Abstract
Sulfolobus spindle-shaped virus 1 (SSV1) and its fusellovirus homologues can be found in many acidic (pH ≤ 4.0) hot springs (≥70°C) around the world. SSV1 contains a 15.5-kb double-stranded DNA genome that encodes 34 proteins with greater than 50 amino acids. A site-specific integrase and a DnaA-like protein have been previously identified by sequence homology, and three structural proteins have been isolated from purified virus and identified by N-terminal sequencing (VP1, VP2, and VP3). The functions of the remaining 29 proteins are currently unknown. To assign functions to these proteins, we have initiated biochemical and structural studies on the SSV1 proteome. Here we report the structure of SSV1 D-63. The structure reveals a helix-turn-helix motif that dimerizes to form an antiparallel four-helix bundle. Mapping residues conserved among three fusellovirus isolates onto the structure shows that one face of the rod-shaped molecule is highly conserved. This conserved surface spans the dimer axis and thus exhibits 2-fold symmetry. Two smaller conserved patches, also related by 2-fold symmetry, are found on the opposite face of the molecule. All of these conserved surfaces are devoid of clefts or pockets typically used to bind small molecules, suggesting that D-63 may function as an adaptor protein in macromolecular assembly.
Sulfolobus spindle-shaped virus 1 (SSV1) was the first crenarchaeal virus, and the first high-temperature virus, to be described in detail. Originally isolated from Sulfolobus shibatae (6, 12), it has also been shown to be lysogenic in other species of Sulfolobus that thrive in high-temperature (>70°C), acidic hot springs (pH < 4.0). SSV1 is characterized by a 60- by 100-nm lemon-shaped virion with tail fibers emanating from one end (30). Packaged within the virion is a 15.5-kb circular double-stranded DNA genome (30). The complete nucleotide sequence for SSV1 identified 34 open reading frames (ORFs) (17). With the exception of other fusellovirus genomes, only limited similarity is found between the SSV1 ORFs and sequences in the public database.
Functions have been suggested for only 5 of the 34 ORFs. Sequence analysis reveals similarity between D-335 and the integrase proteins belonging to the type I tyrosine recombinase family (1). D-335 has been expressed, purified, and characterized, confirming this activity (15). B-251 shows limited sequence similarity to the nucleotide binding protein DnaA (9), a multifunctional protein that binds to specific sites in bacterial DNA and that promotes local unwinding of double-stranded DNA in an ATP-dependent manner. DnaA in Escherichia coli serves two purposes: it promotes initiation of replication and it provides for transcriptional regulation of specific genes. B-251 might participate in either of these functions. Three structural proteins, VP1, VP2, and VP3, have been isolated from purified virus particles and identified by N-terminal sequencing (18). The functions of the remaining 29 proteins remain unknown.
Identification of SSV1 necessitated the creation of a new viral family, the Fuselloviridae, and five additional viruses are now tentatively assigned to this family (23, 26, 29). The sequencing of three of these genomes, those of SSV2 (23), SSV RH (26), and SSV KM (26), is now complete. Stedman et al. find that the SSV1 and SSV2 genomes have 55% identity at the nucleotide level and that each has 34 putative ORFs, with 27 ORFs common to both genomes (23). Sequence identity at the amino acid level for the products of these 27 ORFs varies from 75.9 to 16.3%. Analysis of the SSV RH and SSV KM genomes by Wiedenheft et al. showed that 18 of the 34 ORFs found in SSV1 are conserved across all four SSV genomes (26). In addition, the overall organization of ORFs within SSV2, SSV RH, and SSV KM is very similar to that found in SSV1, and nucleotide sequences corresponding to many of the putative promoters are also conserved (23, 26).
Relative to the large numbers of eukaryotic viruses and bacterial phages that are known (>5,000), only a handful of archaeal viruses have been discovered (<35). Of these, SSV1 is by far the best studied. In addition to serving as a paradigm for crenarchaeal viruses, studies of SSV1 have been instrumental in the acceptance of Archaea as a separate domain of life (27, 31) and in revealing similarities between transcription in archaeal and eukaryotic organisms (19). Nevertheless, the functions of the SSV1 genome and proteome remain largely unknown, and direct genetic and biochemical examination will be required to elucidate their functions.
Structural analysis of SSV gene products is likely to provide valuable clues to their functions. Towards this goal, we have undertaken structural analysis of the SSV1 proteome. Here we report the initial biochemical characterization and crystal structure of SSV1 D-63.
MATERIALS AND METHODS
Cloning.
The D-63 ORF was amplified by nested PCR from SSV1 genomic DNA prepared as described previously (21, 23, 26, 28). The PCR primers added a Shine-Dalgarno sequence, a C-terminal His6 tag, and attB sites to facilitate ligase-free cloning with the Gateway cloning system (Invitrogen). The internal forward and reverse primers were 5′AAAAAGCAGGCTTCGAAGGAGATAGAACCATGAGTAAAGAAGTCTT3′, and 5′GAAAGCTGGGTCCTAGTGATGGTGATGGTGATGGCTCACCTTAATGAGCT3′, respectively, while the external forward and reverse primers were 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACC3′ and 5′GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGTGATGGTGATGGTGATG3′, respectively. The sequence of the entry clone was verified by sequencing. The His-tagged D-63 construct was then inserted into the E. coli expression vector pDEST14 (Invitrogen), yielding pDEST14/D-63.
Expression and purification.
Typically, pDEST14/D-63 was transformed into E. coli BL21(DE3)-RIL (Novagen), and a single colony was used to inoculate a 5-ml overnight Luria broth culture, with subsequent expansion to 1 liter. Cells were grown to an optical density at 600 nm (OD600) of 0.7, and protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. The cells were harvested by centrifugation and stored frozen at −80°C until needed. Cell pellets were then thawed and resuspended in lysis buffer: 400 mM NaCl, 20 mM Tris-Cl, 0.02% NaN3, and 10 mM imidazole adjusted to pH 8.0 with freshly added phenylmethylsulfonyl fluoride (PMSF) at a final concentration of 0.1 mM. The ratio of lysis buffer volume to cell pellet mass was a minimum of 5 ml/g. The cell suspension was then passed once through a microfluidizer (Microfluidics Corporation, Newton, Mass.), and an additional aliquot of PMSF was added. Cell lysates were spun at 15,000 × g for 20 min. The supernatant was applied to a column containing 3 ml of Ni-nitrilotriacetic acid resin (QIAGEN), washed with lysis buffer, and eluted with approximately 10 ml of 400 mM NaCl-20 mM Tris-Cl-0.02% NaN3-400 mM imidazole adjusted to pH 8.0. Eluted D-63 was then diluted 10-fold with H20 and applied to a gravity column containing 2 ml of Q anion-exchange resin (Amersham). The bound protein was washed with 10 ml of 20 mM Tris-Cl-50 mM NaCl, pH 8.0, and eluted with 10 ml of 20 mM Tris-Cl-500 mM NaCl, pH 8.0. Protein yield was typically 20 to 30 mg/liter of cell culture. Anion-exchange-purified D-63 was concentrated to 3.5 mg/ml with Millipore spin concentrators (5,000-molecular-weight cutoff), as measured by Bradford assay (3) using standardized Bradford reagent (Bio-Rad) and bovine serum albumin as the protein standard. For expression and purification of D-63 incorporating selenomethionine, pDEST14/D-63 was freshly transformed into E. coli strain B834(DE3) (Novagen), a methionine auxotroph. Cell growth was in minimal M9 medium utilizing a glucose carbon source and supplemented with biotin and thiamine at 1 mg/liter. Methionine was added to a final concentration of 50 μg/ml for an overnight culture. The overnight culture was added to 1 liter of an identical medium except that selenomethionine was substituted for methionine. After growth to OD600 of 0.7, protein expression was induced, and the protein was purified as described for native protein (above). Selenium incorporation was monitored by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS). Molecular weight and purity were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (16.5% acrylamide, Tris-Tricine buffer system) and MALDI-TOF MS. MALDI-TOF MS was performed with a Bruker Biflex III in the matrix α-cyano-4-hydroxycinnamic acid. Size exclusion chromatography was performed on calibrated Superdex S-75 columns; buffer composition was 200 mM NaCl-50 mM ammonium acetate, pH 5.0, the same pH used for crystal growth.
Crystallization and data collection.
D-63 was crystallized by hanging-drop vapor diffusion. Drops were assembled with 2 μl of D-63 mixed with 2 μl of well solution: 0.2 M (NH4)2SO4, 0.2 M ammonium acetate, and 25% polyethylene glycol 4000, pH 4.7 to 5.0. Plates were incubated at 18°C, and crystals appeared in 4 to 6 weeks. Single crystals were placed in dialysis buttons and moved at 2-h intervals through a series of well solutions supplemented with increasing glycerol concentration in steps of 5% to a final concentration of 25% glycerol. Crystals were then plunge-frozen in liquid nitrogen. A three-wavelength multiwavelength anomalous diffraction data set to 2.9 Å was collected at the selenium K edge (edge, peak, and remote wavelengths) at BioCARS beamline 14-BM-D at the Advanced Photon Source (Tables 1 and 2). Data were integrated and reduced in space group P62 with the HKL software package (16).
TABLE 1.
Data collectiona
| Wavelength (Å) | Resolution rangec | No. of observations | Unique reflections possible/observed | Redundancy | Average I/sigma I | Rsymd | |
|---|---|---|---|---|---|---|---|
| Inflection | 0.9790 | 20-2.60 (2.69-2.60) | 78,562 | 19,885/19,307 | 3.95 | 34.0 | 3.8 (23.8) |
| Peak | 0.9787 | 20-2.60 (2.69-2.60) | 72,103 | 19,083/18,431 | 3.64 | 30.4 | 4.1 (25.7) |
| Remote | 0.9562 | 20-2.60 (2.69-2.60) | 86,028 | 19,907/19,613 | 4.32 | 40.4 | 3.4 (19.3) |
Space group P62. Unit cell parameters: a = 110.65 Å, b = 110.65 Å, c = 47.45 Å, α = β = γ = 90.0°.
SeMet MAD, selenomethionine multiwavelength anomalous diffraction.
Numbers in parentheses refer to the highest-resolution shell.
Rsym = 100·ΣhΣi|Ii (h) − <I (h)>|/ΣhI (h), where Ii (h) is the ith measurement of the h reflection and <I (h)> is the average value of the reflection intensity.
TABLE 2.
Refinement statistics
| Parameter | Value |
|---|---|
| Resolution range (Å) | 15-2.6 |
| Rcryst (%) / Rfree (%)a | 0.225/0.270 |
| No. of protein atoms/no. of water molecules | 2,082/10 |
| Coordinate error (Å)b | 0.212 |
| Real space CCc | 0.920 |
| RMSDe from ideality for bonds (Å)/angles (°) | 0.016/1.534 |
| Ramachandran plotd of most favored, additional, generously allowed (%) | 97.3/2.7/0 |
| Average B value (Å2) | 54.02 |
Rcryst = Σ||Fo|−Fc||/ΣFo| where Fo and Fc are the structure factor amplitudes from the data and the model, respectively. Rfree is Rcryst with 5% of test set structure factors.
Based on maximum liklihood.
Correlation coefficient (CC) is between the model and 2mFo − DFc density map.
Calculated with PROCHECK (10).
RMSD, root mean square difference.
Structure determination and refinement.
SOLVE (24) was used to locate six of eight expected selenomethionine sites corresponding to four monomers per asymmetric unit; the two additional selenomethionine residues were disordered. The initial phases were improved with density modification, including fourfold NCS averaging over residues 8 to 28 and 40 to 61 (2, 5). The resulting electron density map was of excellent quality and was used to build the initial model with the program O (8). Iterative rounds of refinement with Refmac5 (2, 14) and manual rebuilding with O yielded a final model with an R factor of 22.5% (Rfree = 27.0%). The model has good stereochemistry, with no residues in the disallowed regions of the Ramachandran plot (10). Structural comparisons were performed with the DALI server (http://www.ebi.ac.uk/dali) (7). Figures were generated with PYMOL (http://www.pymol.org). Atomic coordinates and structure factors have been deposited into the Protein Data Bank under accession code 1SKV.
RESULTS AND DISCUSSION
The SSV1 D-63 construct used in this work codes for the 63 amino acids of native D-63, plus an additional C-terminal His6 tag, for a total of 69 residues. MALDI-TOF indicates that the N-terminal methionine has been cleaved. Purified D-63 migrates at a calculated molecular mass of 17,100 Da (results not shown) on a calibrated Superdex S-75 size exclusion column, suggesting that D-63 assembles to form a homodimer at pH 5.0, the same pH at which crystals are grown. D-63 crystallizes in space group P62, and the crystals diffract to 2.6-Å resolution. Details of crystal growth, data collection, phasing, and refinement are summarized in Tables 1 and 2 and detailed in Materials and Methods. Four copies of the D-63 polypeptide, designated chains A through D, are present in the asymmetric unit, providing the opportunity to observe multiple conformations of D-63. Each of the four identical chains folds into a helix-turn-helix motif with the two helices running antiparallel to each other. The connecting loop is composed of residues 35 through 39.
The amino- and carboxy-terminal ends of the structure show various amounts of well defined electron density, and within chain D, density at the C-terminal end of the first helix and the connecting loop is also poorly ordered. Well-ordered density is seen for chain A, residues 2 through 65; chain B, residues 6 through 66; chain C, residues 7 through 64; and chain D, residues 2 through 29 and 39 through 66 (residue numbers greater than 63 correspond to the C-terminal His6 tag).
Structural comparison of the four identical chains shows that the conformations are generally similar, though there are significant conformational differences for the C terminus of the first helix and the residues comprising the loop (not shown). The disorder present through this region in chain D further highlights the conformational flexibility within this part of the structure.
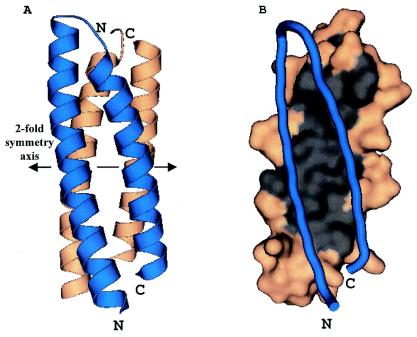
The A and B monomers are related to each other by a 2-fold symmetry axis such that the chains pack against each other to form an antiparallel four-helix bundle (Fig. 1A). An identical relationship is seen for chains C and D. In each case, the 2-fold axes run perpendicular to the long axis of the four-helix bundle. Thus, as one looks down the dimer axis, the four-helix bundle presents one face, composed of the two N-terminal helices (N-terminal face), or the opposite face, composed of two C-terminal helices (C-terminal face). There is extensive contact between the monomers at the dimer interface. The calculated surface area for the D-63 monomer is 5,346 Å2 (2, 11). Formation of the AB dimer buries 1,300 Å2, or 24%, of the solvent-accessible surface area per monomer. The dimer interface is largely hydrophobic and is composed of residues Phe11, Leu14, Val18, Leu21, Ile25, Ile28, Leu42, Ala45, Val49, Ile52, Leu56, and Leu59 (Fig. 1B). The presence of the hydrophobic core is discernible in the primary sequence as a clear heptad repeat (Fig. 2A). The dimer is also stabilized by intermolecular salt bridges and hydrogen bonds. These include HisA26 to AspB15, ArgA32 to GluB7, TyrA46 to GluB53, GluA53 to GlnB50, SerA63 to ArgB32, and their five symmetry-related equivalents between chain B and chain A. Thus the packing of D-63 within the crystal strongly suggests that D-63 is a homodimer, a finding that is consistent with the behavior of the protein on size exclusion columns.
FIG. 1.
(A) The D-63 homodimer forms an antiparallel four-helix bundle. Monomers A and B are each shown as a ribbon diagram depicting secondary structure elements. Blue, monomer A; amber, monomer B. The 2-fold symmetry axis (double-headed arrow) runs horizontally between the two monomers relating the N and C termini of the blue monomer at the bottom to the N and C termini of the amber monomer at the top. (B) The dimer interface is hydrophobic. Monomer A is depicted as a blue worm, while monomer B is shown in amber and gray. All surface residues are amber, with the exception of residues Phe11, Leu14, Val18, Leu21, Ile25, Ile28, Leu42, Ala45, Val49, Ile52, Leu56, and Leu59. These residues are gray and illustrate the extent of the hydrophobic surface found at the dimer interface.
FIG. 2.
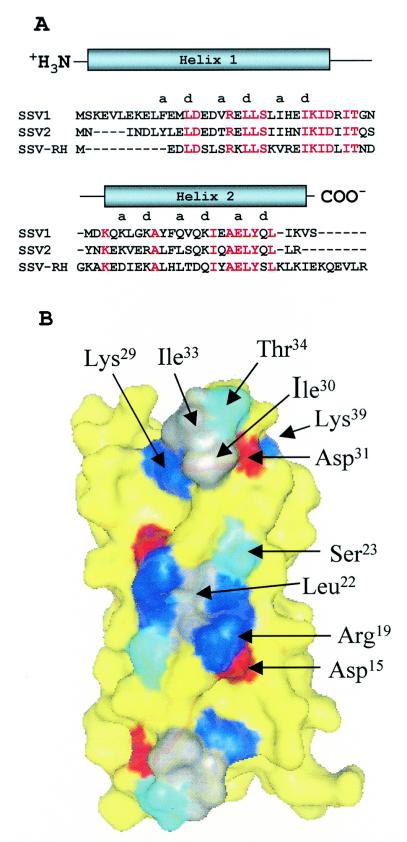
(A) Sequence alignments of D-63 fusellovirus homologues. Strictly conserved residues are red, and nonconserved residues are black. Secondary structure assignments, i.e., the extent of the alpha helices, are indicated above the sequence. The hydrophobic residues occur within the alpha helices in a clear heptad repeat, (ABCDEFG)n, with hydrophobic residues at positions A and D. SSV2 was isolated from Sulfolobus solfataricus strain REY15/4 from Iceland (23). SSV RH was isolated from Yellowstone National Park (26). (B) Conserved surface residues among D-63 homologues. Conserved basic, acidic, polar, and nonpolar residues are blue, red, cyan, and gray, respectively. Nonconserved residues are yellow. Strong conservation of SSV1, SSV2, and SSV RH surface residues is seen for the N-terminal face of the four-helix bundle. The twofold symmetry axis runs perpendicular to this face through the center of the four-helix bundle. Thus the conserved surface exhibits 2-fold symmetry. Conserved residues contributing to this surface are labeled for one of the two monomers. The second monomer is related to the first by the 2-fold symmetry axis. This putative ligand binding surface is further enlarged when conservative substitutions are also mapped to the surface (not shown). The surface is a mixture of both polar and nonpolar residues.
Meaningful sequence similarity between D-63 and nonfusselovirus proteins has not been found. However, D-63 can be aligned with its fusselovirus homologues from SSV2 (D-57) and SSV RH (F-61), though a D-63 homologue is not found in SSV KM. Alignment of the D-63 homologues with ClustalW (25) shows 51% sequence identity between the SSV1 and SSV2 proteins and 37% sequence identity between the SSV1 and SSV RH proteins (Fig. 2A). The three-way alignment shows that ∼32% of the residues, 20 of 63, are strictly conserved. We also note additional similarity between the N-terminal end of SSV1 and the C-terminal end of SSV RH. The N terminus of SSV1 contains three glutamate residues, two basic residues (both Lys), and a Leu-Val pair; while the C terminus of SSV RH contains two glutamate residues (plus one glutamine), two basic residues (Lys and Arg), and a Val-Leu pair. While these residues are at opposite ends of the linear sequence, the hairpin fold of the helix-turn-helix motif places these residues at similar positions within the three-dimensional structure of D-63.
Biological implications.
It is well documented that tertiary (three-dimensional structural) similarities persist far longer on the evolutionary time scale than either primary (amino acid) or genomic sequence (DNA) similarities (4, 13, 20). Thus, prior to structure determination, we hypothesized that the lack of significant sequence similarity between D-63 and other proteins in the public database did not indicate a unique fold for this protein. Rather, we expected that the structure of D-63 would reveal a familiar fold and hoped that this fold would suggest a function for D-63. However, the D-63 helix-turn-helix motif is present in many larger proteins as a supersecondary structural element, while the four-helix bundle of dimeric D-63 is an extremely common fold that has been adapted to a multitude of functions. Thus, a search of the Protein Data Bank for nearest structural neighbors using the DALI structural biology server (7) returns a long list of proteins with statistically significant structural similarities and a long list of functions that might be considered for D-63. However, as each protein on the list is examined in detail, the match is found to be problematic in one way or another. As examples, consider the two best matches to the helix-turn helix motif. The greatest similarity (highest Z score) is to a similar supersecondary structural element present at the dimer interface of valyl-tRNA synthetase, where, in addition to dimerization, the element also plays a critical role in the binding of tRNA to the synthetase. But a role for D-63 in RNA binding is easily discounted; its calculated pI is 5.07. The second best match is to a helix-turn-helix present as the “finger” of GreA (22), a bacterial transcription factor that suppresses elongation arrest. However, crenarchaeal RNA polymerases are more eukaryotic-like than prokaryotic, and one might expect the associated transcription factors to be eukaryotic in nature as well. In these cases, and for all the others on the list of similar structures, it is difficult to rationalize the suggested function based on the constraints of the D-63 structure, knowledge of the SSV1 life cycle, and crenarchaeal biochemistry. Thus, even though there are now numerous examples of structure suggesting function (13), it is clear that, for D-63, the commonality of the helix-turn-helix motif precludes any reliable assignment.
In sharp contrast to the search for function by structural homology, analysis of the surface properties of D-63 is quite enlightening. Generally speaking, residues on the surface of a protein are less likely to be conserved than residues buried within the core of the protein. However, exceptions to this rule occur when surface residues are required for the activity of the protein (13). In such cases, conservation of these residues suggests the location of ligand binding sites, or, for an enzyme, the active site. Sequence alignments of D-63 with its homologues from SSV2 (D-57) and SSV RH (F61) are shown in Fig. 2A. When the strictly conserved residues are mapped to the surface of D-63, a large patch of conserved surface area is found on the N-terminal face of the four-helix bundle (Fig. 2B). The strictly conserved residues comprising this surface (Asp15, Arg19, Leu22, Ser23, His26, Glu27, Lys29, Ile30, Asp31, Ile33, and Thr34) are found primarily within the first (N-terminal) helix. Because the dimer axis passes through the N-terminal face of the four-helix bundle, this conserved surface patch is necessarily 2-fold symmetric. It is also devoid of obvious clefts or pockets that might accommodate a small molecule, suggesting that the conserved surface features are responsible for binding a macromolecule. Smaller patches of strictly conserved surface are seen on the opposite face (C-terminal face; not shown), with significant sequence similarity for the surrounding surface-exposed residues. These areas might also represent conserved ligand binding sites that, like the N-terminal face, are also 2-fold symmetric.
It is possible that the D-63 dimer might dissociate to monomers and that the monomer might bind to the target macromolecules. However, if D-63 is active as the dimeric four-helix bundle, the conserved surface features would serve to recognize macromolecules possessing 2-fold symmetry or would function to bind two copies of a monomeric entity, effectively serving to dimerize the molecule (D-63-induced dimerization). In either event, the conserved D-63 surface features strongly suggest that D-63 functions in macromolecular assembly and that the binding of an unknown ligand(s) by D-63 serves to regulate or modify the behavior of the ligand, thus assisting in maintenance of the viral life cycle. From this perspective, experiments aimed at identification of potential binding partners are indicated. Possible approaches include two-hybrid screens and pull-down assays using coimmunoprecipitation or immobilization of D-63 via the C-terminal His6 tag.
From the structural analysis presented here, a preliminary understanding of the function of D-63 has emerged. These results will guide future biochemical and genetic studies, eventually leading to a more detailed understanding of D-63. As this occurs, the structure will only grow in relevance and will provide additional insight into the means through which D-63 exerts its action. Thus it is clear that the structural analysis of D-63 has been a worthwhile undertaking. Further, it demonstrates the utility of structure determination for proteins of unknown function.
Acknowledgments
We thank the BioCARS staff for their excellent assistance during data collection at sector 14 of the Advanced Photon Source. We are grateful to Philippe Benas, Ryan Todorovich, Cinnimon Spear, and John Gilmore for their able technical assistance.
This work was supported by grants from the National Science Foundation (MCB-0236344) and the National Aeronautics and Space Administration (NAG5-8807). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR07707.
REFERENCES
- 1.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. Narayana, L. S. Pierson III, et al. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, S. 1994. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Buehner, M., G. C. Ford, D. Moras, K. W. Olsen, and M. G. Rossman. 1973. D-glyceraldehyde-3-phosphate dehydrogenase: three-dimensional structure and evolutionary significance. Proc. Natl. Acad. Sci. USA 70:3052-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowtan, K. 1994. DM: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31:34-38. [Google Scholar]
- 6.Grogan, D., P. Palm, and W. Zillig. 1990. Isolate B12, which harbours a virus-like element, represents a new species of the archaebacterial genus Sulfolobus, Sulfolobus shibatae, sp. nov. Arch. Microbiol. 154:594-599. [DOI] [PubMed] [Google Scholar]
- 7.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 8.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A Found. Crystallogr. 47:110-119. [DOI] [PubMed] [Google Scholar]
- 9.Koonin, E. V. 1992. Archaebacterial virus SSV1 encodes a putative DnaA-like protein. Nucleic Acids Res. 20:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 11.Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55:379-400. [DOI] [PubMed] [Google Scholar]
- 12.Martin, A., S. Yeats, D. Janekovic, W.-D. Reiter, W. Aicher, and W. Zillig. 1984. SAV-1, a temperate UV inducible DNA virus like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 3:2165-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moult, J., and E. Melamud. 2000. From fold to function. Curr. Opin. Struct. Biol. 10:384-389. [DOI] [PubMed] [Google Scholar]
- 14.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 15.Muskhelishvili, G., P. Palm, and W. Zillig. 1993. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol. Gen. Genet. 237:334-342. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276(Part A):307-326. [DOI] [PubMed] [Google Scholar]
- 17.Palm, P., C. Schleper, B. Grampp, S. Yeats, P. McWilliam, W. D. Reiter, and W. Zillig. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242-250. [DOI] [PubMed] [Google Scholar]
- 18.Reiter, W. D., P. Palm, A. Henschen, F. Lottspeich, W. Zillig, and B. Grampp. 1987. Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol. Gen. Genet. 206:144-153. [Google Scholar]
- 19.Reiter, W. D., P. Palm, and W. Zillig. 1988. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res. 16:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossman, M. G. 1981. Evolution of glycolytic enzymes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 293:191-203. [DOI] [PubMed] [Google Scholar]
- 21.Schleper, C., K. Kubo, and W. Zillig. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. USA 89:7645-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stebbins, C. E., S. Borukhov, M. Orlova, A. Polyakov, A. Goldfarb, and S. A. Darst. 1995. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature 373:636-640. [DOI] [PubMed] [Google Scholar]
- 23.Stedman, K. M., Q. She, H. Phan, H. P. Arnold, I. Holz, R. A. Garrett, and W. Zillig. 2003. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 154:295-302. [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiedenheft, B., K. Stedman, F. Roberto, D. Willits, A.-K. Gleske, L. Zoeller, J. Snyder, T. Douglas, and M. J. Young. 2004. Comparative genomic analysis of hyperthermophilic archaeal Fusselloviridae viruses. J. Virol. 78:1954-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeats, S., P. McWilliam, and W. Zillig. 1982. A plasmid in the archaebacterium Sulfolobus solfataricus. EMBO J. 1:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zillig, W., H. P. Arnold, I. Holz, D. Prangishvili, A. Schweier, K. Stedman, Q. She, H. Phan, R. Garrett, and J. K. Kristjansson. 1998. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2:131-140. [DOI] [PubMed] [Google Scholar]
- 30.Zillig, W., D. Prangishvilli, C. Schleper, M. Elferink, I. Holz, S. Albers, D. Janekovic, and D. Gotz. 1996. Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol. Rev. 18:225-236. [DOI] [PubMed] [Google Scholar]
- 31.Zillig, W., R. Schnabel, and K. O. Stetter. 1985. Archaebacteria and the origin of the eukaryotic cytoplasm. Curr. Top. Microbiol. Immunol. 114:1-18. [DOI] [PubMed] [Google Scholar]