Abstract
In an effort to identify host proteins involved in herpes simplex virus type 1 replication, monkey and human cellular protein activities (called OF-1) that bind the viral replication origin, oriS, have been described. We show by mass spectrometry that the DNA-binding component of human OF-1 contains Ku70 and Ku80 proteins.
Host cell factors are essential for the replication of many viruses. The minimal in vivo requirements for replication of herpes simplex virus type 1 (HSV-1) are a viral replication origin (oriS) and several virally encoded proteins (3, 7). However, the failure to reconstitute oriS-dependent replication in vitro with purified viral proteins (7) suggests that host proteins are also required. A protein activity from Vero cells, called OF-1, specifically binds oligomeric sequences from oriS (5, 6). As shown in Fig. 1, these sequences overlap the box I site recognized by the HSV-1 replication initiator protein, UL9 (7), suggesting that OF-1 participates in HSV-1 replication. We have partially purified a similar activity from HeLa cells (2) that specifically binds oligomers from the box I region. The DNA-binding component of this activity consists of two subunits of 73 and 90 kDa. In the present study, we have used matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry to identify these subunits as Ku70 and Ku80.
FIG. 1.
Box I region of oriS and oligonucleotide probe used for gel shift assays. A portion of the HSV-1 oriS sequence and the duplex oligonucleotide probe used for OF-1 gel shift assays are shown. The box I binding site for the HSV-1 replication initiator, UL9, in oriS is shown in uppercase letters.
Protein purification.
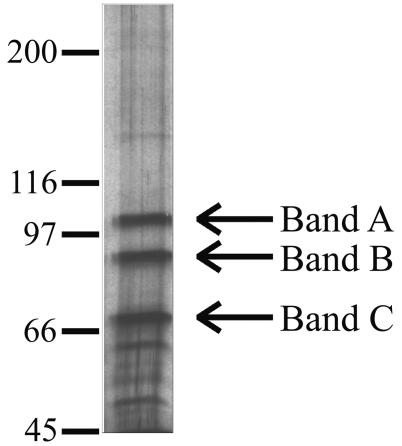
OF-1 was partially purified from HeLa cells as described previously (2), based on its ability to bind the duplex oligonucleotide shown in Fig. 1 in a gel mobility shift assay. As shown in Fig. 2, this preparation contained three major proteins (A, B, and C), as detected by electrophoresis on a 6% polyacrylamide gel containing sodium dodecyl sulfate (SDS) (13) followed by silver staining (16, 20). Bands B and C correspond to the 90- and 73-kDa DNA-binding proteins described previously (2).
FIG. 2.
Separation of protein components in the OF-1 preparation. Purified OF-1 was subjected to SDS-polyacrylamide gel electrophoresis, and the gel was silver stained. Molecular weight markers (in thousands; shown at left) were used to estimate the sizes of bands B and C (designated with arrows), which were excised for analysis by mass spectrometry.
Mass spectrometry.
Sixty picomoles of OF-1 was subjected to SDS-7.5% polyacrylamide gel electrophoresis. Designated silver-stained bands were excised and digested in situ with trypsin (Roche Molecular Biochemicals, Mannheim, Germany). The resultant peptides were extracted as described previously (16) and purified by using C18ZipTips (Millipore, Bedford, Mass.). MALDI-TOF mass spectrometry was performed by the mass spectrometry facility of the University of Arizona Department of Chemistry, with a Reflex-III MALDI-TOF instrument (Bruker-Franzen Analytik, Bremen, Germany) equipped with a delayed-extraction ion source. Samples were dried onto matrix films produced from saturated 4-hydroxy-α-cyanocinnamic acid (Sigma, St. Louis, Mo.). Acceleration and reflector (ion mirror) voltages were 20 and 21 kV, respectively. Spectra were generated by pulsed irradiation from a 337-nm-wavelength N2 laser and internally calibrated. A control spectrum was generated from polyacrylamide that lacked protein.
Analysis of m/z peaks from each MALDI-TOF spectrum identified band B from OF-1 as the 83-kDa subunit (called Ku80) of the human Ku heterodimer and band C as the 70-kDa subunit. Each data set was used to search the GenPept database with the MS-Fit program of ProteinProspector (http://prospector.ucsf.edu/), allowing for oxidation of methionine and phosphorylation of threonine, serine, and tyrosine. This program compares the experimental m/z peaks to those expected for trypsin digests of all database proteins. For band B (Table 1), 21 of 58 observed peaks matched those predicted for tryptic digestion of Ku80, with 34% of the sequence covered by the matches. For band C (Table 2), 16 of 38 peaks matched those predicted for Ku70, with 33% sequence coverage. We also performed a database search using only the unmatched peaks from each spectrum. No other significant matches were obtained, indicating that the matched proteins (Ku80 and Ku70) were the only detectable polypeptides.
TABLE 1.
Masses for tryptic peptides that identify band B of OF-1 as the human Ku80 polypeptide (GenPept accession no. 307094)a
| Measured mass (Da)b | Predicted mass (Da)c | Δ Mass (ppm)j | Positionsd | Fraction of matched fragments in PSD analysise | Peptide sequencef |
|---|---|---|---|---|---|
| 963.53 | 963.5264 | +3.78 | 641-648 | (R)AFREEAIK(F) | |
| 977.52 | 977.5056 | +14.71 | 308-315 | (K)EDIIQGFR(Y) | |
| 1,100.59 | 1,100.6039 | −12.62 | 490-497 | (R)LFQCLLHR(A) | |
| 1,121.62 | 1,121.6505 | −27.20 | 36-44 | (K)KVITMFVQR(Q) | |
| 1,239.64 | 1,239.6672 | −21.96 | 354-363 | (R)RFFMGNQVLK(V) | |
| 1,239.64 | 1,239.6754 | −28.56 | 535-544 | (K)TLFPLIEAKK(K)h | |
| 1,317.64i | 1,317.6803 | −30.58 | 131-141 | 10/19 | (R)HIEIFTDLSSR(F) |
| 1,364.54 | 1,364.5831 | −31.58 | 172-184 | (K)EDGSGDRGDGPFR(L) | |
| 1,377.72 | 1,377.7643 | −32.15 | 401-413 | (R)ANPQVGVAFPHIK(H) | |
| 1,434.71 | 1,434.6799 | +20.97 | 272-282 | (R)VKKTWTVVDAK(T)h | |
| 1,533.81 | 1,533.8654 | −36.12 | 400-413 | (K)RANPQVGVAFPHIK(H) | |
| 1,739.85 | 1,739.8717 | −12.46 | 641-654 | (R)AFREEAIKFSEEQR(F) | |
| 1,915.95 | 1,915.9431 | +3.58 | 130-144 | (K)RHIEIFTDLSSRFSK(S)h | |
| 1,931.02 | 1,930.9472 | +37.70 | 82-97 | (R)HLMLPDFDLLEDIESK(I)g | |
| 2,242.94 | 2,243.0216 | −36.40 | 546-565 | (K)DQVTAQEIFQDNHEDGPTAK(K) | |
| 2,321.23 | 2,321.1699 | +25.90 | 444-465 | (K)YAPTEAQLNAVDALIDSMSLAK(K) | |
| 2,465.28 | 2,465.2598 | +8.21 | 443-465 | (K)KYAPTEAQLNAVDALIDSMSLAK(K)g | |
| 2,465.28 | 2,465.2598 | +8.21 | 444-466 | (K)YAPTEAQLNAVDALIDSMSLAKK(D)g | |
| 2,499.17 | 2,499.2116 | −16.63 | 544-565 | (K)KKDQVTAQEIFQDNHEDGPTAK(K) | |
| 2,499.17 | 2,499.2116 | −16.63 | 545-566 | (K)KDQVTAQEIFQDNHEDGPTAKK(L) | |
| 2,620.21 | 2,620.2162 | −2.38 | 210-232 | (K)MVMISLEGEDGLDEIYSFSESLR(K) | |
| 2,653.35 | 2,653.3812 | −11.77 | 503-525 | (R)EPLPPIQQHIWNMLNPPAEVTTK(S) | |
| 2,669.36 | 2,669.3761 | −6.04 | 503-525 | (R)EPLPPIQQHIWNMLNPPAEVTTK(S)g | |
| 3,121.42i | 3,121.4827 | −20.07 | 569-599 | 33/34 | (K)TEQGGAHFSVSSLAEGSVTSVGSVNPAENFR(V) |
Peptides resulting from tryptic digestion of band B in Fig. 2 were subjected to MALDI-TOF mass spectrometry, and peak masses from the spectrum were used to search the GenPept database. Twenty-one of 58 observed peptide peaks matched the predicted trypsin fragments for Ku80 (within 40 ppm). Matched peptides cover 34% of the protein sequence, with a mean mass error of −9.17 ppm. The mean number of predicted trypsin cleavage sites not cleaved in the matching peptides was 0.8. The calculated mass of Ku80 was 82.7 kDa.
Observed monoisotopic (+), singly charged mass from MALDI-TOF mass spectrometry.
Predicted monoisotopic (+), singly charged mass of the matching tryptic fragment of Ku80. If two different tryptic peptides of Ku80 are possible matches, both matching peptides are shown.
Amino acids defining the beginning and end positions of the matched tryptic fragment.
Fraction of observed peaks in the PSD spectrum that match those predicted by theoretical fragmentation of the corresponding database peptide, allowing a fragment ion mass error of 2,500 ppm.
Sequence of peptides with predicted masses matching the observed masses and, as indicated, with predicted PSD fragmentation spectra matching the observed patterns.
Predicted mass includes one methionine oxidation.
Predicted mass includes one or two phosphorylations.
Sample was selected for PSD analysis.
Difference between predicted and measured mass of each fragment.
TABLE 2.
Masses for tryptic peptides that identify band C of OF-1 as the human Ku70 polypeptide (GenPept accession no. 307095)a
| Measured mass (Da)b | Predicted mass (Da)c | Δ Mass (ppm)j | Positiond | Fraction of matched fragments in PSD analysise | Protein sequencef |
|---|---|---|---|---|---|
| 1,161.67 | 1,161.6632 | +5.87 | 115-123 | (K)RILELDQFK(G) | |
| 1,388.68 | 1,388.6909 | −7.86 | 219-230 | (R)DIISIAEDEDLR(V) | |
| 1,399.74i | 1,399.7374 | +1.85 | 207-218 | 34/38 | (K)KPGGFDISLFYR(D) |
| 1,418.72 | 1,418.7354 | −10.83 | 195-206 | (R)DTGIFLDLMHLK(K)g | |
| 1,445.70 | 1,445.6686 | +21.71 | 576-586 | (K)FTVPMLKEACR(A)h | |
| 1,573.81 | 1,573.8226 | −8.00 | 101-114 | (K)NIYVLQELDNPGAK(R) | |
| 1,701.90 | 1,701.8812 | +11.06 | 231-244 | (R)VHFEESSKLEDLLR(K) | |
| 1,703.84 | 1,703.8142 | +15.17 | 475-488 | (R)SDSFENPVLQQHFR(N) | |
| 1,731.81 | 1,731.8206 | −6.13 | 302-317 | (R)TFNTSTGGLLLPSDTK(R)h | |
| 1,807.95 | 1,807.9554 | −2.99 | 302-318 | (R)TFNTSTGGLLLPSDTKR(S) | |
| 2,026.11i | 2,026.0861 | +11.81 | 75-92 | 20/29 | (K)IISSDRDLLAVVFYGTEK(D) |
| 2,269.24 | 2,269.2080 | +14.11 | 75-94 | (K)IISSDRDLLAVVFYGTEKDK(N) | |
| 2,288.24i | 2,288.2443 | −1.89 | 425-444 | 35/39 | (K)IQVTPPGFQLVFLPFADDKR(K) |
| 2,343.09 | 2,343.1386 | −20.74 | 95-114 | (K)NSVNFKNIYVLQELDNPGAK(R)h | |
| 2,500.24 | 2,500.2724 | −12.95 | 404-424 | (R)RNIPPYFVALVPQEEELDDQK(I) | |
| 2,501.08 | 2,500.9963 | +33.45 | 10-31 | (K)TEGDEEAEEEQEENLEASGDYK(Y) |
Polypeptides were processed and analyzed as described in footnote a in Table 1. Sixteen of 38 observed peptide peaks matched predicted trypsin fragments for Ku70 (within 40 ppm). Matched peptides cover 33% of the sequence, with a mean mass error of +2.73 ppm. The mean number of predicted trypsin cleavage sites not cleaved in the matching peptides was 0.6. The calculated mass of Ku70 was 69.8 kDa.
Observed monoisotopic (+), singly charged masses.
Predicted monoisotopic (+), singly charged mass of the matching tryptic fragment of Ku70.
Amino acids defining the beginning and end positions of the matched tryptic fragment.
Fraction of observed peaks in the PSD spectrum that match those predicted by theoretical fragmentation of the corresponding database peptide, allowing a fragment ion mass error of 2,500 ppm.
Sequence of peptides with predicted masses matching the observed masses and, as indicated, with predicted PSD fragmentation spectra matching the observed patterns.
Predicted mass includes one methionine oxidation.
Predicted mass includes one or two phosphorylations.
Sample was selected for PSD analysis.
Difference between predicted and measured mass of each fragment.
Identifications were confirmed by fragmentation of selected m/z peaks (parent ions) from the MALDI-TOF spectra by postsource decay (PSD) (18). This process generates a spectrum of ionized fragments that is predictable based on the sequence of the original parent ion peptide. Selected parent ion peaks were isolated by a 700-V pulse, and fragmentation was achieved by gradually stepping down the reflectron voltages from 21 kV. Spectra were internally calibrated using adrenocorticotropic hormone (Sigma) and analyzed with FAST and XACq software (Bruker Daltronics, Billerica, Mass.).
Mass peaks resulting from PSD were used to search the GenPept database with the MS-Tag program of ProteinProspector. This program compares the fragmentation pattern of the parent ion to that predicted for each tryptic fragment of all database proteins. For band B, 10 (of 19) and 33 (of 34) PSD peaks produced from parent ion peaks at m/z 1,317.64 and 3,121.42, respectively, matched predicted fragmentation products for Ku80 tryptic peptides (Table 1). For band C, 34 (of 35), 20 (of 29), and 35 (of 39) PSD peaks produced from parent ion peaks at m/z 1,399.74, 2,026.11, and 2,288.24, respectively, matched predicted products for Ku70 tryptic peptides (Table 2).
A possible role for Ku-associated OF-1 in HSV-1 replication.
The Ku70/Ku80 heterodimer participates in several cellular processes, including DNA strand break repair, rearrangements of immunoglobulin and T-cell receptor gene segments, telomere maintenance, transcription regulation, and DNA replication (1, 4, 8, 10, 12, 15, 19). A role for Ku70/Ku80 in DNA replication is indicated by its presence within origin-specific DNA-binding complexes in primates and yeast (Saccharomyces cerevisiae) (1, 9, 10, 12, 14, 15), by its cyclical association with monkey cell origins during late G1 and early S phases (12), by a requirement for origin binding during in vitro replication assays (9, 14), and by its regulation of origin firing in yeast telomeric regions (4). Although Ku70/Ku80 can bind avidly to DNA ends without regard to sequence (e.g., during DNA repair), it also binds specifically to sequences such as replication origins (1, 10, 12, 14). In addition, specificity can be imposed by associated proteins, as observed for a Ku-associated origin-binding complex in S. cerevisiae (15).
Ku70/Ku80 has been implicated in transcription regulation in herpesviruses (11, 17). A role for this complex in HSV-1 replication is now suggested by our finding that it constitutes a component of human OF-1. OF-1 activities from primate cells bind specifically to HSV-1 oriS sequences (2, 5, 6), as shown by preferential binding in gel mobility shift assays to oriS oligomers, compared to oligomers with randomized or mutant sequences. Thus, sequence-specific (or structure-specific) binding, rather than end binding, appears to account for the observed specificity. This specificity might be imparted by Ku70/Ku80 itself or by other OF-1-associated proteins.
Taken together, the findings that OF-1 binds specifically to HSV-1 oriS sequences, that OF-1 contains Ku70/Ku80, and that Ku70/Ku80 is involved in cellular DNA replication argue that OF-1 plays a role in viral replication. In addition, it was previously shown that human OF-1 facilitates binding of UL9, the HSV-1 initiator protein, to oriS oligomers (2), further suggesting that virus-host protein interactions are involved in Ku-associated OF-1 function.
Acknowledgments
L. B. Murata was supported in part by NIH grant T32CA09213. The Bruker Reflex-III MALDI-TOF instrument was provided by NIH grant S10RR1388.
We thank Arpad Somogyi and Linda Breci for expert assistance with mass spectrometry data acquisition and interpretation.
REFERENCES
- 1.Araujo, F. D., J. D. Knox, S. Ramchandani, R. Pelletier, P. Bigey, G. Price, M. Szyf, and M. Zannis-Hadjopoulos. 1999. Identification of initiation sites for DNA replication in the human dnmt1 (DNA-methyltransferase) locus. J. Biol. Chem. 274:9335-9341. [DOI] [PubMed] [Google Scholar]
- 2.Baker, R. O., L. B. Murata, M. S. Dodson, and J. D. Hall. 2000. Purification and characterization of OF-1, a host factor implicated in herpes simplex replication. J. Biol. Chem. 275:30050-30057. [DOI] [PubMed] [Google Scholar]
- 3.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347-384. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove, A. J., C. A. Nieduszynski, and A. D. Donaldson. 2002. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 16:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dabrowski, C. E., P. J. Carmillo, and P. A. Schaffer. 1994. Cellular protein interactions with herpes simplex virus type 1 oriS. Mol. Cell. Biol. 14:2545-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabrowski, C. E., and P. A. Schaffer. 1991. Herpes simplex virus type 1 origin-specific binding protein: oriS-binding properties and effects of cellular proteins. J. Virol. 65:3140-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehman, I. R., and P. E. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059-28062. [DOI] [PubMed] [Google Scholar]
- 8.Lieber, M. R., Y. Ma, U. Pannicke, and K. Schwarz. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 4:712-720. [DOI] [PubMed] [Google Scholar]
- 9.Matheos, D., M. T. Ruiz, G. B. Price, and M. Zannis-Hadjopoulos. 2002. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochim. Biophys. Acta 1578:59-72. [DOI] [PubMed] [Google Scholar]
- 10.Novac, O., D. Matheos, F. D. Araujo, G. B. Price, and M. Zannis-Hadjopoulos. 2001. In vivo association of Ku with mammalian origins of DNA replication. Mol. Biol. Cell. 12:3386-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petroski, M. D., and E. K. Wagner. 1998. Purification and characterization of a cellular protein that binds to the downstream activation sequence of the strict late UL38 promoter of herpes simplex virus type 1. J. Virol. 72:8181-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz, M. T., D. Matheos, G. B. Price, and M. Zannis-Hadjopoulos. 1999. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol. Biol. Cell. 10:567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Schild-Poulter, C., D. Matheos, O. Novac, B. Cui, W. Giffin, M. T. Ruiz, G. B. Price, M. Zannis-Hadjopoulos, and R. J. Hache. 2003. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 22:65-78. [DOI] [PubMed] [Google Scholar]
- 15.Shakibai, N., V. Kumar, and S. Eisenberg. 1996. The Ku-like protein from Saccharomyces cerevisiae is required in vitro for the assembly of a stable multiprotein complex at a eukaryotic origin of replication. Proc. Natl. Acad. Sci. USA 93:11569-11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 17.Shieh, B., J. Schultz, M. Guinness, and J. Lacy. 1997. Regulation of the human IgE receptor (Fc epsilonRII/CD23) by Epstein-Barr virus (EBV): Ku autoantigen binds specifically to an EBV-responsive enhancer of CD23. Int. Immunol. 9:1885-1895. [DOI] [PubMed] [Google Scholar]
- 18.Spengler, B. 1997. Post-source decay analysis in matrix-assisted laser desorption/ionization mass spectrometry of biomolecules. J. Mass Spectrom. 32:1019-1036. [Google Scholar]
- 19.Tuteja, R., and N. Tuteja. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35:1-33. [DOI] [PubMed] [Google Scholar]
- 20.University of Washington Yeast Resource Center. 2001. Silver stain protocol for SDS-PAGE gels. [Online]. http://depts.washington.edu/∼yeastrc/ms_silver.htm.