SUMMARY
Induced CREB activity is a hallmark of long-term memory, but the full repertoire of CREB transcriptional targets required specifically for memory is not known in any system. To obtain a more complete picture of the mechanisms involved in memory, we combined memory training with genome-wide transcriptional analysis of C. elegans CREB mutants. This approach identified 757 significant CREB/memory-induced targets and confirmed the involvement of known memory genes from other organisms, but also suggested new mechanisms and novel components that may be conserved through mammals. CREB mediates distinct basal and memory transcriptional programs at least partially through spatial restriction of CREB activity: basal targets are regulated primarily in nonneuronal tissues, while memory targets are enriched for neuronal expression, emanating from CREB activity in AIM neurons. This suite of novel memory-associated genes will provide a platform for the discovery of orthologous mammalian long-term memory components.
INTRODUCTION
Transcription factors are critical components in the cellular response to the environment. The human genome encodes roughly 1,500 transcription factors (TFs) (Vaquerizas et al., 2009), yet many individual transcriptional regulators are multifunctional, with roles in diverse biological processes throughout the body. TFs employ a variety of strategies to mediate different biological outcomes, including cell type, subcellular spatial restriction, and temporal control. TF activity is also governed by cellular context, which is mediated by the presence of cell-specific cofactors and signaling pathways that ultimately regulate the selection of target genes.
The cAMP-response element-binding protein (CREB) is an example of an evolutionarily conserved bZIP TF that influences distinct cellular processes in different contexts, including long-term memory, cell survival and proliferation, apoptosis, differentiation, metabolism, hematopoiesis, and immune activity (Mayr and Montminy, 2001). CREB activation is induced by growth factors, neurotransmitters, and stress signals that promote CREB phosphorylation. Activated CREB binds to the cAMP-response element (CRE) (TGACGTCA) of target gene promoter regions and induces gene expression through the recruitment of transcriptional cofactors such as CBP/p300 and TORCs (Chrivia et al., 1993; Conkright et al., 2003). C. elegans CREB is required for long-term memory (Kauffman et al., 2010). CREB has also been implicated in the regulation of metabolism and longevity, as reduction of the CREB-regulated transcriptional coactivator CRTC-1 increases C. elegans wild-type median lifespan by 53% (Mair et al., 2011). Presumably the CREB targets that mediate such different biological processes are distinct, but these targets, as well as the mechanism for CREB’s differential selection of targets, are unknown in any system.
Several studies have characterized individual and genome-wide CREB targets using a range of cultured cell types to understand how CREB mediates such diverse phenotypes. Analysis of basal and artificially induced CREB activity identified thousands of CREB binding sites (Impey et al., 2004; Zhang et al., 2005), and CREB is suggested to bind to ~19,000 regions of the human genome (Euskirchen et al., 2004). However, CREB target gene selection varies greatly among different cell types (Cha-Molstad et al., 2004). For example, external stimuli such as in vitro forskolin activation of adenylate cyclase alter CREB target specificity relative to unstimulated controls (Cha-Molstad et al., 2004; Euskirchen et al., 2004; Zhang et al., 2005). Cell type and context clearly influence CREB’s transcriptional program, suggesting that in vivo analyses of CREB targets are needed to further refine the role of CREB in specific biological processes.
CREB is a conserved regulator of long-term memory in a variety of organisms, including Aplysia, worms, flies, mice, and humans (Silva et al., 1998; Yin et al., 1995). While a number of resting-state or artificially induced CREB target genes are known from in vitro and in vivo neuron studies, it is unclear how these targets specifically relate to the in vivo CREB transcriptional program that is activated in the context of a long-term memory training paradigm (Barco et al., 2005; Impey et al., 2004; Lesiak et al., 2013; Ploski et al., 2010). A microarray study of transgenic mice expressing a constitutively active form of CREB in the hippocampus revealed only four potential “memory genes,” including CREB (Barco et al., 2005), and in vitro stimulation of long-term potentiation, a cellular correlate of long-term memory, in hippocampal slices uncovered 41 “plasticity-associated genes” in the mouse dentate gyrus (Ploski et al., 2010). An in vivo genome-wide study of direct and indirect CREB targets induced by long-term memory training of an animal has yet to be performed, suggesting that additional CREB-dependent, long-term memory genes and molecular mechanisms remain unknown. The use of model organisms to study the in vivo CREB-dependent changes that occur directly in response to memory training could address this knowledge gap.
To enable a rapid, high-throughput system for the identification and functional testing of CREB’s long-term memory targets, we developed an assay of olfactory long-term associative memory (LTAM) in C. elegans (Kauffman et al., 2010). Repeated presentation of the neutral odorant butanone (conditioned stimulus) paired with food (unconditioned stimulus) results in a long-term increase in C. elegans’ attraction to butanone (Kauffman et al., 2011, 2010). Testing chemotaxis to butanone immediately after training (0 hr) measures memory acquisition, while testing at 16 hr after training is a measure of long-term memory (Kauffman et al., 2010). As in other organisms, C. elegans’ long-term memory requires protein synthesis and CREB-dependent gene transcription, while learning and short-term memory do not require CREB or transcription (Kauffman et al., 2010). Although CREB is expressed ubiquitously (Kimura et al., 2002), we found that neuronal expression of CREB rescues the long-term memory ability of CREB null worms, and neuronal CREB overexpression increases the duration of long-term memory (Kauffman et al., 2010). Thus, as in humans, neuronal CREB’s transcriptional targets must be important for memory in C. elegans. The fact that the major regulators of long-term memory are conserved between worms and mammals suggests that core memory components may be shared evolutionarily as well.
In this study, we identified the global direct and indirect CREB target genes expressed in the basal, resting state and those induced upon long-term memory training. The basal and memory-induced gene sets are distinct in composition, function, and spatial enrichment: the CREB/basal genes mediate growth and development functions primarily in nonneuronal tissues, while the CREB/LTAM-dependent genes are expressed primarily in neurons, and we show that these targets, including several novel genes, regulate long-term memory. We also found that CREB activity in the AIM interneurons is required for multiple forms of olfactory associative long-term memory. Because CREB activity is only induced in one pair of interneurons upon long-term memory training, current direct binding approaches likely could not have identified this set of CREB-regulated memory genes, highlighting the efficacy of our transcriptional profiling approach. Many CREB/LTAM targets are expressed in neurons other than AIM, suggesting that CREB activation in AIM leads to transcriptional changes in other cells of the downstream long-term memory network. Many of the novel and functionally verified CREB/LTAM genes found here possess mammalian orthologs, providing a unique resource for investigators to explore the roles of these memory genes in higher organisms.
RESULTS
Basal CREB Activity Regulates Growth and Development Independent of Lifespan
CREB is required for long-term memory in C. elegans (Kauffman et al., 2010), while the reduction of the CREB-regulated transcriptional coactivator CRTC-1 increases C. elegans wild-type median lifespan by 53% (Mair et al., 2011). Because C. elegans CREB is implicated in these distinct biological processes, we sought to identify targets of CREB in the naive state by comparing expression profiles of CREB null (crh-1(tz2)) mutants to wild-type animals in normal (basal) conditions (Figure 1) and upon memory conditioning (Figure 2A). At a false discovery rate (FDR) of 0.27%, 652 and 1,033 genes were up- and down-regulated, respectively, by crh-1 loss (Table S2). However, we found no enrichment of neuronal GO terms in the basal CREB set (Figure 1A), no enrichment for neuronally expressed genes (p = 0.98; Figure 2D), and no bias toward expression in neurons (Figure 2E), suggesting that basal targets of CREB are not involved in memory function. Instead, basal CREB-upregulated targets are enriched for GO terms associated with growth and development, such as chromosome function, cell migration, reproduction, and morphogenesis, while repressed genes are associated with cuticle growth and metabolism (Figures 1A). Therefore, simple comparison of CREB mutant and wild-type gene expression did not yield obvious memory genes and instead indicated that CREB plays a role in growth and development. To test this hypothesis, we measured the size and growth rates of CREB mutants. We found that CREB mutants are significantly smaller than wild-type animals (Figure 1B) and develop more slowly (Figure 1C).
Figure 1. Basal CREB Activity Regulates Metabolism Independent of Effects on Lifespan.
(A) Gene Ontology (GO)-term analysis of CREB basal targets identified by one-class SAM analysis.
(B) Worms carrying mutations of crh-1 are significantly smaller than N2 animals of the same stage. Mean ± SEM, n ≥ 50 per strain, ***p < 0.001.
(C) Worms carrying mutations in crh-1 develop more slowly than N2 animals. The distribution of developmental stages of crh-1 mutants was determined to be significantly different from N2 animals at all time points measured by chi-square tests. n > 600 per strain, p < 0.0001.
(D) Pearson correlation of whole-genome expression data shows that while long-lived daf-2 and daf-7 mutants are correlated (0.34), crh-1 mutants appear uncorrelated with the long-lived mutants.
(E) daf-2 and daf-7 mutants increase expression of the top 100 daf-2-dependent/DAF-16-induced (Class I) lifespan-regulating genes, but these are not upregulated in crh-1 mutants.
(F) GSEA analysis shows that lifespan-regulating genes are enriched in several long-lived mutants, but not in crh-1 mutants. A heatmap of gene clustering is displayed on the y axis while gene enrichment scores compared to daf-2/daf-16 gold-standard genes are shown on the x axis. Gold standard genes are enriched in daf-2 mutants (positive control) with a nominal p value of 0.0, FDR q value of 0.0, and Normalized Enrichment Score (NES) of 3.28. daf-7 versus daf-7;daf-3 transcriptional data yielded a positive NES of 2.70; atp-2 RNAi versus vector RNAi yielded a positive NES of 2.40; dauer versus 12 hr post-dauer yielded a NES of 2.31 for the gold standard gene set. All enrichment analyses yielded a nominal p value of 0.0 and FDR q value of 0.0, and no significant negative enrichment was detected in any data set. No significant positive enrichment was observed in the crh-1 versus wild-type transcriptome (NES = −1.2433004, nominal p value = 0.0, FDR q value = 0.0).
(G) crh-1 mutant worms (crh-1(tz2) and crh-1(n3315) alleles) are not long-lived relative to wild-type (p = 0.25 and 0.4, respectively).
Figure 2. LTAM Training Induces CREB Activation of a Distinct Neuron-Enriched Memory Gene Program.
(A) Overview of the long-term memory assay and collection scheme for CREB-dependent LTAM (“CREB/LTAM”) microarray analysis.
(B) 1 and 2: CREB-independent up- and downregulated genes are largely starvation associated (Figure S5 and Table S4). 3 and 4: CREB/LTAM down- and upregulated genes (Table S5).
(C) GO-term analysis of CREB/LTAM upregulated genes identified by two-class SAM analysis (see Table S5) and basal CREB-dependent upregulated genes shows that the two gene sets are largely nonoverlapping; the LTAM-dependent set is associated with behavior, synapse, and signaling, while basal gargets are associated with growth.
(D) Most CREB/LTAM-induced genes are expressed in neurons and are distinct from CREB’s basal targets (black circles). A hypergeometric test showed that CREB/LTAM-induced, but not basal CREB targets, exhibit significant overlap with neuronal-expressed genes (p value = 1.7 × 10−15 and 0.983, respectively). Genes whose promoters contain CRE sites are shown in red. Asterisk indicates two neuronally expressed genes that are regulated by CREB both basally and upon LTAM training.
(E) CREB/LTAM genes are primarily associated with neuronal expression, while CREB basal genes are mostly expressed in the intestine and hypodermis. Expression patterns of the top 50 genes in each set whose expression is known were compared.
(F) Heatmap showing the expression of the 722 genes that exhibited significantly increased expression in wild-type but not crh-1 mutants upon LTAM training in different genetic contexts.
(G) Heatmap showing the expression of the 35 genes upregulated by training in wild-type animals that were dramatically downregulated in untrained as well as trained crh-1 mutants. CREB/LTAM-induced genes are not upregulated in untrained wild-type worms and LTAM-trained crh-1 mutants (F and G). Neuronal CREB rescue in crh-1 mutants and CREB overexpression in wild-type animals recapitulates LTAM gene expression. Black bars indicate untrained, naive animals and red bars indicate LTAM-trained worms.
To test whether crh-1 mutants increase expression of prolongevity genes, as suggested by the role of CRTC-1 in lifespan (Mair et al., 2011), we compared the basal transcriptome of crh-1 mutants to that of the well-characterized, long-lived daf-2 and daf-7 mutants. daf-2 insulin receptor mutants live twice as long as wild-type animals and require the DAF-16 forkhead transcription factor for their lifespan extension (Kenyon et al., 1993; Ogg et al., 1997). DAF-16 influences aging by increasing the expression of a wide variety of cellular stress-response, antimicrobial, and metabolic genes in daf-2 mutants (Murphy et al., 2003; Tepper et al., 2013). Mutations in the TGF-β-like ligand daf-7 extend lifespan by about 30%, and this lifespan extension requires both the downstream SMAD effector, DAF-3, and DAF-16 activity, and the expression profiles of TGF-β dauer and IIS mutants are similar (Shaw et al., 2007). However, crh-1 expression profiles did not correlate with profiles of these longevity mutants (Figures 1D and 1E; Table S3). Unsupervised hierarchical clustering of daf-2 versus daf-16;daf-2, daf-7 versus daf-7;daf-3, and crh-1 versus N2 revealed no correlation between crh-1 and longevity mutants (0.09), unlike the high correlation between the two longevity pathways (0.34) (Figure 1D), as we previously found (Shaw et al., 2007).
We then assessed transcriptional differences between crh-1 and long-lived mutants using Gene Set Enrichment Analysis (GSEA), which identifies statistically significant, concordant differences between two genotypes. A gold standard gene set, which contains 477 significantly overexpressed genes in daf-2 versus daf-2;daf-16 mutants, is robustly enriched in daf-2 and daf-7 mutants (Figure 1F), as well as in two other longevity paradigms, dauer diapause (McElwee et al., 2004; Wang and Kim, 2003) and mitochondrial dysfunction (Dillin et al., 2002). By contrast, no significant positive enrichment of shared longevity genes was observed among crh-1 mutants. Consistent with these gene expression data, lifespan analysis using multiple alleles of crh-1 revealed no difference in mean lifespan compared to wild-type animals (Figure 1G), in agreement with crh-1(n3315) mutant results (Mair et al., 2011). Together, these data demonstrate that CREB and its basal transcriptional targets primarily regulate development and growth independent of lifespan-regulating genes or memory genes.
Long-Term Memory Training Induces CREB-Mediated Activation of a Distinct Neuron-Enriched Memory Gene Program
Because transcriptional analysis under naive conditions (Figure 1) did not identify CREB-induced memory genes, we used our olfactory LTAM training paradigm (Kauffman et al., 2010) in combination with transcriptional analysis to identify CREB-dependent target genes in the context of long-term memory. Briefly, repeated presentation of the neutral odorant butanone (conditioned stimulus) paired with food (unconditioned stimulus) results in a long-term (>16 hr) increase in C. elegans’ attraction to butanone (Kauffman et al., 2010, 2011). We compared the transcriptional profiles of wild-type (N2) worms and CREB null (crh-1(tz2)) mutants before and after butanone/food LTAM training (Figure 2A). Using one-class SAM analysis, we identified four nonoverlapping sets of genes whose expression is altered by the presence of CREB and/or memory training (Figure 2B). A total of 1,575 genes increased expression (FDR = 1.23%) in both wild-type and crh-1 mutants upon LTAM training. These shared genes were termed “CREB-independent/LTAM-induced” (Figure 2B, #1; Table S4). GO-term analysis of these genes indicates that they are primarily associated with transcription, intracellular signaling, and development (Figure S1A). This set is also enriched for neuronal genes (~55% expressed in neurons, p = 3.4 × 10−13), including neuronal secreted proteins: the neurotrophic factor ndnf-1, FMRF-like peptides (flp-4, flp-6, flp-12, flp-14, flp-16, flp-18, flp-19, flp-21, flp-27), neuropeptides (nlp-2, nlp-5, nlp-9, nlp-12, nlp-14, nlp-15, nlp-20, nlp-25, nlp-47), and insulin-like peptides (ins-7, ins-17, ins-34) are upregulated with training in a CREB-independent manner. Insulin-like peptides ins-6 and ins-7 have been previously shown to mediate aversive olfactory learning in specific neurons via paracrine signaling (Chen et al., 2013), and it is possible that the secreted peptides that we identified similarly influence long-term memory formation. Because their transcription is CREB independent, these genes may act upstream of CREB to induce CREB’s activation upon LTAM training, or in pathways parallel to CREB.
A second set of CREB-independent genes reduced expression upon LTAM training (Figure 2B, #2; Table S4). Many of the CREB-independent/LTAM genes are associated with metabolic processes of the starvation response (Figure S2; Van Gilst et al., 2005), likely due to the repeated cycles of starvation and feeding used in our training paradigm.
Two-class SAM analysis revealed 722 genes (FDR = 1.83%) that exhibited significantly increased expression in wild-type worms, but not in crh-1 mutants, upon LTAM training (Figure 2B, #4, Figure 2F; Table S5). These are the CREB- and LTAM-dependent genes that we set out to identify. An additional 35 genes were initially omitted due to microarray data filtering resulting from undetectable expression in CREB mutant arrays (Figure 2G). However, these genes showed increased expression upon training in wild-type, CREB rescue, and CREB-overexpressing animals, with undetectable expression in crh-1 mutants both pre- and post-training (Figure 2G; Table S5). Together, these 757 genes were designated “CREB-dependent/LTAM-induced” (or “CREB/LTAM”) genes. Additionally, 307 CREB-dependent genes were significantly downregulated in wild-type worms with LTAM training (Figure 2B, #3). In a separate series of LTAM training and microarray analysis, the direct comparison of trained wild-type and trained crh-1 worms confirmed a substantial subset of the induced CREB/LTAM genes (Figures S1B and S1C; Table S1).
We performed quantitative RT-PCR analysis of a subset of the LTAM-dependent memory-tested genes, confirming their CREB dependence (Figure S3). To test whether the increased expression of CREB/LTAM genes was specific to the pairing of odorant and feeding state, we examined the gene expression of 25 CREB/LTAM genes selected from a wide spectrum of fold changes and significance scores. We compared mock-trained (spaced-training in the absence of odorant) with LTAM-trained (Figure S4) or unpaired odorant exposure versus control (Figure S5). Induction of gene expression was significantly increased in LTAM training compared to mock training for 23 of 25 genes (Figure S4), and only one of the 25 showed increased expression upon odorant exposure (Figure S5).
Hypergeometric analysis of the CREB/LTAM genes showed that the set is enriched for neuronal transcripts (p value = 1.7 × 10−15) identified using adult neuron-specific RNA-sequencing (Figure 2D) (R.K., unpublished data). Genes that are memory induced are distinct from CREB basal targets, even among neuron-expressed genes; only 12 genes overlap between the 463 neuronally enriched CREB/LTAM genes and 281 CREB neuronal basal genes (Figure 2D).
To further examine the tissue distribution of CREB basal and CREB/LTAM genes, we curated the expression patterns of the top 50 genes in each set whose expression is known. CREB basal targets are predominantly expressed in the intestine and hypodermis, while CREB/LTAM genes are detected primarily in neurons (Figure 2E). Furthermore, GO-term analysis showed that CREB/LTAM genes are associated with behavior, ion channel activity, and intracellular signaling and do not significantly overlap with the GO-terms of CREB basal genes (Figure 2C). These data demonstrate that CREB activates a neuron-enriched transcriptional program upon long-term memory formation that is distinct from its growth-regulating basal state.
CREB itself is expressed ubiquitously (Reece-Hoyes et al., 2007), so it is possible that many of the basal and LTAM-induced genes are direct CREB targets. Indeed, the CREB binding sequence CRE site (TGACGTCA) is significantly enriched in the promoters of both the CREB basal and CREB/LTAM genes (basal set e-value 2.4 × 10−24; LTAM set e-value 1.5 × 10−14). However, only 37 of the 757 (4.9%) of LTAM genes have full-length CREs in their upstream 1,000 bp promoter regions (Figure 2D; Table S5), suggesting that very few of the memory targets’ promoters could be directly bound by CREB. Similarly, only 55 of the 652 (8.4%) basal targets have CREs in their promoters (Table S2), suggesting that the bulk of the expression changes we have identified in both naive and trained conditions are indirect targets of CREB and thus would not have been identified through direct binding experiments (Figure 2D). CREB occupies half-sites (CGTCA) in human promoters (Euskirchen et al., 2004), which are present in 64% of CREB/LTAM gene promoters, although no enrichment of half CRE sites was observed (e-value = 0.18).
To test the hypothesis that neuronal CREB expression regulates the long-term memory transcriptional program, we rescued CREB activity only in neurons of crh-1 mutants by expressing the pan-neuronal Pcmk-1::crh-1b transgene (Kimura et al., 2002). Neuronal expression of CREB is sufficient to restore long-term memory in crh-1 mutants (Kauffman et al., 2010). We find that expression of this transgene also rescues the expression of CREB/LTAM-dependent genes upon training (Figures 2F and 2G), suggesting the CREB/LTAM-induced targets in wild-type worms are downstream of neuronal CREB activity. Neuronal overexpression of CREB in wild-type animals extends long-term memory up to at least 24 hr (Kauffman et al., 2010), and correspondingly also upregulates the transcription of CREB/LTAM genes (Figures 2F and 2G).
CREB/LTAM-Induced Genes Have Known Neuronal and Memory Functions in Mammals
The known genes regulated by CREB after memory training are largely associated with neuronal functions (Table 1). Proteins required for long-term potentiation, including those involved in vesicle trafficking, formation, and docking; synaptogenesis and synaptic plasticity; calcium signaling, post-synaptic scaffolding, and RNA binding (Dubnau et al., 2003; Renner et al., 2008), are upregulated, as are ion channels (Voglis and Tavernarakis, 2006), predicted kinases (Mayford, 2007), seven-transmembrane receptors (MacDonald et al., 2007), neurotransmitter secretion, and neuropeptide signaling genes (Feany, 1996). A striking number of processes that were previously implicated in memory were enriched in our CREB/LTAM set, indicating not only that our microarray approach successfully identified known memory-associated genes, but also that CREB achieves its effects in memory through the concerted upregulation of a large set of conserved components that are required for memory in many organisms (Table 1). Many of these memory genes were not previously known to be downstream targets of CREB signaling. Together these data highlight the specificity of CREB’s transcriptional activity in memory and suggest that CREB executes its roles in memory and in metabolism through separate molecular mechanisms.
Table 1.
CREB/LTAM Genes Fall into Categories that Are Associated with Memory in Mammalian Models
| Neurotransmitter/neuropeptide receptor-related | Acetylcholine receptors: unc-38, acr-7, acr-8, lev-1 | Drd3d, Grinbf, Grin2af, Grm3f, Grfa1c, Htr2cc, Chrna1c, Fgfr2c, Glur1g, Glur2g, Nr2a, Nr2bg, Trkbg, Gabrdg, M1Rg, 2ARg, D5rg, mGlur5,7 g, nAchrg, Chrnb4h, Chrna3h, Grik1h, Clcn5h |
| Glutamate receptors: glr-6 | ||
| Dopamine receptors: dop-2 | ||
| Other: srw-85 | ||
| Peptide neurotransmitters/insulins/secreted molecules | Lipocalins: lpr-3, lpr-6, lpr-1, lpr-4, lpr-7, lpr-5 | Vgfc, Penkc, Tac1c, Ecel1c, Npyc, Scg2c, Sstc, Cartptc, Bdnf, Orexing, Npsg, Igf2g, Mchg, Dyng, Galg, Nocicepting, Angg, Insh |
| Insulins: ins-22, ins-6a, ins-2 | ||
| Neuropeptides: nlp-15 | ||
| Synaptogenesis | Agrin: agr-1 | Nptx-2c, Nlgn1g, Nlgn3g |
| Neuroligin: nlg-1 | ||
| Neurexin: nrx-1 | ||
| Other: syd-1 | ||
| Synaptic vesicle function | SNARE complex: snt-1, ric-4 | Slc17a6c, SynJ2c, Rim1ag, Syt5h |
| VGAT: unc-47 | ||
| Other: rab-3 | ||
| Synaptic scaffolding | MAGUKS: magu-3, magu-2, magu-4 | Cd9c, Homer1c, Shank3g, Akap5g, Psd95g, Ccam2ah |
| Shank: shn-1 | ||
| Other: tsp-13, casy-1a, lad-2 | ||
| Kinase/phosphatase signaling | MAPK signaling: mnk-1, mpk-2, sma-5, jnk-1, jkk-1 | Dusp1c, Sik1c, Sik3c, Prkacgf, Akap13c, Sgk1c, Ptpn4e, Mapk7 e, CamkIIg, CamkIVg, Camkkg, Erk1/2g, Mek1/2g, Pkag, Pkgg, Pkcg, PKMzg, fyng, PAKg, CK2g, Msk1g, Cdk5g, Rsk2g, Ptp1bg, Pteng, Stepg |
| PKA: kin-2 | ||
| Tyrosine kinases: scd-2, ddr-1 | ||
| Phosphatases: ptp-1 | ||
| G protein signaling | G subunit: egl-30a, gpa-17 | Gna11d, Rab11be, Gpr3c, Prkcaf, S1pr3c, Rasgef1bg, Gpr123g, Gemg, Rgs14g, Rasgrf1g |
| GEF: rhgf-1 | ||
| GTP binding proteins: rab-37, rab11.2 | ||
| RhoGAP: rrc-1 | ||
| Ion channels/transporters | K+ channels: twk-28, twk-13, twk-24, slo-1 | Cav3.2g, Kcnh5h, Kcnah |
| Innexins: inx-15, inx-17 | ||
| TRP channels: trpl-2, trpa-1 | ||
| Cl− channels: glc-4, avr-14 | ||
| Other: unc-2, lgc-27 | ||
| Cytoskeletal remodeling | Filamin: fln-2 | Cyfip2c,Vcle, Spnb2c, Vimc |
| Vinculin: deb-1 | ||
| Coronin: cor-1 | ||
| Other: eps-8, jac-1 | ||
| Calcium signaling | CAMKII: unc-43 | Cnn1b, Taglne, Camk2gd, Ncs1g, Cng, Ryrg, Insp3Rg |
| Calponin: cpn-2 | ||
| Calmodulin: cal-3 | ||
| cAMP/GMP signaling | Adenylate cyclase: acy-1a | Pde6de, Adcy8b, Adcy1b, Pde4g |
| Phosphodiesterase: pdl-1 | ||
| RNA binding proteins | pab-2, sym-2 | Fmrpg, Adarg, Cpebg |
| Transcription factors | bZIP: atf-2, fos-1, zip-1 | Nfil3b, Fosb, FosBb, Stat3b, Npas4c, Cremc, Rfx4c, Runx1e, FosL2e, c-jung, junBh, junDg, C/EBPdg, c-forg, Nfg, Egr3g, zif268(Egr1)g, Mef2cg, Pparag, Mecp2g, Srfg, Npas4g, Crtc1g, Foxo6g, Nfato4g, Lhx1g, GluA1g, AhRg, Stat4h |
| Homeodomain: ceh-48, ceh-5, egl-5, unc- 42, unc-30, unc-86 | ||
| ETS: ast-1, ets-5 | ||
| STAT: sta-1 | ||
| bHLH: lin-32 | ||
| Runx: rnt-1 | ||
| Nuclear hormone receptors | nhr-203, nhr-120, nhr-177, nhr-59, nhr-21, nhr- 44, nhr-213, nhr-98, nhr-127, nhr-8, nhr-201 | Nr4a2c, Nr4a1c, Rarbg, Grg, Rxryg, Mrg, Trag, Erag, Erbg, Nr1h2h |
| Chromatin remodeling | set-30 | Nap1l5c, Sirt1g, Hdac3g, Setbd1g, CBP/p300g, Hdac1g, Hdac4g, Hdac2g, Tet1g, Baf53bg, Rbap48g |
| Neural migration and development | Hedgehog family: wrt-1, hog-1 | Pcdh11xb, Nid1e, Nnatc, Efnb1e, Ppf1b1e, Rek7g, Wnt3ag, Disc1g, Ngr1g, Pdgfrg, Cmp1g |
| Ankyrin-like: unc-44 | ||
| Nidogen: nid-1 | ||
| Ephrin: efn-2 | ||
| Syntrophin: stn-2 | ||
| Cadherin: fmi-1 | ||
| Plexin: plx-2 | ||
| Other: eva-1, max-1, dyf-1, che-11, hlb-1, nphp-1, dex-1 | ||
| Synaptogenesis | pqn-73, pqn-13, pqn-32, pqn-37 | Cpebg, Prpcg |
Genes identified by SAM analysis to be CREB and LTAM training dependent were manually curated into functional categories involved in learning and memory in higher organisms. Mammalian genes with similar functions are also listed, displaying a conservation of processes that regulate memory. Bold text indicates genes with LTAM effects.
Genes with known roles in learning or other associative behaviors in C. elegans
Mammalian neuronal CREB- and activity-dependent genes with C. elegans orthologs/paralogs on CREB/LTAM-dependent list
Neuronal CREB- and activity-dependent genes that fall into gene category
Human genes linked to memory with direct orthologs
CREB/activity-dependent genes in other tissues
Human genes linked to memory that fall into gene category
Mammalian genes linked to memory that fall into gene category
Mammalian memory training-induced genes
Known and Novel CREB/LTAM-Induced Genes Are Required for Long-Term Memory
To assess the contributions of CREB/LTAM candidate genes to long-term memory, we reduced activity of selected genes, either with loss-of-function mutants or by RNA interference, and assayed memory performance measured immediately after training (0 hr; further referred to as “learning”) and 16 hr after training (long-term memory) (Figure 4), in addition to motility and chemotaxis (Figure S6). We tested a selection of CREB-dependent LTAM-upregulated genes whose homologs are known to play a role in long-term memory in other organisms (Figures 3A–3C), neuronal genes with no previously known role in long-term memory (Figures 3D and 3E), and novel genes with no known function (Figures 3F and 3G).
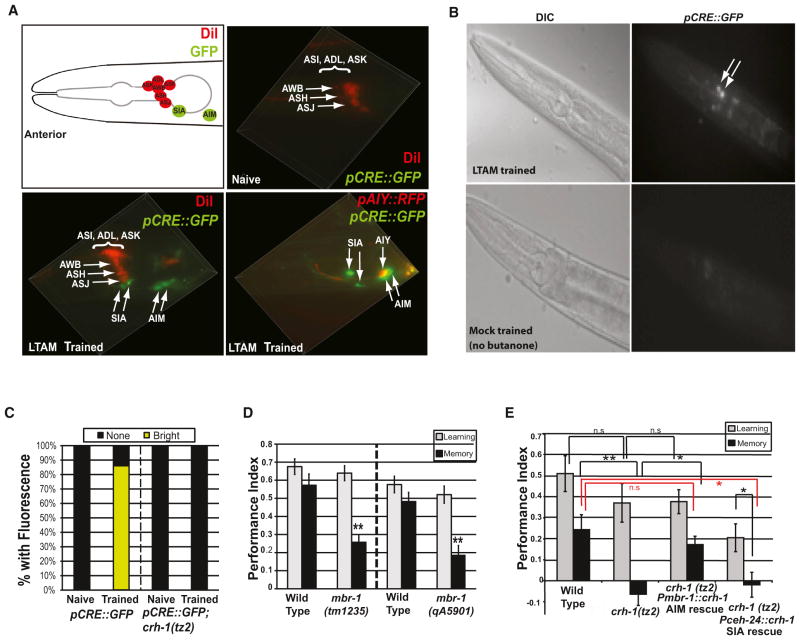
Figure 4. CREB Is Selectively Activated in the AIM Interneurons upon Memory Training.
(A and B) LTAM training, but not mock training, induces CREB in the AIM and SIA neurons. (A) Naive, untrained pCRE::GFP worms display only dim GFP expression.
(C) LTAM training-induced CREB activation in the AIM neurons is eliminated in a crh-1 mutant (n > 20 for each condition). (D) Perturbing the AIM interneuron through mbr-1 mutation reduces 16 hr memory. Mean ± SEM.
(E) Expressing CREB activity in the AIM, but not SIA neuron, rescues memory to a level comparable to wild-type’s. Mean ± SEM.
Figure 3. CREB/LTAM-Dependent Genes Are Required for LTAM.
(A–C) Animals with reduction of function (mutations or RNAi treatment) of CREB/LTAM-induced genes with known memory roles in other organisms have defective C. elegans LTAM at 16 hr post-training.
(D) LTAM tests of CREB/LTAM-induced neuronal genes with no known memory roles.
(E) Temperature-sensitive alleles of unc-86 demonstrate a defect in long-term memory when treated at the restrictive temperature (25°C).
(F and G) Reduction of CREB/LTAM-induced genes with unknown functions causes defects in LTAM. Mean ± SEM, n ≥ 4–6 for each time point, *p < 0.05, **p < 0.01, ***p < 0.001.
The expression of the cAMP, Jun/JNK/JKK, and calcium-signaling regulators upstream of CREB increased upon LTAM training in a CREB-dependent manner (Table S5). These signaling proteins are required for long-term memory in other systems (Abel and Nguyen, 2008). We found that CaMKII/unc-43(gk452), jnk-1(gk7), and jkk-1(km2) mutants were also defective specifically in long-term memory (Figure 3A), suggesting that their roles in memory are conserved. Signaling downstream to the insulin receptor has been implicated in synaptic plasticity and cognitive function in mammals, including humans (Huang et al., 2010). The expression of ins-22 increased in a CREB/LTAM-dependent manner, and reducing ins-22 levels using RNA interference decreased both learning and LTAM performance (Figure 3B).
Histone methylation and acetylation and subsequent modification of expression have been implicated in learning and memory in several systems (Zovkic et al., 2013). The SET family histone methylase set-30 is upregulated with LTAM training in a CREB-dependent fashion (Table S5), and loss of set-30 affected long-term memory (Figure 3C). Thus, chromatin regulators that are required for long-term memory in other organisms may be conserved in C. elegans long-term memory, and the expression of the genes involved in these processes is regulated by CREB.
Many neuronal proteins that have not been previously implicated in the regulation of long-term memory, such as specific ion channels, synaptic proteins, and synaptogenesis components, were upregulated with LTAM training in a CREB-dependent manner (Table 1). Mutants of slo-1, a calcium-activated BK potassium channel that controls vesicle release at synapses at the C. elegans neuromuscular junction (Wang et al., 2001), were significantly impaired for learning as well as long-term memory (Figure 3D), but also show defects in motility and chemotaxis (Figure S6A). Loss of nid-1 (nidogen) activity causes defects in cell migration, axon guidance, and synapse morphology (Ackley et al., 2005), and nid-1(cg119) animals have decreased memory (Figure 3D). The neuronally expressed GPI-anchored immunoglobulin superfamily protein gene wrk-1 was previously implicated in axon guidance (Boulin et al., 2006); we find that it is also required for memory (Figure 3D).
The POU-domain transcription factors Brn3a/Pou4f1 and Brn3b/Pou4f2 and their C. elegans ortholog UNC-86 are required for neuronal cell fate determination, cell maintenance, and neurite outgrowth (Finney and Ruvkun, 1990; McEvilly and Rosenfeld, 1999; Sze and Ruvkun, 2003; Sze et al., 2002). The expression levels of unc-86 were undetectable in naive crh-1 mutants (Figure 2G) and only marginally increased in crh-1 mutants after training compared to wild-type (Figure S3; Table S5). To avoid the developmental defects (including chemotaxis) caused by embryonic and larval loss of unc-86, we utilized a temperature-sensitive allele, unc-86(n848ts) (Ferguson and Horvitz, 1985), to knock down unc-86 function after development. We compared LTAM performance of mutants with intact or adult-ablated UNC-86 with wild-type worms cultivated at the same temperatures. All animals had normal spaced associative learning (0 hr), but animals with ablated UNC-86 have defective LTAM (Figure 3E), indicating that unc-86 is necessary for long-term memory post-developmentally.
A large portion of the CREB/LTAM-dependent upregulated gene set was comprised of Wnt and Hedgehog signaling pathway components, immunoglobulins, ubiquitination/protein turnover genes, and axon guidance/neuronal migration components (Table 1 and Table S5), suggesting that cellular remodeling processes may occur during memory formation. The human agrin homolog, agr-1, acts in synaptic clefts (Hrus et al., 2007), and its loss disrupts C. elegans LTAM (Figure 3G). Loss of hst-3.1, a heparin sulfate sulfanotransferase similar to those with roles in axon guidance (Rhiner et al., 2005), and ptr-15, a Patched homolog, also severely impaired memory (Figure 3G). The requirement for cell adhesion and neural migration genes in LTAM suggests that dynamic cell organization changes are important for plasticity in C. elegans memory.
Novel CREB/LTAM-Dependent Genes Suggest New Long-Term Memory Mechanisms
The CREB/LTAM-dependent upregulated gene set also includes many uncharacterized genes and genes not known to function in neurons (Table S5) that may encode previously undiscovered components of memory. Reducing the levels of some of the top-ranked, previously uncharacterized LTAM/CREB genes (Figures 3F and 3G) caused significant defects in memory. Several genes appear to be specific for long-term memory: knockdown of F29G6.2, tag-234, tag-273, mig-6, insc-1, and lpr-5 did not affect motility (Figure S6) or spaced learning, but specifically impaired 16 hr long-term memory (Figures 3F and 3G). While many of these new memory genes had no previously described function, several have orthologs in flies and mice (Table S5), and some have neuron-associated functions. The learning and memory roles of these novel LTAM genes indicate that they comprise new memory components that may be conserved in higher organisms. Thus, our CREB/LTAM gene set may contain many more novel memory components that remain to be tested.
CREB Is Selectively Activated in the AIM Interneurons upon Memory Training
Although CREB is expressed ubiquitously (Kimura et al., 2002), its activation in particular neurons has been associated with specific behaviors. CREB activity in the reversal interneurons AVA and AVD is required for habituation, a nonassociative tap withdrawal behavior (Timbers and Rankin, 2011), and in the AFD neuron for thermotaxis (Nishida et al., 2011). Our microarray data suggest that CREB activity in neurons is sufficient to rescue LTAM and the transcriptional program specific to memory. To identify the site of CREB activation upon positive olfactory LTAM training (Kauffman et al., 2010), we used reporter transgenic worms in which GFP expression is controlled by the CRE reporter (pCRE::GFP) (Kimura et al., 2002). In untrained worms, CREB expression is barely detectible (Figure 4A). After spaced butanone/food long-term memory training, we observed fluorescence in two sets of neurons (Figures 4A and 4B); this fluorescence increase is abrogated in a crh-1 mutant (Figure 4C). pCRE::GFP expression in the SIA, where CREB is activated upon extended starvation (Suo et al., 2006) (Figure 4A), was expected, since our training paradigm involves cycling between starvation and food paired with butanone. Fluorescence in the SIA neurons was also observed upon mock training in the absence of butanone (Figure 4B). However, CREB rescue in the SIA neurons does not rescue LTAM (Figure 4E), suggesting that CREB activation in the SIA is not specific to or required for long-term memory.
We also observed prominent GFP expression in the AIM interneurons (Figure 4A). To distinguish the AIM from the neighboring AIY neuron, we crossed pCRE::GFP worms with pAIY::RFP (Wenick and Hobert, 2004) and found that GFP and RFP do not overlap in the bigenic animals after training (Figure 4A), confirming CREB expression in the AIM. The activation of pCRE::GFP in the AIM is specific to memory training, since GFP expression is not detected in the AIM upon mock training (Figure 4B). Furthermore, mutations in mbr-1, an ortholog of a honeybee mushroom body transcription factor that is required for normal AIM development (Kage et al., 2005), abrogate long-term memory (Figure 4D) but have no effect on chemotaxis, learning, or short-term memory (Figure S8B). Finally, rescue of CREB under the mbr-1 promoter rescues long-term memory (Figure 4E). Together, these data show that CREB is selectively activated in the AIM interneuron upon associative long-term memory training.
To examine the dynamics of CREB activation, we quantified the levels of pCRE::GFP fluorescence in populations of LTAM-trained worms. While phosphorylated CREB levels rise fairly linearly with training, pCRE::GFP fluorescence increased markedly after 3–4 training sessions, paralleling the increase in LTAM performance (Figures 5A and 5D). Mutants of unc-43 and jnk-1, both activators of CREB (Johannessen et al., 2004) and downstream targets of CREB (Table S5), blocked the pCRE::GFP fluorescence increase (Figures 5B–5D), suggesting that a feedforward mechanism of CREB activation of its own upstream activators occurs with every food/butanone training cycle.
Figure 5. CREB Is Activated in the AIM Interneuron upon Different Olfactory-Associative Memory Training Paradigms.
(A) Levels of CREB activity increase with training cycles in pCRE::GFP worms and correlate with both the 16 hr performance index and the level of activated (phosphorylated) CREB protein. p values are based on comparisons of individual training blocks relative to the naive of the same strain. Chi-square tests were performed on distributions.
(B and C) Loss of function of the CREB-activating CaMKII (unc-43) and JNK signaling (jnk-1) pathways reduces LTAM training-induced CREB activation. p values are based on comparisons of individual training blocks relative to the naive of the same strain. Chi-square tests were performed on distributions.
(D) Levels of CREB activity increase with training cycles in pCRE::GFP worms and correlate with both the 16 hr performance index and the level of activated (phosphorylated) CREB protein. Loss of function of the CREB-activating CaMKII (unc-43) and JNK signaling (jnk-1) pathways reduces LTAM training-induced CREB activation. p values are based on comparisons of individual mutant training blocks (B and C) to analogous wild-type (A) controls. Error bars are ± SEM.
(E) GFP expression is observed in the AIM (white arrows) in pCRE::GFP worms upon spaced memory training using various associative olfactory paradigms.
(F) Levels of bright fluorescence in AIM increase upon associative training as in (E) above. Chi-square tests were performed on distributions. p values are relative to mock-trained animals. **p< 0.01, ***p< 0.001 ****p< 0.0001.
(G) CREB is activated in the AIM (white arrow) upon aversive associative long-term memory training using the attractive odorant diacetyl.
Many CREB/LTAM-induced genes that are required for long-term memory (Figure 3) are not detected in AIM neurons (Figure S7). Rather, their expression is prominent in neurons of the nerve ring, or in socket cell glia in the case of F21G4.1, or both (ins-22). Knockdown of F21G4.1 blocks 16 hr memory (Figure 3G), suggesting that socket cell glia may play a role in LTAM. The PQN-73 prion-like protein is expressed in the arcade cells and is also required for LTAM (Figures S7 and 3G). While it is possible that transcriptional GFP fusions fail to capture expression of these genes in the AIM, it is likely that downstream CREB/LTAM transcriptional targets are expressed in cells other than the AIM, supporting a model in which CREB activates gene expression in the AIM, which then induces the expression of genes that signal to other cells to induce additional expression changes.
If the AIM neuron acts as a general memory center for the integration of paired stimuli, one would expect that CREB activation should occur upon formation of associations, independent of the particular conditioned or unconditioned stimuli presented. To test this hypothesis, we trained pCRE::GFP worms using several alternative olfactory spaced conditioning paradigms, including pairing of butanone with heat shock, butanone with starvation, and pyrazine (sensed by the AWA neuron) with food (Figures 5E and 5F). While spaced food/starvation without odor pairing modestly increased fluorescence, spaced training with all of the associative conditioning paradigms induced bright fluorescence in the AIM. Similarly, LTAM training with a diacetyl-starvation LTAM paradigm (Vukojevic et al., 2012) also induces CREB activity in the AIM and SIA (Figure 5G). Together, these results demonstrate that CREB is activated in the AIM in response to olfactory associative conditioning and is not specific to butanone, sensing through the AWC, or food/starvation training. General, nonassociative stress, such as heat shock without butanone pairing, induced a distributed pattern of nerve ring and pharyngeal neuron fluorescence (Figure S8A), suggesting that CREB activation in the AIM is not due to nonassociative stress conditions, but that there may be a second type of CREB response to heat stress. Together, these results show that CREB activation in the AIM is specific for associative memory formation.
DISCUSSION
The role of CREB in long-term memory has been studied at great length for the past 20 years. Remarkably, the full spectrum of CREB-induced gene expression changes that result from long-term memory training remains largely unexplored. Using an olfactory LTAM paradigm in C. elegans, we identified genome-wide direct and indirect neuronal CREB target genes that are induced upon memory conditioning. While previous studies of basal CREB targets have been performed in vivo and in vitro, with artificial CREB activation, or ChIP-seq analysis limited to direct, basal CREB targets (Barco et al., 2005; Impey et al., 2004; Lesiak et al., 2013; Ploski et al., 2010), to our knowledge, this study is the first unbiased analysis of global CREB activity induced in the context of a physiologically relevant memory training task. It is worth noting that direct binding approaches such as ChIP-seq may miss 95% of the CREB-regulated LTAM genes, because (1) CREB activity is only induced in one pair of interneurons, and currently no cell-specific ChIP-seq approaches exist in adult C. elegans, and (2) only 37 of the LTAM-regulated genes have full-length CRE sites. By instead using a comparative transcriptional approach that incorporated the relevant biological context, long-term memory formation, we were able to successfully identify the larger set of CREB-dependent memory genes. Importantly, our analysis revealed novel and evolutionarily conserved genes, biological processes, and sites of CREB activity during LTAM training, setting the framework for the study of these memory processes in mammals.
In vitro analysis of CREB binding sites demonstrates that cell type and context influence target gene selection (Cha-Molstad et al., 2004). Our finding that basal CREB targets are distinct from the CREB transcriptional program induced by memory training further supports the multifunctional nature of CREB in vivo. Only ~2.5% of CREB/LTAM target genes are expressed in the basal state, and while CREB/LTAM targets are primarily neuronal in location and function, the basal CREB target genes are enriched in growth and development processes in nonneuronal tissues. (Interestingly, however, a third of the presumed direct CREB targets in basal conditions are neuronal, suggesting that CREB activity in some neurons may be specific). Basal CREB activity regulates the expression of genes throughout the worm, as CREB is ubiquitously expressed, and a GFP reporter showed no distinct tissues with CREB activity in basal conditions. Conversely, acute induction of CREB in the AIM interneurons occurs following several olfactory memory training paradigms (Figure 3), demonstrating that the different outcomes of CREB signaling are mediated by tissue and cell-specific regulation of target gene selection.
How does CREB activity in the AIM neurons completely alter the transcriptional outcome of CREB signaling? Little is known regarding additional functions and the expression profile of the AIM neurons, but the activity of transcriptional cofactors and/or downstream signaling components specific to the AIM could explain how CREB mediates such distinct target gene regulation in the context of LTAM. The CREB/LTAM gene set is enriched for full-length and not half CRE sites, possibly indicating that a small set of direct CREB targets are induced by CREB in the AIM. A caveat of this approach is that in the absence of ChIP-seq analysis of CREB binding sites, direct and indirect CREB targets are not easily distinguished, and the optimal CRE site in C. elegans is currently unknown. As ~35% of CREB/LTAM promoters lack any discernable CRE site, additional TFs, signaling pathways, and neurons downstream of in parallel of CREB and AIM are likely required for CREB’s behavioral output.
The expression of 40 TFs (Reece-Hoyes et al., 2011) is regulated by CREB/LTAM training. These TFs may be critical but unappreciated components of the long-term memory regulatory machinery. UNC-86 is among the many evolutionarily conserved TFs that we find downstream of CREB signaling and is required for LTAM. UNC-86 is a POU family TF that is highly conserved in structure and function with respect to its Brn3a/Pou4f1 and Brn3b/Pou4f2 mammalian orthologs, which are required for neuronal cell fate determination, cell maintenance, and neurite outgrowth (Finney and Ruvkun, 1990; McEvilly and Rosenfeld, 1999; Sze and Ruvkun, 2003; Sze et al., 2002). Interestingly, CRE sites are present in the Brn3b promoter, and Brn3a sequences were present in a ChIP-seq study of basal, neuronal CREB targets (Lesiak et al., 2013). This suggests that unc-86 ortholog expression is similarly regulated by CREB and may be part of a conserved transcriptional program regulating long-term memory. This is the first report of a role for the POU family of TFs in long-term memory, and the previously demonstrated conservation of UNC-86 function warrants its investigation in mammalian memory.
The CREB/LTAM list is significantly enriched in mammalian orthologs with known roles in memory (Cnn1, Nfil3, Fos, Stat3, Pcdh11x, Adcy1,8), and comparison of worm LTAM genes with vertebrate memory profiles reveals a high level of functional overlap. Cavallaro et al. (2002) identified rat hippocampal long-term memory genes induced by spatial learning, including insulin, nuclear hormone receptors, K+ and Cl− channels, acetylcholine and kainate glutamate receptors, STAT and bZIP TFs, etc., all of which are induced with LTAM training in worms (Table 1). Conservation of known memory pathways further validates C. elegans as a model for the discovery of novel mammalian memory components.
Among the 757 CREB/LTAM genes identified by this study, many genes comprising several functional categories are interesting areas for follow-up studies in mammals. Mammalian TFs, nuclear hormone receptors (NHRs), and chromatin remodeling factors directly influence the gene expression that underlies long-term memory (Table 1). It is likely that additional uncharacterized mammalian memory components within these categories affect memory, making the orthologs of the C. elegans CREB/LTAM genes identified here exciting candidates to test. CREB/LTAM induces the expression of 11 NHR genes, and nhr-120 was functionally shown to be required for LTAM. The nhr-120 ortholog, RXR, is necessary for hippocampal CA1 long-term depression (Chiang et al., 1998). Additionally, the nuclear hormone receptors Nr4a1 and Nr4a2 are necessary for hippocampal dependent object location memory (McNulty et al., 2012), promoting the examination of the remaining NHR candidates and orthologs for memory functions. Additional regulators of memory-related gene expression are prion-forming proteins, such as the cytoplasmic polyadenylation element-binding protein (CPEB), whose synaptic activity-induced prion-like aggregation is critical for long-term memory stabilization (Si et al., 2003). We identified four prion domain-containing CREB/LTAM genes in this study, and pqn-73 is required for LTAM. It is possible that these potential prion-like proteins and their orthologs may reveal novel roles for prions in memory processes.
By combining functional memory training with transcriptional analysis, we uncovered new genes, mechanisms, and cells underlying long-term memory. Using functional memory assays, we verified the requirement of many of these components in LTAM, finding that the reduction of many individual proteins abrogated memory. This suggests that a plastic phenotype such as memory might require the activity of a predetermined set of proteins, without much allowance for compensatory mechanisms. This aspect of LTAM is in sharp contrast to aging, where individual downstream targets of DAF-16 have a relatively small effect on lifespan and act cumulatively (Murphy et al., 2003). The fragility of long-term memory, as illustrated by our studies of LTAM in C. elegans, underscores the need to fully characterize memory pathways to understand declines of memory with age and neuropathological disease.
EXPERIMENTAL PROCEDURES
RNA Collection, Microarray Hybridization, RT-PCR, and qRT-PCR
Worms were collected and RNA was hybridized on 4x44K C. elegans arrays (Agilent) at 60°C overnight, as previously described (Shaw et al., 2007). Four biological replicates of crh-1(tz2) versus wild-type N2 arrays and naive versus LTAM-trained worms of three wild-type N2, five crh-1(tz2), two Pcmk-1::crh-1β, and two crh-1(tz2);Pcmk-1::crh-1β were hybridized. qRT-PCR reactions were run followed by a dissociation reaction to determine specificity of the amplified product; gene expression was quantified using the ΔΔCt method using pmp-3 as a reference gene.
Microarray Analysis
Data were analyzed as previously described (Shaw et al., 2007); additional details available in Supplemental Experimental Procedures. Data are available at PUMAdb (http://puma.princeton.edu).
Ortholog Analysis
Human orthologs of C. elegans CREB/LTAM-dependent upregulated genes were determined using OrthoList (http://www.greenwaldlab.org/ortholist/) (Shaye and Greenwald, 2011).
Developmental Analysis
Worms were bleached onto NGM plates with OP50 and incubated at 20°C (~600 eggs per genotype). Developmental stages were determined at time points post-synchronization. Differences in distribution of developmental stages were analyzed by chi-square tests.
Lifespan Analysis
Worms were bleached onto HG plates with OP50, incubated at 20°C, analyzed as previously described (Shaw et al., 2007), and analyzed using OASIS (Yang et al., 2011).
Butanone/Food LTAM Assay
Wild-type and mutant animals were trained and tested as previously described (Kauffman et al., 2010). For RNAi tests, after 1 hr of starvation, eri-1(mg366);lin-15B(n744) worms were conditioned in liquid for 30 min (3 ml of 1:1,000 LB + OP50:100% butanone), then washed twice with M9 and starved in M9 for 30 min.
Variations of the Associative Spaced Training Protocol
Mock training was performed with seven cycles of 30 min OP50 with no odorant followed by 30 min periods on unseeded plates. Pyrazine training was performed using 2 μl of 10% pyrazine on the plate lid with food cycles. Butanone/starvation was performed using 2 μl of 10% butanone on the plate lid during starvation cycles. Diacetyl LTAM training was performed as described in Vukojevic et al., 2012.
unc-86 Temperature-Sensitive LTAMs
unc-86(n848ts) were grown at 15°C until L4 and then transferred to 25°C. Control or ablated animals were starved and underwent 7 cycles of training at 15°C or 25°C, respectively. Animals were also held at 15°C or 25°C for 16 hr before assessment of long-term memory.
Supplementary Material
Acknowledgments
We thank M. Barr, the C. elegans Genetics Center, C. elegans Gene Expression Consortium, M. Costa, C. Kenyon, A.V. Mariqc, I. Hope, S. Mitani, M. Hagiwara, M. Alkema, and Q.L. Ch’ng for strains. This work was funded by an NIH Cognitive Aging R01, Keck Young Scholars, and McKnight Scholars awards to C.T.M. and an NRSA Fellowship to R.K. C.T.M. is the director of the Paul F. Glenn Laboratory for Aging Research at Princeton University, which supported this work.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2014.12.029.
AUTHOR CONTRIBUTIONS
V.L., R.N.A., R.K., A.K., and C.T.M. participated in research design. V.L., R.N.A., R.K., A.K., G.S., W.K., and D.X. conducted experiments. A.K. performed microarrays, and V.L., R.N.A., and R.K. performed statistical analyses of microarray and behavioral data. R.K., C.T.M., and V.L. wrote the manuscript.
References
- Abel T, Nguyen PV. Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Prog Brain Res. 2008;169:97–115. doi: 10.1016/S0079-6123(07)00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Patterson SL, Alarcon JM, Gromova P, Mata-Roig M, Morozov A, Kandel ER. Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron. 2005;48:123–137. doi: 10.1016/j.neuron.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Boulin T, Pocock R, Hobert O. A novel Eph receptor-interacting IgSF protein provides C. elegans motoneurons with midline guidepost function. Curr Biol. 2006;16:1871–1883. doi: 10.1016/j.cub.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Cavallaro S, D’Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proc Natl Acad Sci USA. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc Natl Acad Sci USA. 2004;101:13572–13577. doi: 10.1073/pnas.0405587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hendricks M, Cornils A, Maier W, Alcedo J, Zhang Y. Two insulin-like peptides antagonistically regulate aversive olfactory learning in C. elegans. Neuron. 2013;77:572–585. doi: 10.1016/j.neuron.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Misner D, Kempermann G, Schikorski T, Giguère V, Sucov HM, Gage FH, Stevens CF, Evans RM. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron. 1998;21:1353–1361. doi: 10.1016/s0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Tully T. Neural substrates of memory: from synapse to system. J Neurobiol. 2003;54:238–253. doi: 10.1002/neu.10170. [DOI] [PubMed] [Google Scholar]
- Euskirchen G, Royce TE, Bertone P, Martone R, Rinn JL, Nelson FK, Sayward F, Luscombe NM, Miller P, Gerstein M, et al. CREB binds to multiple loci on human chromosome 22. Mol Cell Biol. 2004;24:3804–3814. doi: 10.1128/MCB.24.9.3804-3814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB. Neuropeptide modulation of learning and memory processes. Rev Neurosci. 1996;7:151–164. doi: 10.1515/revneuro.1996.7.2.151. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Hrus A, Lau G, Hutter H, Schenk S, Ferralli J, Brown-Luedi M, Chiquet-Ehrismann R, Canevascini S. C. elegans agrin is expressed in pharynx, IL1 neurons and distal tip cells and does not genetically interact with genes involved in synaptogenesis or muscle function. PLoS ONE. 2007;2:e731. doi: 10.1371/journal.pone.0000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Lee CC, Hsu KS. The role of insulin receptor signaling in synaptic plasticity and cognitive function. Chang Gung Med J. 2010;33:115–125. [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kage E, Hayashi Y, Takeuchi H, Hirotsu T, Kunitomo H, Inoue T, Arai H, Iino Y, Kubo T. MBR-1, a novel helix-turn-helix transcription factor, is required for pruning excessive neurites in Caenorhabditis elegans. Curr Biol. 2005;15:1554–1559. doi: 10.1016/j.cub.2005.07.057. [DOI] [PubMed] [Google Scholar]
- Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp. 2011:49. doi: 10.3791/2490. http://dx.doi.org/10.3791/2490. [DOI] [PMC free article] [PubMed]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Corcoran EE, Eto K, Gengyo-Ando K, Muramatsu MA, Kobayashi R, Freedman JH, Mitani S, Hagiwara M, Means AR, Tokumitsu H. A CaMK cascade activates CRE-mediated transcription in neurons of Caenorhabditis elegans. EMBO Rep. 2002;3:962–966. doi: 10.1093/embo-reports/kvf191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak A, Pelz C, Ando H, Zhu M, Davare M, Lambert TJ, Hansen KF, Obrietan K, Appleyard SM, Impey S, Wayman GA. A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PLoS ONE. 2013;8:e64658. doi: 10.1371/journal.pone.0064658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JF, Jackson MF, Beazely MA. G protein-coupled receptors control NMDARs and metaplasticity in the hippocampus. Biochim Biophys Acta. 2007;1768:941–951. doi: 10.1016/j.bbamem.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues APC, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M. Protein kinase signaling in synaptic plasticity and memory. Curr Opin Neurobiol. 2007;17:313–317. doi: 10.1016/j.conb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, Rosenfeld MG. The role of POU domain proteins in the regulation of mammalian pituitary and nervous system development. Prog Nucleic Acid Res Mol Biol. 1999;63:223–255. doi: 10.1016/s0079-6603(08)60724-2. [DOI] [PubMed] [Google Scholar]
- McNulty SE, Barrett RM, Vogel-Ciernia A, Malvaez M, Hernandez N, Davatolhagh MF, Matheos DP, Schiffman A, Wood MA. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn Mem. 2012;19:588–592. doi: 10.1101/lm.026385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Sugi T, Nonomura M, Mori I. Identification of the AFD neuron as the site of action of the CREB protein in Caenorhabditis elegans thermotaxis. EMBO Rep. 2011;12:855–862. doi: 10.1038/embor.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Ploski JE, Park KW, Ping J, Monsey MS, Schafe GE. Identification of plasticity-associated genes regulated by Pavlovian fear conditioning in the lateral amygdala. J Neurochem. 2010;112:636–650. doi: 10.1111/j.1471-4159.2009.06491.x. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJM, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, Walhout AJM. Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat Methods. 2011;8:1059–1064. doi: 10.1038/nmeth.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M, Specht CG, Triller A. Molecular dynamics of post-synaptic receptors and scaffold proteins. Curr Opin Neurobiol. 2008;18:532–540. doi: 10.1016/j.conb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Rhiner C, Gysi S, Fröhli E, Hengartner MO, Hajnal A. Syndecan regulates cell migration and axon guidance in C. elegans. Development. 2005;132:4621–4633. doi: 10.1242/dev.02042. [DOI] [PubMed] [Google Scholar]
- Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr Biol. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Suo S, Kimura Y, Van Tol HHM. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J Neurosci. 2006;26:10082–10090. doi: 10.1523/JNEUROSCI.0819-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Ruvkun G. Activity of the Caenorhabditis elegans UNC-86 POU transcription factor modulates olfactory sensitivity. Proc Natl Acad Sci USA. 2003;100:9560–9565. doi: 10.1073/pnas.1530752100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Zhang S, Li J, Ruvkun G. The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development. 2002;129:3901–3911. doi: 10.1242/dev.129.16.3901. [DOI] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timbers TA, Rankin CH. Tap withdrawal circuit interneurons require CREB for long-term habituation in Caenorhabditis elegans. Behav Neurosci. 2011;125:560–566. doi: 10.1037/a0024370. [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci USA. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Voglis G, Tavernarakis N. The role of synaptic ion channels in synaptic plasticity. EMBO Rep. 2006;7:1104–1110. doi: 10.1038/sj.embor.7400830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V, Gschwind L, Vogler C, Demougin P, de Quervain DJF, Papassotiropoulos A, Stetak A. A role for α-adducin (ADD-1) in nematode and human memory. EMBO J. 2012;31:1453–1466. doi: 10.1038/emboj.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Wenick AS, Hobert O. Genomiccis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE. 2011;6:e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.