both SOSIP gp140 trimers and gp120 monomers.
Electron microscopy
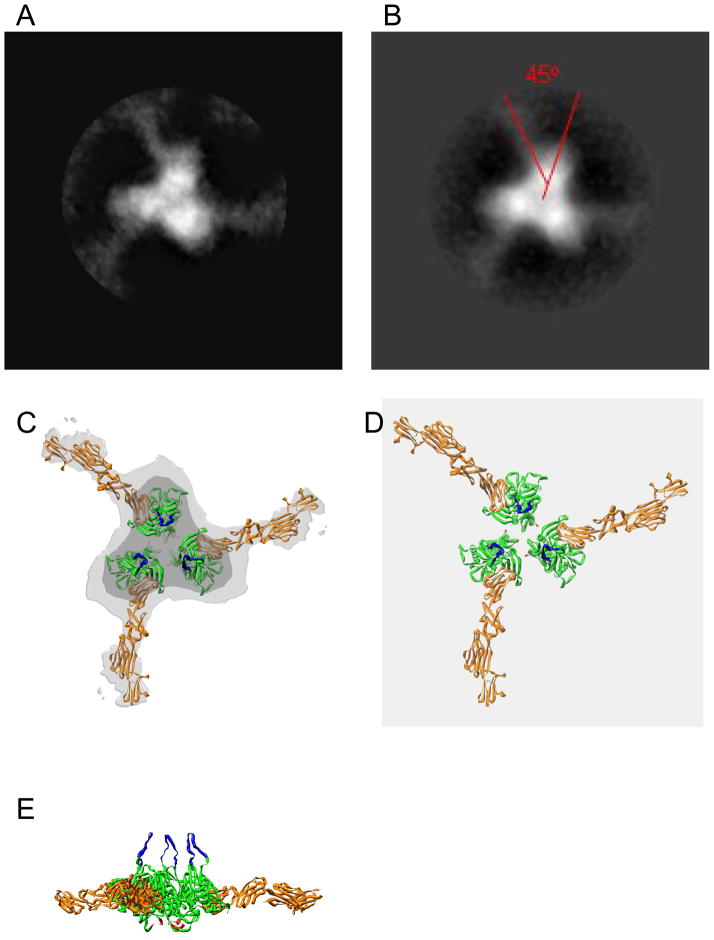
To expand upon prior EM studies of unliganded SOSIP trimers [29], trimer complexes with four-domain sCD4 were analyzed by negative stain EM (Fig. 3). Following incubation with sCD4 at saturating concentrations, three sCD4 molecules were engaged with most trimers (Fig. 3), consistent with the BN-PAGE results (Fig. 2). The sCD4 molecules attached to trimers in a symmetrical manner, each binding laterally to one side of each gp120 subunit. An analysis of 86 averaged sCD4-SOSIP complexes (Fig. 4A and B) indicates that sCD4 bound at about 45 degrees laterally with respect to the radial axis of the gp120 subunits. Due to the flattening effect resulting from the adherence of the complexes to the carbon substrate and the subsequent drying prior to EM analysis, there could be some unobserved degree of deflection of the projecting sCD4 molecules from the plane of the sCD4-SOSIP complexes. The imposition of a counterclockwise chirality (as viewed from above) on the SOSIP images (Fig 4A and 4B) places the CD4 molecules in an orientation very similar to that observed for native gp120 on virions [67]), and allowed the atomic structure of the CD4-liganded gp120 complex [50] to be fitted into the negative stain EM density map (Fig 4C). Top and side views of the fitted atomic models are also shown following removal of the EM image densities (Fig 4D and 4E, respectively).
Fig. 3. Negative stain electron micrograph gallery and interpretive diagram.
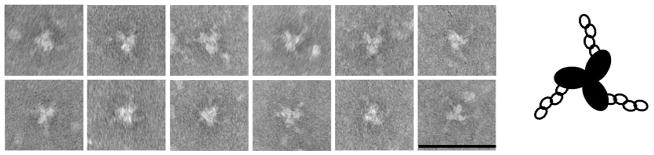
KNH1144 SOSIP gp140 trimers complexed with four-domain sCD4. Bar = 50 nm.
Fig. 4. Averaged sCD4-SOSIP complexes.
(A) Raw averaged complexes showing three sCD4 molecules binding per trimer. (B) Trimerized (rotationally averaged) sCD4-SOSIP complexes with sCD4 and gp120 axes for one subunit indicated by red lines. sCD4 appears less dense due to its narrower cross-sectional dimensions displacing less negative stain than that of the thicker SOSIP subunits. (C) Trimerized sCD4-SOSIP averaged image fitted with 2D-sCD4-gp120 crystal structure (pdb code: 2B4C) as modified to incorporate 4-domain sCD4 (pdb code:lwio). Green indicates the bulk of gp120 and gold indicates sCD4. The Light grey profile represents relaxed density threshold profile in which sCD4 densities are apparent, while the dark gray inner profile represents a more stringent density threshold profiling the major density of the SOSIP subunits. The atomic models were placed to maximize correspondence to relevant EM image densities. V3 loops (blue) and N- and C-termini (red) were positioned to approximate their expected positions within the Env spike (gp120 in gold). (D) Atomic model from (C) without density. (E) Atomic model from (D) rotated 90° for a side view.
Anti-gp120 titers of rabbit antisera
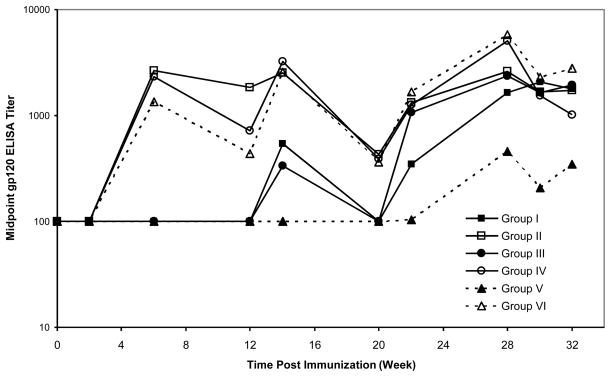
The sera from KNH1144 SOSIP and gp120 immunized rabbits were analyzed for anti-KNH1144 gp120 binding Abs by ELISA. Mean midpoint titers from each group depicted in Fig. 1 are presented in Fig. 5. Anti-gp120 Ab titers in SOSIP-immunized animals (groups II, IV and VI) reached high levels after only two immunizations. Titers displayed a saw-tooth pattern reflecting a rise and subsequent fall in titers between boosts. Gp120 titers developed more slowly in the gp120-immunized compared to SOSIP-immunized animals, with measurable levels of Abs requiring three (groups I and III) or four (group V) immunizations. At the end of study, mean titers for groups I and III were similar to those of the matched SOSIP groups, but group V titers remained significantly lower (p=0.05). Collectively, these results suggest that Ribi adjuvant was less effective than Quil A for stimulating responses to gp120, but the two adjuvants were comparable in stimulating gp120 Ab titers in response to SOSIP.
Fig. 5. Anti-gp120 binding Ab elicited by SOSIP trimer immunization.
Rabbits were immunized with SOSIP trimers (open symbols) or gp120 monomers (filled symbols) as follows: 30 μg of SOSIP (group II) or gp120 (group I) using Quil A in the presence of Tween 20 (square), 30 μg of SOSIP (group IV) or gp120 (group III) using Quil A in the absence of Tween 20 (circle), and 100 μg followed by 30 μg boosts of SOSIP (group VI) or gp120 (group V) using Ribi in the presence of Tween 20 (square) at the times indicated. The arrows indicate the time of immunizations. The anti-gp120 antibody responses were measured by ELISA. Each a point represents the mean midpoint anti-gp120 binding titer for each immunization group (n=4 animals).
Neutralization of HIV-1 Env-pseudotyped viruses by SOSIP/Ribi antisera
Rabbit sera from week 30 of the immunizations were analyzed in the PhenoSense™ neutralization assay against homologous (KNH1144) and heterologous viruses, using MLV glycoprotein pseudotyped virus as a control. Results for Groups V and VI are shown in Table 1. The inherent susceptibility of each virus to neutralization is indicated by the activity of the reference HIV-infected patient plasma Z23 (see bottom row of Table 1). Significant neutralization of the homologous KNH1144 virus was observed for one SOSIP antisera and none of the gp120 antisera. Heterologous neutralization was observed for some subtype B and C viruses. Antisera from all four SOSIP animals showed activity against neutralizing-sensitive viruses MB C8, SF162, MN and NL4-3. In contrast, gp120 elicited minimal neutralizing activity against these viruses (Table 1). The difference in titers of SOSIP and gp120 antisera was significant for SF162 (p = 0.04) and MN (p = 0.002). No intra-subtype neutralization was observed for the other subtype A viruses tested (ARW92020 and AUG93077), thus highlighting the relative resistance of these viruses. Collectively, these results suggest improved neutralization, especially of neutralization-sensitive viruses by SOSIP antisera relative to gp120 antisera. Given the differences in overall anti-KNH1144 gp120 ELISA titers between the gp120 and SOSIP immunized groups, it is impossible to infer whether NAbs were detected in the SOSIP serum due simply to an increased binding titer, or to a difference in overall specificity profile of the serum. We then measured the binding in ELISA of SOSIP antisera (animal 6-4) and gp120 antisera (animal 5-1) to gp120s derived from HIV-1 strains MN and HXB2. For MN gp120, the binding titers were 1,200 for the SOSIP antisera and 13,000 for the gp120 antisera. For HXB2, the corresponding titers were 450 and 3,200, respectively. Therefore, the SOSIP antisera contained lower amounts of antibodies that bound to these gp120s. MN virus was included in our virus neutralization panel, and HXB2 is highly similar in sequence to the NL4-3 virus that was included in the neutralization panel. MN and NL4-3 viruses were neutralized by SOSIP but not gp120 antisera (Table 1) despite the fact that the SOSIP antisera contained lower titers of antibodies to the corresponding gp120s. Therefore, no direct correlation between anti-gp120 titers and neutralization titers was observed for heterologous isolates MN and NL4-3.
Table 1. Neutralization of Env-pseudotyped viruses by SOSIP and gp120 antisera.
Sera were evaluated at week 30 for neutralization of different Env-pseudotyped viruses as indicated. The results represent the reciprocal of the dilution that resulted in 50% neutralization. A neutralization titer of 10 (1:10 dilution of sera) represents the lower detection limit of the assay for rabbit sera. HIV-1 pseudoviruses were not neutralized by pre-immune sera. Serum titers are color-coded as follows: white boxes (titer < 10 for rabbit sera or <40 for control HIV+ patient plasma), green boxes (titer = 11-100), yellow boxes (titer = 101-1000), and red boxes (titer > 1,000). The subtype of each HIV-1 virus is indicated in parentheses.
| Group | Immunogen | Animal | 50% Neutralization of env-pseudotyped viruses | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KNH1144 (A) | ARW92020 (A) | AUG93077 (A) | MBC8 (C) | MN (B) | SF162 (B) | NL43 (B) | MLV | |||
| V | gp120 | 5-1 | <10 | <10 | <10 | 11 | <10 | <10 | <10 | <10 |
| 5-2 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | ||
| 5-3 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | ||
| 5-4 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | ||
| VI | SOSIP trimer | 6-1 | <10 | <10 | <10 | 40 | 58 | 235 | 39 | <10 |
| 6-2 | <10 | <10 | <10 | 12 | 35 | 32 | 16 | <10 | ||
| 6-3 | <10 | <10 | <10 | 37 | 70 | 114 | 75 | <10 | ||
| 6-4 | 31 | <10 | <10 | 18 | 42 | 274 | 12 | <10 | ||
| Pre-immune | N/A | NA | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| Control Z23 | HIV+ patient plasma | N/A | 151 | 219 | 327 | 2,276 | 6,833 | 18,533 | 3,175 | <40 |
Enhancement of pseudovirus infectivity by Env/Quil A antisera
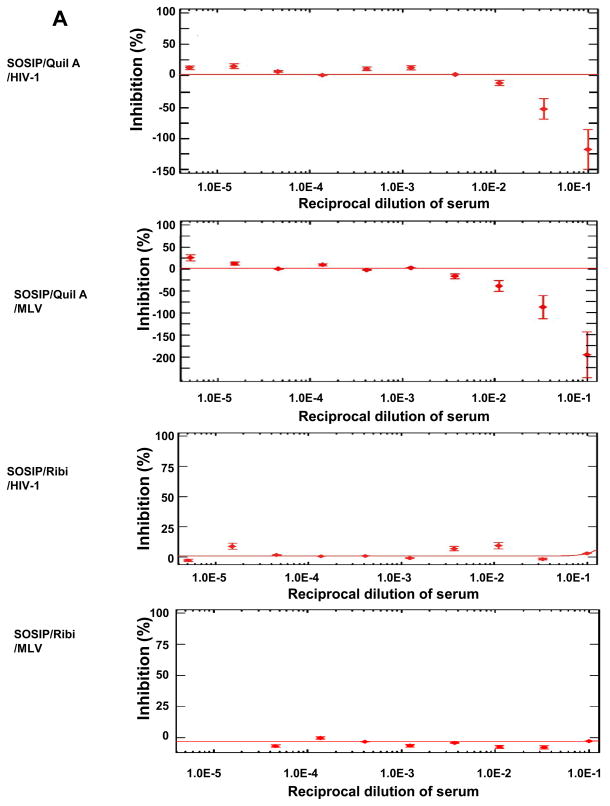
The sera from rabbits immunized with SOSIP in Quil A (groups II and IV) increased HIV-1 pseudovirus infectivity for U87-CD4-CCR5/CXCR4 cells. Data for subtype C 93IN905 are shown in Fig. 6A (top panel), and similar results were observed for other HIV-1 isolates (not shown). The same phenomenon was also observed for MLV pseudovirus (Fig. 6A, second panel), which was a control in the PhenoSense™ neutralization assay. However, this effect was not observed with sera from SOSIP/Ribi immunized animals (group VI) (Fig. 6A, third and bottom panels). Furthermore, no enhancement was found for monomeric gp120 antisera with either Quil A (Group I and III) or Ribi adjuvant (Group V) or pre-immune sera (not shown). The increased infectivity of both HIV-1 and MLV pseudoviruses in the presence of SOSIP/Quil A antisera suggested that anti-HIV Env antibodies in sera were unlikely to be responsible.
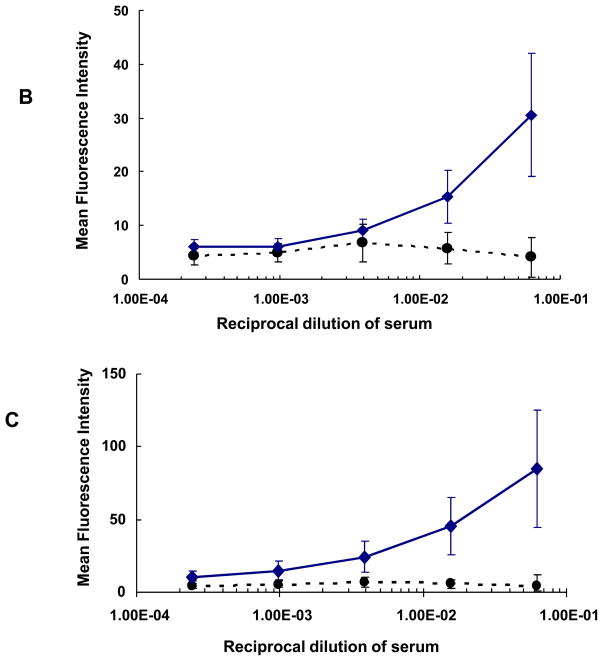
Fig. 6. Sera anti-cell reactivity and its effect on neutralization assay.
(A) Enhancement (as reflected in negative percent inhibition) of 93IN905 pseudovirus infection by SOSIP/Quil A antisera but not by SOSIP/Ribi or any gp120 antisera. Week 30 rabbit sera were evaluated for HIV-1 pseudovirus neutralization. MLV was used as the control. Data points in each figure indicate mean reciprocal dilutions of the serum from a representative animal (3 replicates) in each group that resulted in percent inhibition or enhancement of infectivity of pseudovirus on U87.CD4.CCR5 cells. The error bars represent one standard deviation. (B) Binding of week 30 rabbit sera to U87-CD4-CCR5 cells by flow cytometry. Circles indicate the mean of the mean fluorescence intensity (MFI) values for all pre-immune sera at different dilutions. The error bars represent one standard deviation. Diamonds represent the mean MFI values of eight SOSIP/Quil A antisera in Groups II and IV at different dilutions, and the error bars represent one standard deviation. (C) Binding of rabbit sera at week 30 to HEK 293T cells in flow cytometry analysis. Symbols have the same meaning as in (B).
We suspected that anti-HCP antibodies might explain the non-specific virus enhancement observed with SOSIP/Quil A sera. Flow cytometry experiments were performed to determine whether Abs elicited by SOSIP trimers in Quil A were capable of binding to U87-CD4-CCR5 cells (which were used in the neutralization assay at Monogram), and HEK293T cells (which were used for the production of pseudoviruses at Monogram). Sera from SOSIP/Quil A immunized animals bound avidly to U87-CD4-CCR5 cells (Fig. 6B) and HEK293T cells (Fig. 6C), suggesting the presence of anti-HCP Abs. Anti-HCP Abs were not detected in sera from the other groups (not shown). Thus, when Quil A (but not Ribi) was used as adjuvant, the residual HCPs in SOSIP trimers (10.8 ppm) appear to have elicited anti-HCP Abs that increased pseudovirus infectivity in the PhenoSense™ neutralization assay.
Dissecting the specificity of serum responses
i) Analysis of binding Abs
Virus capture competition measures the ability of serum to inhibit capture of pseudovirions by various mAbs [62–64]. Terminal sera from two rabbits, one immunized with gp120/Ribi and one with SOSIP/Ribi, were assessed by virus capture competition for the presence of Abs to defined epitopes exposed on KNH1144 pseudovirus particles (Table 2). Several well-characterized mAbs isolated from subtype B-infected individuals (namely b12, 2G12 and several V3 mAbs) were unable to capture KNH1144 efficiently and/or to self-compete effectively, limiting a complete analysis. However, 15e, E51 (in the presence of sCD4) and 7B2 captured the virus effectively. CD4bs mAbs, detected via competition with mAb 15e, were only observed in SOSIP serum. Conversely, titers to CD4i epitopes, as judged by E51 competition, were low or undetectable in both cases. Neither sera reacted to the immunodominant (cluster I) epitope of gp41, suggesting that this non-neutralizing epitope is buried or otherwise poorly immunogenic in SOSIP trimers, although the epitope might also be directly altered by SOSIP mutations.
Table 2. Binding analysis of sera and virus capture competition assay.
The sera at Week 30 from rabbits immunized with KNH1144 immunogens were analyzed. The result represents the reciprocal of the dilution that inhibits mAb-mediated virus by 50%. The serum titers are color-coded as in Table 1. Titers for self-competition Ab represent the concentration which inhibits mAb-mediated virus by 50%. The Ab titers are color-coded as follows: white boxes (titer > 10 μg/ml for mAbs or < 20 for sera), yellow boxes (titer = 0.11-10 μg/ml for mAbs or 20-1000 for sera), and red boxes (titer ≤ 0.1 μg/ml for mAbs and > 1000 for sera).
| Serum | Immunogen | Virus capture competition | ||
|---|---|---|---|---|
| 15e (CD4bs) | E51/sCD4 (CD4i) | 7B2 (cluster I) | ||
| 5-1 | KNH1144 gp120 | <20 | <20 | <20 |
| 6-4 | KNH1144 SOSIP | 180 | <20 | <20 |
| self-competition (μg/ml) | 0.14 | 0.011 | 0.037 | |
ii) Analysis of neutralization activity in different assay formats
Sera were analyzed in alternative assay formats to probe whether the NAbs inhibited early or late stages of viral entry (Table 3). The standard format provided a reference for neutralization in other formats. Post-CD4 format measured neutralizing activity between the CD4 and CCR5 binding steps while a post-CD4/CCR5 format measured the neutralizing activity after CCR5 binding. SOSIP gp140 serum (animal 6-4) neutralized the homologous virus, but the gp120 control serum (animal 5-1) did not detectably neutralize the homologous virus in the standard assay (Table 1). In the post-CD4/CCR5 assay format for detecting MPER activity, neither sera showed detectable activity. However, mAb 2F5 is active against KNH1144 (Table 3) and relevant subtype B viruses in both standard and post-CD4/CCR5 formats [37;61]. Therefore, MPER NAbs do not appear to mediate the neutralizing activity found in the SOSIP sera.
Table 3. Neutralization activities of KNH1144 antisera in different formats.
Neutralization titers for the KNH1144 sera were measured in the CF2-based assays. V3 and CD4i activity was assessed in a post-CD4 format and MPER titers were measured in post-CD4/CCR5 assays. Results are expressed as ID50s or IC50s corresponding to dilutions or concentrations in μg/ml. The titers are color coded for sera and Abs as described for Tables 1 and 2, respectively.
| Serum | Immunogen (Protein only) | KNH1144 | KNH1144 SOS |
|---|---|---|---|
| CF2 post-CD4 | CF2 post-CD4/CCR5 | ||
| 5-1 | KNH1144 gp120 | 500 | <10 |
| 6-4 | KNH1144 SOSIP | 12,000 | <10 |
| 2F5 (μg/ml) | 0.1 | 0.1 | |
| b12 (μg/ml) | >10 | >10 |
iii) Mapping of NAb specificity by BN-PAGE
We next investigated Ab binding to native, virion-derived trimers by BN-PAGE gel-shift assays, as described previously for subtype B Envs [37;62;63;66]. Adapting this assay successfully to subtype A isolates depends on several criteria, including efficient Env expression, migration of trimers as a well-defined band in BN-PAGE and efficient gp120/gp41 processing[66]. Initial studies with KNH1144 codon-optimized gp160 (KNH1144opt) revealed poorly expressed and diffuse trimer bands in BN-PAGE (data not shown). However, gp160 from the wild-type clone, KNH1144ec1, expressed more efficiently and formed a sharp trimer band. We therefore selected this KNH1144ec1 gp160 clone for mapping. Since anti-HCP Abs do not interfere in this Env binding assay, Env/Quil A antisera from SOSIP rabbit 4-3 and gp120 rabbit 3-2 were used in the experiments.
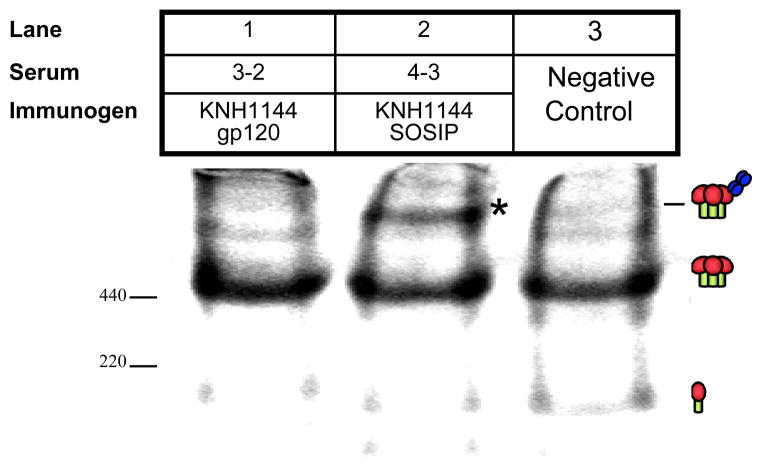
SOSIP serum 4-3 induced a perceptible shift of KNH1144ec1 trimers (Fig. 7, lane 2; marked with an asterisk). Although a faint band appeared to be present above the trimer band in lane 1 in which the gp120 serum was assessed, this band was much weaker and moreover, was comparable to the faint bands above the trimer band in the control lane 3. Thus, the SOSIP serum appears better able to recognize native Env trimers than the gp120 serum. The shift induced by SOSIP serum was consistent with the binding of a single Ab to the trimers, as estimated by UN-SCAN-IT software, according to our previous analysis [68]. An analysis of neutralizing plasmas from chronically infected patients with broad neutralizing activity in this BN-PAGE assay [68] revealed a largely consistent pattern: that trimer binding begins with the appearance of trimer-IgG complexes at low IgG concentrations. Then, at higher IgG concentrations, the unliganded trimer band becomes depleted as more IgG binds. The significant amount of remaining unliganded trimer (Fig. 7, lane 2), coupled with the presence of prominent trimer-IgG complexes, therefore suggests partial trimer complexing by the SOSIP, but not the gp120, serum (Fig. 7, lane 1).
Fig. 7. Native gel shift mobility assay to detect KNH1144 trimer-binding Abs.
Sera from KNH1144 SOSIP trimer and gp120 monomer immunized rabbits (#4-3 and # 3-2, respectively) were evaluated for binding to the WT KNH1144 trimer in BN-PAGE. The trimer shift is highlighted by *.
We next investigated whether the trimer-binding Abs in the SOSIP sera were directed to the CD4bs. Soluble CD4 bound to wild-type KNH1144 trimers, but mAb b12 did not bind effectively (data not shown) in keeping with b12’s known inability to neutralize this isolate [26]. In addition, D368R and D368G mutations, which selectively eliminate sCD4 binding in the context of subtype B trimers [68], led to poor expression of KNH1144 trimers. These factors precluded a meaningful analysis of the potential presence of CD4bs Abs in KNH144 SOSIP sera by BN-PAGE gel-shift assays.
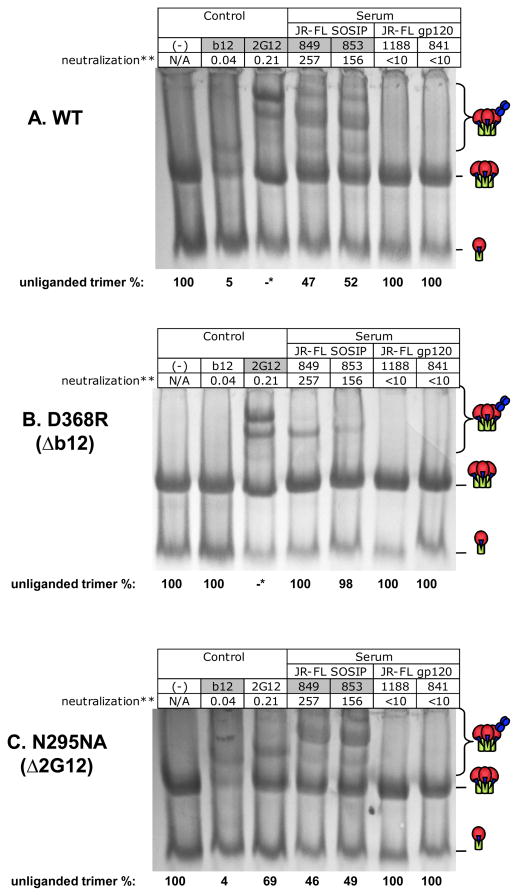
Considering the difficulty of mapping the neutralizing activity of the KNH1144 vaccine sera, we sought instead to investigate the basis for the neutralizing activity previously reported with JR-FL SOSIP sera [30]. However, we do not consider JR-FL as a surrogate for our clade A immunogen in the epitope-specificity mapping. In this JR-FL study, rabbits were immunized with JR-FL SOSIP trimers (rabbits 849 and 853) or JR-FL gp120 (rabbits 841 and 1188) using DNA-prime/protein-boost (rabbits 841 and 849) or protein-only (rabbits 853 and 1188) immunization regimens. Two JR-FL SOSIP antisera from rabbits 849 and 853 and two JR-FL antisera from Rabbits 841 and 1188 were used in this experiment. MAbs b12 and 2G12 were used as controls. Trimer shifts were quantified by densitometry based on the changes of the unliganded trimer band intensity [68]. In Fig. 8, trimer binding is measured as a depletion of unliganded trimer, indicated by cartoons on the right of each panel. In each lane, the percentage residual unliganded trimer, measured by densitometry, is indicated at the bottom. As an additional guide to comprehending of Fig 8, the box labels at the top of any lane in which trimer-Ab binding occurs are shaded gray.
Fig. 8. BN-PAGE trimer shift mapping sera generated against JR-FL immunogens.
JR-FL sera were analyzed for binding to (A) wild-type (WT), (B) 386D/R mutant (CD4bs knockout mutant) and (C) 295 N/A (2G12 knockout mutant) JR-FL SOSIP trimers. MAbs 2G12 and b12 were used as controls. Percent unliganded trimer is estimated at the bottom of each gel. In the case of 2G12 shifts of WT and 368 mutant (*), interpretation was complicated by the prominent band representing monomer+ 2G12, as reported previously [66]. **50% neutralization titers from the PhenoSense assay are given in μg/ml (b12 and 2G12) or titers for sera.
The JR-FL SOSIP antisera (849 and 853) bound wild-type JR-FL trimers, resulting in 47% and 52% remaining unliganded trimer, respectively. However, the gp120 antisera did not bind wild-type JR-FL trimers (Fig. 8A). As reported previously [66], 2G12 shifts were complicated by a dominant band representing monomer + 2G12 that migrates in these gels only slightly faster than the unliganded trimer (Fig. 8A, compare lanes 1 and 3). Effective 2G12 binding to trimers is evidenced, however, by the appearance of high molecular weight 2G12-Env trimer complexes (Fig. 8A, upper part of lane 3). To minimize the possible confounding effects of monomer + IgG on measuring levels of unliganded trimers (as in Fig. 8A, lane 3), we used protein A Dynabeads to remove IgG-bound Env and confirmed by Western blot analysis that 2G12 depleted unliganded trimer (data not shown).
IgG1b12 was unable to bind to the D386R mutant (Fig. 8B, lane 2), as expected [68]. Interestingly, this mutant also largely eliminated trimer shifts by the SOSIP antisera (compare Fig. 8A and 8B, lanes 4 and 5 relative to lane 1). In contrast, the N295A mutant eliminated 2G12 binding (Fig. 8C, compare lanes 1 and 3), but not b12 binding (Fig. 8C, compare lanes 1 and 2), but did not affect binding of SOSIP antisera (46–49% unliganded trimer, Fig. 8C lanes 4 and 5). Collectively, these data suggest that JR-FL SOSIP NAbs were specific for an epitope sensitive to residue D368 in the CD4bs. In additional experiments using the post-CD4 neutralization format, we found that the SOSIP sera 849 and 853 had approximately ~4 fold lower titers (ID50 titers of 3,000 and 5,000, respectively) than the gp120 sera 841 and 1188 (ID50 titers of 12,000 and 20,000, respectively). Previous studies have shown that the post-CD4 assay measures V3 loop and CD4i antibodies. Therefore, it appears that while JR-FL SOSIP effectively elicits neutralizing CD4bs NAbs, JR-FL gp120 preferentially elicits V3 loop and/or CD4i Abs. In post-CD4/CCR5 neutralization assays, none of the sera were effective (not shown), suggesting that MPER NAbs did not contribute appreciably to the neutralizing activity of JR-FL SOSIP sera.
DISCUSSION
Building on our previous work describing HIV-1 SOSIP trimers [24–30], we investigated the ligand-bound structure and immunogenicity of proteolytically cleaved gp140 trimers derived from a subtype A virus. SOSIP gp140 trimers were observed to bind three molecules of sCD4 in a symmetrical configuration similar to that observed for virus-associated Env spikes. Compared to matched gp120 monomers, KNH1144 SOSIP trimers elicited modest, but consistently higher levels of NAbs to homologous and heterologous HIV-1 isolates. The specificity of the neutralizing activity elicited here and that previously elicited by our clade B prototype SOSIP gp140 [30] mapped in part to the CD4bs. Our studies also demonstrate that low levels (parts per million) of HCP in subunit vaccines can under certain conditions induce Abs. Methods are described for determining the presence of such Abs and their potential interference in serological assays. Overall, these findings provide guidance for the further development of subunit vaccines for HIV-1 infection and other HIV-1 related diseases.
Electron microscopy indicated that SOSIP trimer binds sCD4 molecules in a manner similar to that seen for native Env [67]. Most trimers were observed to bind sCD4 trivalently, indicating that each SOSIP gp140 subunit was competent in binding CD4 and that all three CD4 binding sites could be occupied simultaneously. The sCD4 molecules bind to the SOSIP trimer at about 45 degrees laterally with respect to the radial axis of gp120 subunits in a symmetrical orientation. Together, these data support the contention that the general configuration of SOSIP trimer reflects that of the native Env spike of HIV-1. The stability and authenticity of KNH1144 SOSIP trimers would appear to make them particularly relevant candidates for ongoing attempts to crystallize Env trimers.
Compared to gp120, KNH1144 SOSIP elicited NAb activity that was modestly higher in terms of magnitude and breadth, and these findings are in agreement with those of our previous studies for JR-FL SOSIP [27;30]. The group of animals receiving KNH1144 gp120 formulated in Ribi generated relatively lower binding antibody titers to KNH1144 gp120 (group V) compared to the SOSIP group, rendering it difficult to be sure whether any neutralizing activity measured in group VI animals receiving SOSIP in Ribi emerged due to a shift in specificity towards neutralizing epitopes or is simply a result of higher overall Ab binding titers. However, as mentioned above, MN and NL4-3 viruses were neutralized by SOSIP but not gp120 antisera (Table 1) despite the fact that the SOSIP antisera contained lower titers of antibodies to the corresponding gp120s. There is no direct correlation between the binding antibody titers and neutralization titers. Historically, monomeric gp120 has rarely, if ever, been reported to elicit neutralizing responses effective against a homologous primary isolate, so therefore any such activity elicited by a vaccine candidate is of interest.
Mapping studies revealed the presence of Abs in SOSIP serum that was effective in the post-CD4 neutralization format (Table 3), namely anti-V3 loop and CD4i Abs. However, the titers were much lower than those found in infected patient sera [37;68]. Since CD4i Abs were undetectable in virus capture assays (Table 2), it appears likely that anti-V3 Abs are responsible for the low post-CD4 titers observed, as this is the only other specificity known to have a strong activity in this assay format [37]. The neutralization of SF162, MN and other relatively neutralization-sensitive isolates could be explained by broadly reactive V3 loop-specific Abs [69–75]. However, anti-V3 Abs are unlikely to explain neutralization activity against KNH1144, a relatively resistant primary isolate. We ruled out a contribution of MPER NAbs using the post-CD4/CCR5 assay (Table 3). This left CD4bs Abs as the only remaining specificity we measured (Table 2) that might conceivably explain the homologous neutralization we observed with some sera. However, we were unable in the present study to incontrovertibly demonstrate that CD4bs Abs explained neutralization by KNH1144 SOSIP sera since mutant forms of the KNH1144 Env trimer (Fig 7) did not express effectively for use in mapping by BN-PAGE. However, we were able to show that the homologous NAbs elicited by JR-FL SOSIP gp140 were directed to a D368R sensitive epitope on the trimers.
The residual HCPs in the purified SOSIP immunogen, even at a level of 10.8 ppm, in some cases generated Abs that measurably bound cells and enhanced the infection of pseudoviruses used in neutralization assays. Responses were adjuvant-dependent. The same preparation of SOSIP gave rise to HCP Abs in the presence of Quil A, but not Ribi, adjuvant. The mechanism by which the anti-HCP Abs could enhance the infectivity of HIV is not completely understood. Conceptually, anti-HCP Abs could enhance infectivity by binding to either the target cell for infection or to the virus, since virions contain HCP on their surface. Antibody-dependent enhancement (ADE) of HIV infectivity was reported by Homsy [76] and its significance in vaccine development was discussed by others [77–80]. ADE of infection can be mediated by Fc and complement receptors on cells. Theoretically, anti-HCP Abs could cross-link virus and increase the rate of virus adsorption and infectivity. Our findings further highlight the need to measure and remove HCP in subunit vaccines, and to test for HCP Abs when their presence in immune sera could potentially interfere with serological analyses and potentiate viral infectivity.
In summary, results from the present and prior studies [27;30] have shown that SOSIP trimers mimic native Env protein configuration and are superior to matched gp120 monomers in eliciting NAbs. Studies are ongoing to generate SOSIP trimers to protect humans from HIV-1 infection and to elicit a substantially stronger and more robust NAb response against HIV-1 isolates, taking into account the results observed using the KNH1144 construct [5;81–83]. One particularly promising result here was the induction of NAbs that target the CD4bs, a neutralization target on gp120 that is also recognized by the broadly neutralizing NAb IgG1b12. Although the activity we observed was relatively limited to the vaccine strain, it remains possible that with appropriate adjustments, the breadth of this neutralizing activity can be increased. Studies are ongoing to map additional contact points in the CD4bs for SOSIP NAbs. This information may assist in formulating approaches to better elicit CD4bs NAbs. SOSIP mutations provide a useful means to generate purified, stable, cleaved HIV-1 Env trimers for structural studies and for further development of HIV-1 vaccines designed to elicit NAbs effective against relevant HIV-1 field isolates.
Acknowledgments
This study was supported by NIH contract N01 AI30030. Additional support came from the Bill and Melinda Gates Collaboration for AIDS Vaccine Discovery Vaccine Immune Monitoring Consortium grant #38619 (JMB). We thank Drs. D. Burton and R. Pantophlet for providing mAbs b12 and X5, Dr. H. Katinger for providing mAbs 2G12, 2F5 and 4E10, Dr. R. Pejchal for providing 2D-sCD4, Dr. S. Zolla-Pazner for providing mAb 447-52D, Dr. M. Zwick for providing mAb Z13e1, Dr. Robert Doms for vaccinia-expressed gp120s, and F. McCutchan for providing the KNH1144ec1 plasmid. We would like to thank J. Gao, A. M. Cupo, A. A. Ouattara, V. M. Seetharaman, D. Fisch, L. Krawiec and G. Yiannoulos for their exceptional contributions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Letvin NL, Walker BD. Immunopathogenesis and immunotherapy in AIDS virus infections. Nat Med. 2003 Jul;9(7):861–6. doi: 10.1038/nm0703-861. [DOI] [PubMed] [Google Scholar]
- 2.Conley AJ, Kessler JA, II, Boots LJ, et al. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J Virol. 1996 Oct;70(10):6751–8. doi: 10.1128/jvi.70.10.6751-6758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emini EA, Schleif WA, Nunberg JH, et al. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain- specific monoclonal antibody. Nature. 1992 Feb 20;355(6362):728–30. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- 4.Mascola JR, Lewis MG, Stiegler G, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999 May;73(5):4009–18. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascola JR, Stiegler G, VanCott TC, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000 Feb;6(2):207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 6.Phogat S, Wyatt RT, Karlsson Hedestam GB. Inhibition of HIV-1 entry by antibodies: potential viral and cellular targets. J Intern Med. 2007 Jul;262(1):26–43. doi: 10.1111/j.1365-2796.2007.01820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006 Aug;5(4):579–95. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- 8.Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006 Jun;5(3):347–63. doi: 10.1586/14760584.5.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–69. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 10.Kowalski M, Potz J, Basiripour L, et al. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–5. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 11.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995 Dec;2(12):1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 12.Belshe RB, Gorse GJ, Mulligan MJ, et al. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-I and gp 120 SF-2 recombinant vaccines in uninfected volunteers. NIAD AIDS Vaccine Evaluation Group. AIDS. 1998 Dec;12(18):2407–15. doi: 10.1097/00002030-199818000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Connor RI, Korber BT, Graham BS, et al. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J Virol. 1998 Feb;72(2):1552–76. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert PB, Peterson ML, Follmann D, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005 Mar 1;191(5):666–77. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 15.Mascola JR, Snyder SW, Weislow OS, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996;173(2):340–8. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 16.Matthews TJ. Dilemma of neutralization resistance of HIV-1 field isolates and vaccine development. AIDS Res Hum Retroviruses. 1994 Jun;10(6):631–2. doi: 10.1089/aid.1994.10.631. [DOI] [PubMed] [Google Scholar]
- 17.McElrath MJ, Corey L, Montefiori D, et al. A phase II study of two HIV type 1 envelope vaccines, comparing their immunogenicity in populations at risk for acquiring HIV type 1 infection. AIDS Vaccine Evaluation Group. AIDS Res Hum Retroviruses. 2000 Jun 10;16(9):907–19. doi: 10.1089/08892220050042846. [DOI] [PubMed] [Google Scholar]
- 18.Migasena S, Suntharasamai P, Pitisuttithum P, et al. AIDSVAX (MN) in Bangkok injecting drug users: a report on safety and immunogenicity, including macrophage-tropic virus neutralization. AIDS Res Hum Retroviruses. 2000 May 1;16(7):655–63. doi: 10.1089/088922200308882. [DOI] [PubMed] [Google Scholar]
- 19.Parren PW, Mondor I, Naniche D, et al. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998 May;72(5):3512–9. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell ND, Graham BS, Keefer MC, et al. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J Acquir Immune Defic Syndr. 2007 Feb 1;44(2):203–12. doi: 10.1097/01.qai.0000248356.48501.ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanCott TC, Mascola JR, Loomis-Price LD, et al. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary human immunodeficiency virus type 1 envelope. J Virol. 1999 Jun;73(6):4640–50. doi: 10.1128/jvi.73.6.4640-4650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrin T, Nunberg JH. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS. 1994 Nov;8(11):1622–3. doi: 10.1097/00002030-199411000-00017. [DOI] [PubMed] [Google Scholar]
- 23.Nelson JD, Kinkead H, Brunel FM, et al. Antibody elicited against the gp41 N-heptad repeat (NHR) coiled-coil can neutralize HIV-1 with modest potency but non-neutralizing antibodies also bind to NHR mimetics. Virology. 2008 Jul 20;377(1):170–83. doi: 10.1016/j.virol.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binley JM, Sanders RW, Master A, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002 Mar;76(6):2606–16. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders RW, Vesanen M, Schuelke N, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002 Sep;76(17):8875–89. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beddows S, Kirschner M, Campbell-Gardener L, et al. Construction and characterization of soluble, cleaved, and stabilized trimeric Env proteins based on HIV type 1 Env subtype A. AIDS Res Hum Retroviruses. 2006 Jun;22(6):569–79. doi: 10.1089/aid.2006.22.569. [DOI] [PubMed] [Google Scholar]
- 27.Beddows S, Schulke N, Kirschner M, et al. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. The Journal of Virology. 2005 Jul 15;79(14):8812–27. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirschner M, Monrose V, Paluch M, Techodamrongsin N, Rethwilm A, Moore JP. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr Purif. 2006 Jul;48(1):61–8. doi: 10.1016/j.pep.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Iyer SP, Franti M, Krauchuk AA, et al. Purified, proteolytically mature HIV type 1 SOSIP gp140 envelope trimers. AIDS Res Hum Retroviruses. 2007 Jun;23(6):817–28. doi: 10.1089/aid.2006.0261. [DOI] [PubMed] [Google Scholar]
- 30.Beddows S, Franti M, Dey AK, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007 Apr 10;360(2):329–40. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Schulke N, Vesanen MS, Sanders RW, et al. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76(15):7760–76. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burton DR, Pyati J, Koduri R, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–7. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 33.Zhou T, Xu L, Dey B, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007 Feb 15;445(7129):732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders RW, de Jong EC, Baldwin CE, Schuitemaker JH, Kapsenberg ML, Berkhout B. Differential transmission of human immunodeficiency virus type 1 by distinct subsets of effector dendritic cells. J Virol. 2002;76(15):7812–21. doi: 10.1128/JVI.76.15.7812-7821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scanlan CN, Pantophlet R, Wormald MR, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002 Jul;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan N, Sun Y, Sattentau Q, et al. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998 Jun;72(6):4694–703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks ET, Moore PL, Richman D, et al. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum Antibodies. 2005;14(3–4):101–13. [PMC free article] [PubMed] [Google Scholar]
- 38.Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure. 2003 Feb;11(2):225–36. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 39.Pantophlet R, Aguilar-Sino RO, Wrin T, Cavacini LA, Burton DR. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology. 2007 Aug 1;364(2):441–53. doi: 10.1016/j.virol.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muster T, Steindl F, Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993 Nov;67(11):6642–7. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zwick MB, Labrijn AF, Wang M, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001 Nov;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Binley JM, Sanders RW, Clas B, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binley JM, Wrin T, Korber B, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004 Dec 1;78(23):13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008 May 27;105(21):7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004 May;78(10):5205–15. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maddon PJ, Molineaux SM, Maddon DE, et al. Structure and expression of the human and mouse T4 genes. Proc Nat Acad Sci USA. 1987;84:9155–9. doi: 10.1073/pnas.84.24.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allaway GP, Davis-Bruno KL, Beaudry GA, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer which neutralizes primary HIV-1 isolates. AIDS Res Hum Retroviruses. 1995;11:533–9. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- 48.Trkola A, Dragic T, Arthos J, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–7. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 49.Frank J, Radermacher M, Penczek P, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996 Jan;116(1):190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 50.Huang CC, Tang M, Zhang MY, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005 Nov 11;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu H, Kwong PD, Hendrickson WA. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997 May 29;387(6632):527–30. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 52.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996 Mar;70(3):1863–72. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore JP, Cao Y, Leu J, Qin L, Korber B, Ho DD. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996 Jan;70(1):427–44. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore JP, Trkola A, Korber B, et al. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995 Jan;69(1):122–30. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selvarajah S, Puffer B, Pantophlet R, Law M, Doms RW, Burton DR. Comparing antigenicity and immunogenicity of engineered gp120. J Virol. 2005 Oct;79(19):12148–63. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003 Mar 18;100(7):4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007 Feb;51(2):566–75. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000 Apr;44(4):920–8. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dowling WE, Kim B, Mason CJ, et al. Forty-one near full-length HIV-1 sequences from Kenya reveal an epidemic of subtype A and A-containing recombinants. AIDS. 2002 Sep 6;16(13):1809–20. doi: 10.1097/00002030-200209060-00015. [DOI] [PubMed] [Google Scholar]
- 60.Brown BK, Darden JM, Tovanabutra S, et al. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J Virol. 2005 May;79(10):6089–101. doi: 10.1128/JVI.79.10.6089-6101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Binley JM, Cayanan CS, Wiley C, Schülke N, Olson WC, Burton DR. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J Virol. 2003 May 15;77(10):5678–84. doi: 10.1128/JVI.77.10.5678-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore PL, Crooks ET, Porter L, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006 Mar;80(5):2515–28. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crooks ET, Moore PL, Franti M, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007 Sep 30;366(2):245–62. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derby NR, Kraft Z, Kan E, et al. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol. 2006 Sep;80(17):8745–62. doi: 10.1128/JVI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001 Mar;75(5):2041–50. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crooks ET, Jiang P, Franti M, et al. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology. 2008 Aug 1;377(2):364–78. doi: 10.1016/j.virol.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008 Jul 30;455(7209):109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Binley JM, Lybarger EA, Crooks ET, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008 Dec;82(23):11651–68. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore JP, Sattentau QJ, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68(1):469–84. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Javaherian K, Langlois AJ, LaRosa GJ, et al. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990 Dec 14;250(4987):1590–3. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- 71.Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, Lu S. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology. 2008 Mar 15;372(2):233–46. doi: 10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 72.Gorny MK, Williams C, Volsky B, et al. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002 Sep;76(18):9035–45. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorny MK, Williams C, Volsky B, et al. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006 Jul;80(14):6865–72. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krachmarov C, Pinter A, Honnen WJ, et al. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005 Jan;79(2):780–90. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kraft Z, Strouss K, Sutton WF, et al. Characterization of neutralizing antibody responses elicited by clade A envelope immunogens derived from early transmitted viruses. J Virol. 2008 Jun;82(12):5912–21. doi: 10.1128/JVI.00389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Homsy J, Meyer M, Tateno M, Clarkson S, Levy JA. The Fc and not CD4 receptor mediates antibody enhancement of HIV infection in human cells. Science. 1989 Jun 16;244(4910):1357–60. doi: 10.1126/science.2786647. [DOI] [PubMed] [Google Scholar]
- 77.Langlois AJ, Weinhold KJ, Matthews TJ, Greenberg ML, Bolognesi DP. The ability of certain SIV vaccines to provoke reactions against normal cells. Science. 1992 Jan 17;255(5042):292–3. doi: 10.1126/science.1549775. [DOI] [PubMed] [Google Scholar]
- 78.Mitchell WM, Ding L, Gabriel J. Inactivation of a common epitope responsible for the induction of antibody-dependent enhancement of HIV. AIDS. 1998 Jan 22;12(2):147–56. doi: 10.1097/00002030-199802000-00004. [DOI] [PubMed] [Google Scholar]
- 79.Robinson WE, Jr, Montefiori DC, Mitchell WM, et al. Antibody-dependent enhancement of human immunodeficiency virus type 1 (HIV-1) infection in vitro by serum from HIV-1-infected and passively immunized chimpanzees. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4710–4. doi: 10.1073/pnas.86.12.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas S, Redfern JB, Lidbury BA, Mahalingam S. Antibody-dependent enhancement and vaccine development. Expert Rev Vaccines. 2006 Aug;5(4):409–12. doi: 10.1586/14760584.5.4.409. [DOI] [PubMed] [Google Scholar]
- 81.Nishimura Y, Igarashi T, Haigwood N, et al. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J Virol. 2002 Mar;76(5):2123–30. doi: 10.1128/jvi.76.5.2123-2130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parren PW, Marx PA, Hessell AJ, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001 Sep;75(17):8340–7. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trkola A, Kuster H, Rusert P, et al. In vivo efficacy of human immunodeficiency virus neutralizing antibodies: estimates for protective titers. J Virol. 2008 Feb;82(3):1591–9. doi: 10.1128/JVI.01792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]