Abstract
Synthesis of the relaxed-circular (RC) DNA genomes of hepadnaviruses by reverse transcriptase involves two template switches during plus-strand DNA synthesis. These template switches require repeat sequences (so-called donor and acceptor sites) between which a complementary strand of nucleic acid is transferred. To determine cis-acting elements apart from the donor and acceptor sites that are required for plus-strand RC DNA synthesis by hepatitis B virus (HBV), a series of mutants bearing a small deletion were made and analyzed for their impact on the viral genome synthesis. We found three novel cis-acting elements in the HBV genome: one element, located in the middle of the minus strand, is indispensable, whereas the other two elements, located near either end of the minus strand, contribute modestly to the plus-strand RC DNA synthesis. The data indicated that the first element facilitates plus-strand RNA primer translocation or subsequent elongation during plus-strand RC DNA synthesis, while the last two elements, although distantly located on the minus strand, act at multiple steps to promote plus-strand RC DNA synthesis. The necessity of multiple cis-acting elements on the minus-strand template reflects the complex nature of hepadnavirus reverse transcription.
Hepadnaviruses are small (ca. 3.2-kb), double-stranded DNA viruses that replicate through an RNA intermediate via reverse transcription. Prototypical members of the family include human hepatitis B virus (HBV), woodchuck hepatitis virus, and duck hepatitis B virus (DHBV) (4). Retroid elements, including retroviruses and hepadnaviruses, replicate by converting their single-stranded RNA templates into double-stranded DNAs via reverse transcription. During reverse transcription, a process described as strand transfer or template switching is required for the successful synthesis of a double-stranded DNA genome (5). Template switching, in which the DNA strand undergoing synthesis switches from one template to another, is mediated by complementarity between a donor and an acceptor site. For retroviruses, two template switches are required to generate a linear double-stranded DNA: one for minus-strand DNA synthesis and another for plus-strand DNA synthesis. In hepadnaviruses, in addition to these two template switches, a third template switch, termed circularization, is required to generate a relaxed-circular (RC) DNA genome (4).
Reverse transcription of hepadnaviruses takes place within the viral capsid in the cytoplasm of infected cells (Fig. 1). The first template switch required for the synthesis of the RC DNA genome occurs shortly after the initiation of minus-strand DNA synthesis. Recognition of a stem-loop structure (an encapsidation signal designated ɛ near the 5′ end of the pregenomic RNA [pgRNA]) by P (polymerase) protein directs encapsidation of the pgRNA and P protein into a nascent capsid particle (8, 10, 11). Minus-strand DNA synthesis is initiated by protein priming with P protein as a primer and the bulge region of ɛ as a template (26, 27). Following the synthesis of four nucleotides, the nascent minus-strand DNA switches templates to a position near the 3′ end of the pgRNA, i.e., the 3′ copy direct repeat 1 (DR1*) (Fig. 1B). Minus-strand DNA synthesis resumes with the RNase H activity of the P protein degrading the pgRNA and proceeds to the 5′ end of pgRNA, resulting in a full-length minus-strand DNA. A short segment of RNA, the remnant of the pgRNA cleavage by the RNase H activity, serves as a primer for plus-strand DNA synthesis (13, 17). The second template switch occurs before this RNA remnant serves as a primer (Fig. 1C).
FIG. 1.
Model for hepadnavirus reverse transcription. (A) Initiation of minus-strand DNA synthesis. The thin line represents the pgRNA. The direct repeats, DR1 and DR2, are indicated by boxes. Minus-strand DNA synthesis is templated by the UUCA sequence within the bulge region of the 5′ ɛ. The oval circle represents the P protein. (B) Minus-strand template switch. The 4-nt-linked viral P protein translocates to an acceptor site, the UUCA sequence, overlapping the 3′ copy of DR1 via 4-bp homology. (C) Elongation and completion of minus-strand DNA synthesis. Following the minus-strand transfer, minus-strand synthesis resumes, with concomitant degradation of the pgRNA by RNase H activity encoded by the P protein. (D) In situ priming from DR1. Some of the plus-strand primers do not translocate but are used to initiate plus-strand synthesis from DR1 to generate a DL DNA. (E) Generation of a DL genome. (F) Plus-strand primer translocation to DR2. The RNA primer contains a DR1 sequence complementary to DR2. This complementarity is required for subsequent translocation to DR2. To generate the circular duplex genome, the RNA primer translocates from DR1 to an acceptor site, DR2. (G) Initiation of plus-strand DNA synthesis following translocation. After translocation to DR2, plus-strand DNA synthesis is initiated at DR2. (H) Template switch to circularize the viral genome. The growing point of plus-strand DNA synthesis switches templates from the 5′ end to the 3′ end of minus-strand DNA. (I) Generation of an RC DNA genome.
Depending on whether the second template switch takes place during plus-strand DNA synthesis, one of two distinct double-stranded DNA products, RC DNA (Fig. 1I) or duplex-linear (DL) DNA (Fig. 1E), is generated. The RC DNA, a mature product of viral DNA synthesis found in virions, is generated when the plus-strand primer translocates to an acceptor site, DR2, near the 5′ end of the minus-strand DNA template (Fig. 1F). Following translocation, the plus-strand DNA synthesis initiated from DR2 proceeds to the 5′ end of the minus-strand template (25) (Fig. 1G). For continuation of plus-strand DNA synthesis, an intramolecular template switch must occur (Fig. 1H). A short terminal redundancy on the minus strand, called r, was shown to facilitate this third template switch (16). The third template switch, termed circularization, results in the generation of RC DNA (Fig. 1I). On the other hand, the second form of double-stranded DNA is generated when the plus-strand primers do not translocate and instead initiate from DR1 (Fig. 1D). This type of synthesis, called in situ priming, yields a DL species (Fig. 1E).
Although the donor and acceptor sites used for template switches during hepadnavirus DNA synthesis have been identified (1, 7, 8, 11, 16, 22), it was not determined whether other cis-acting sequences might be involved in this process. Recently, such cis-acting elements (named 3E, M, and 5E), which, when deleted, abolish the RC DNA synthesis, were identified in the DHBV genome (7). Here, we report the identification of three novel cis-acting elements of the HBV genome, termed α, γ, and δ, that are essential for RC DNA synthesis.
MATERIALS AND METHODS
Cell culture and transfection.
Huh7 and HepG2.2.15 cells (23) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 10 μg of gentamicin per ml at 37°C in 5% CO2 and were split every third day. Transfection was performed as described previously (10).
HBV expression constructs.
The nucleotide sequences of the HBV genome were numbered starting at the unique EcoRI site of the HBV ayw subtype (GenBank accession number V01460), according to the method of Galibert et al. (3); nucleotide position 1 of HBV subtype ayw is the A of the EcoRI site (GAATTC). Nucleotide numbers indicate the HBV sequence number. The HBV expression construct (pCMV-HBV/30), referred to here as the wild type, is capable of supporting viral replication upon transfection (10). The P-null version of the wild type was generated by changing the AUG initiation codon of the P open reading frame (ORF) to ACG, leading to a silent mutation to the overlapping core ORF, by using overlap extension PCR (12). A helper plasmid, R063 (pCMV-CP), was made that lacks the 5′ ɛ encapsidation signal but expresses the viral C and P proteins. Standard procedures were used to create the deletion mutants, and details of the construction of PCR primer sequence will be provided upon request. All of the molecular clones were verified by DNA sequencing or restriction enzyme digestion analysis.
Southern blot analysis.
Viral DNAs from cytoplasmic capsids were isolated from Huh7 cells 3 days after transfection as described previously (21). Viral DNAs extracted from the cytoplasmic capsids of transfected Huh7 cells were analyzed by Southern blot analysis to measure the viral replication intermediates. The viral DNA was electrophoresed through a 1.3% agarose gel at 50 V for 2 h in 0.5× Tris-acetate-EDTA buffer. DNA was transferred to a nylon membrane (Hybond-XL; Amersham) in 20× SSC (3.0 M NaCl, 0.3 M sodium acetate [pH 7.0]). The membrane was dried and exposed to 250 mJ of 254-nm-wavelength UV light per cm2. The membrane was prehybridized and hybridized with a 32P-labeled full-length HBV DNA probe in hybridization solution (1% bovine serum albumin, 1 mM EDTA, 0.2 M sodium phosphate [pH 7.2], 7% sodium dodecyl sulfate, 50 mg of sheared salmon sperm DNA per ml) for 12 h at 65°C. Then, the membrane was washed with small volume of 2× SSC containing 0.1% sodium dodecyl sulfate for 4 h at 65°C. After being dried, the membrane was autoradiographed with film (Hyperfilm-MP; Amersham) at −70°C. Phosphorimages of the different forms of viral DNA were quantified with a Bio-imaging analyzer (BAS-2500; Fujifilm).
Primer extension analysis.
Primer extension analysis was performed with viral DNA extracted from cytoplasmic core particles of transfected Huh7 cells. The end-labeled oligonucleotides used in these reactions were M primer (derived from nucleotide [nt] 1764 to1783), B primer (complementary to nt 1661 to 1683), and P primer (complementary to nt 1878 to 1899). Briefly, primer extension with the M primer was used to determine the 5′ ends of minus-strand DNAs from various deletion mutants and to measure the level of minus-strand DNA initiated at DR1* (18). The B primer was used to determine the amount of plus-strand DNA initiated at DR2 before the circularization step, and the P primer was used to determine the amount of 5′ termini of plus-strand DNA located at DR1 and DR2. The plus-strand DNA that was detected at DR2 with the P primer has circularized and synthesized at least 298 nt of plus-strand DNA. Primer extension reaction mixtures contained 1× ThermoPol reaction buffer [20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100], 0.2 mM deoxynucleoside triphosphates, 1 U of Vent (Exo−) DNA polymerase (New England Biolabs), 0.6 pmol of 5′-end-labeled oligonucleotide, and approximately 300 pg of viral DNA. The reaction mixtures were incubated in a thermocycler (Perkin-Elmer GeneAmp PCR system 2400). The thermocycling parameters used for the extension with M primer were 20 cycles of 95°C for 40 s, 46°C for 40 s, and 72°C for 40 s. The annealing temperatures for the P and B primers were 52 and 56°C, respectively. To prepare an internal standard for the analysis with the M primer, approximately 1 ng of the wild-type construct digested with AatII (nt 1419) and BglII (nt 1986) was added to reaction mixtures. Samples were denatured at 95°C for 5 min prior to electrophoresis through a 6% polyacrylamide-8 M urea sequencing gel. The gels were then dried and autoradiographed to Amersham Hyperfilm-MP at −70°C. Phosphorimages of the extended DNA were quantified using by the Bio-imaging analyzer (BAS-2500; Fujifilm).
RESULTS
Identification of three cis-acting elements essential for RC DNA synthesis.
The aim of this study was to identify cis-acting elements that are required for HBV genomic DNA synthesis and are distinct from previously characterized elements such as 5′ ɛ, DR1, DR2, and r (1, 7, 8, 11, 16, 22). To this end, a series of deletion mutants that spanned most of the HBV genome between 5′ ɛ and DR2 were constructed (Fig. 2). First, we made a wild-type HBV expression construct, pCMV-HBV/30 (referred to here as the wild type), that is competent to support viral DNA synthesis (10). Subsequently, a series of deletions made separately in various subcloning plasmids were transferred to the wild-type construct. To determine whether a sequence deleted in each mutant is essential for HBV genome replication, each mutant was transfected into Huh7 cells, along with pR063, a helper plasmid that provided C and P proteins in trans. Viral replication intermediates were extracted from core particles and analyzed by Southern blot analysis (Fig. 3A). Analysis of DNA isolated from cytoplasmic core particles following transfection of a wild-type HBV construct revealed three major replication intermediates, i.e., single-stranded DNA, DL DNA, and RC DNA, as expected (Fig. 3A, lanes 1 and 10). These replication intermediates were synthesized in characteristic proportions: in a typical experiment, DL DNA was the predominant species, whereas RC and single-stranded DNAs were less prominent. It was noted that, unlike the case for DHBV, less RC DNA was detected in nucleocapsids (typically ca. 25%). Here we show that three novel cis-acting elements, termed the α, γ, and δ elements, contribute to plus-strand DNA synthesis.
FIG. 2.
Maps of deletion mutants used in this study. (A) Map of the pgRNA of HBV. The structure of pgRNA is shown with cis-acting elements that are known to be essential for viral genome synthesis; R represents the region of terminal redundancy that includes DR1 and ɛ. The direct repeats, DR1 and DR2, are represented as boxes; the 3′ copy of DR1 is designated DR1*. The encapsidation signal, ɛ, is indicated by the stem-loop structure at the 5′ end of the pgRNA. ORFs for C (core) and P (polymerase) are indicated by boxes. (B) A series of deletion mutants that encompass the genome from 5′ ɛ to DR2 were made. The size of the deletion in each mutant (in nucleotides) is indicated in parentheses after the plasmid name. The nucleotide positions of each deletion mutant are indicated by a line along with nucleotide number, inclusive. Most of the deletion mutants used in this study are defective in the expression of functional C or P protein, which are instead provided in trans by a helper plasmid. None of these deletion mutants exhibited a dominant negative effect on viral genome replication (data not shown).
FIG. 3.
Southern analysis reveals three novel cis-acting elements in the viral genome. (A) Southern analysis of the replication intermediate DNAs isolated from cytoplasmic core particles from cells transfected by each deletion mutant along with a helper plasmid providing C and P proteins. The positions of RC, DL, and single-stranded (SS) DNA forms are indicated. The viral DNAs extracted from HepG2.2.15 cells (an HBV-producing cell line) served as size markers for the three replication intermediates (23). WT, wild type. (B) Relative amount of RC DNA with respect to RC plus DL DNA. The amounts of RC DNA and DL DNA for each mutant as determined from at least six independent transfections were quantified with a Bio-imaging analyzer. The mean ratio and standard deviation for each mutant are shown.
To measure the effect of deletions on the synthesis of RC DNA, the relative ratios of the amount of RC DNA to the amount of RC plus DL DNA were estimated from six independent transfection experiments (Fig. 3B). Overall, reductions of the ratio were found in mutants with deletions in three distinct regions of the viral genome. First, far less RC DNA was detected in the R022 mutant (Fig. 3A, lane 6). To further delineate this region, two additional smaller deletion mutants, R045 and R044, were made (Fig. 2B). Southern blot analysis of core particles from cells transfected by these two smaller deletion mutants indicated that RC DNA was not detected in the R045 mutant but was readily detectable in the R044 mutant (Fig. 3A, lanes 7 and 8). Nonetheless, the relative ratio of RC DNA to RC plus DL DNA produced by the R044 mutant was comparable to that of the wild type (Fig. 3B). Thus, a sequence deleted in R045 but not in R044 was identified as a novel cis-acting element, termed α, that is essential for RC DNA synthesis. Further delineation of the region deleted in the R022 mutant corroborated this conclusion (see Fig. 7). In addition, reduced amounts of RC DNA were detected in the R060, R048, R027, and R053 mutants relative to the wild type (Fig. 3A, lanes 2, 3, 15 and 16). Further characterization of these mutants allowed us to identify two additional cis-acting elements, termed γ and δ (see below).
FIG. 7.
Determination of the boundary of the α region by deletion analysis. (A) Map of the five small deletion mutants within the α region. The nucleotide sequences deleted in each mutant are shown by solid line along with the nucleotide number, inclusive. The size of the deletion in each mutant (in nucleotides) is indicated in parentheses after the plasmid name. (B) Southern blot analysis was performed as described for Fig. 3A. Each mutant generated in the background of a P-null version of the wild type was transfected into cells along with the helper. WT, wild type; SS, single-stranded DNA; S/M, size marker.
HBV mutants with a deletion of the α element are defective in primer translocation to DR2 during plus-strand DNA synthesis.
In mutants lacking the α element, RC DNA was not detectable. Because mutants lacking the α element failed to accumulate RC DNA, we sought to determine the step at which these mutants were defective during plus-strand DNA synthesis (Fig. 1C to I). Lack of RC DNA, the plus-strand DNA initiated at DR2, could be due to failure of the plus-strand primer translocation to DR2. If this is the case, the reduction in the accumulation of RC DNA molecules could be associated with a proportional increase in the number of in situ-primed DNA molecules. To examine this possibility, we performed primer extension analysis with the P primer. Strategies for the primer extension analysis are depicted in Fig. 4; the P primer was designed to identify the 5′ end of the plus-strand DNA. The extension product of DL and RC DNA would map to DR1 or DR2, respectively (Fig. 4B and D). As shown in Fig. 5A, primer extension analysis with the P primer indicated that the plus-strand DNAs initiated at DR2 and DR1 were detected in the wild type, as anticipated (Fig. 5A, lane 1). By contrast, no detectable amount of 5′ termini at DR2 was found in the α element mutants R022 and R045 (Fig. 5A, lanes 4 and 5). Importantly, the amount of plus-strand DNA initiated at DR1 increased in these mutants relative to the wild type (Fig. 5A, lanes 4 and 5). It is possible that an increase in the amount of in situ priming would result in the accumulation of DL DNA at the expense of RC DNA. The data are consistent with the interpretation that the reduction in the relative number of RC DNA molecules was accompanied by a proportional increase in the number of in situ-primed DNA molecules.
FIG. 4.
Strategies used to measure replication intermediates by primer extension analysis. Only part of the plus-strand DNA of the RC genome is shown on the top for clarity; DR1, DR2, and r are boxed or underlined. The positions of the annealing sites for the primers are indicated by arrows, with the nucleotide numbers in parentheses. Four replication intermediates found in cytoplasmic capsids are shown. Each replication intermediate has a full-length minus-strand DNA (thick line) with the P protein (oval) linked to the 5′ end. (A) Single-stranded DNA. The M primer is designed to map the 5′ end of the minus-strand DNA. (B) DL DNA. The DL DNA is generated when the RNA primer initiates synthesis at its site of generation (so-called in situ priming). The P primer is designed to map the 5′ end of the plus-strand DNA. (C) RC DNA before circularization. The RNA primer for the plus-strand synthesis anneals to the DR2 site on the minus strand. Primer extension with the B primer will yield a signal at DR2, while that with the P primer will not. (D) RC DNA after circularization. The B primer is designed to map the 5′ end of the plus-strand DNA before circularization, whereas the P primer is designed to map the 5′ end of the plus-strand DNA after circularization.
FIG. 5.
Primer extension analysis reveals that three regions (the α, γ, and δ elements) in the minus-strand DNA facilitate primer translocation to DR2 during plus-strand DNA synthesis. (A) Primer extension analysis to determine the levels and positions of the 5′ termini of plus-strand DNA for the deletion mutants indicated. The P primer was used to determine the levels and positions of the 5′ ends of the plus-strand DNAs located at DR2 and DR1 for the mutant and wild-type (WT) viruses. A sequencing ladder generated with the P primer flanks the primer extension reactions. Positions of DR1 and DR2 are indicated. (B) Detection of 5′ termini of minus-strand DNA. The M primer was used to determine the level and position of the 5′ end of minus-strand DNA. A sequencing ladder made with primer M is shown for comparison. (C) The amount of plus-strand DNA initiated at DR2 relative to the amount of minus-strand DNA initiated at DR1 was calculated for each mutant after normalization to the extension products detected from the internal standard by the M primer. For each mutant, the mean and standard deviation of the ratio of plus-strand DNA to minus-strand DNA from three independent transfections is shown. (D) The amount of plus-strand DNA initiated by in situ priming relative to the amount of minus-strand DNA was calculated for each mutant. Other details are the same as for panel C.
To substantiate this notion, we examined whether minus-strand DNA synthesis, a step prior to the plus-strand DNA synthesis, proceeds normally in the α element mutants. Thus, we measured minus-stand DNA synthesis by the M primer; the M primer is designed to identify the 5′ end of the minus-strand DNA (Fig. 4A to D). In the wild type, we confirmed that the M primer extension products of the three major replication intermediates map to DR1* (Fig. 5B, lane 1). Similar primer extension analysis of the α element mutants R022 and R045 indicated that these mutants also have 5′ termini of minus-stand DNA at the wild-type position (DR1*) and at a level comparable to that of the wild type (Fig. 5B, lanes 4 and 5). These results indicate that minus-strand DNA synthesis proceeds normally in the α element mutants.
To quantify plus-strand DNA synthesis, the amounts of plus-strand DNAs initiated at DR2 and DR1 were normalized to the amount of minus-strand DNA determined by primer extension with the M primer. As shown in Fig. 5C and D, in the α element mutants, a significant increase in the amount of in situ-primed plus-strand DNA initiated at DR1 was evident, whereas the DR2-primed plus-strand RC DNA was substantially reduced relative to the wild type. In other words, the α element mutants have a phenotype that is consistent with a defect in the template switch that translocates the plus-strand primer to DR2 during plus-strand DNA synthesis. From the collective results, we concluded that the α element facilitates plus-strand primer translocation from the donor site (DR1) to the acceptor site (DR2) or subsequent initiation and/or elongation during plus-strand RC DNA synthesis (Fig. 1C to F).
The possibility that the plus-strand primer (a capped RNA generated by RNase H activity of the viral P protein) is generated incorrectly in the α element mutants has not been excluded (Fig. 1C). However, it seems unlikely that the α element, which is located approximately 0.8 knt upstream from the 3′ end of the minus-strand DNA template, directly affects the specificity of the RNase H activity. In fact, the sites cleaved by the RNase H activity could be inferred from the results of primer extension analysis that detects the 5′ end of in situ-primed plus-strand DNA (17). Primer extension analysis with the P primer indicated that for the wild type, the extension products mapped to few sites within DR1 as well as the 3′ termini of DR1 (Fig. 5A, lane 1). In fact, the sites cleaved by the RNase H activity of P protein in the α element mutants appeared to be normal in comparison to those of the wild type (Fig. 5A, lane 1 versus lanes 4 and 5). These findings suggested that the generation of the RNA primer is not impaired in the α element mutants. Therefore, primer translocation or elongation is the most likely step of plus-strand DNA synthesis that is affected by mutations in the α element.
Lack of plus-strand DNA initiated from DR2 in the α element mutants.
The lack of plus-strand DNA initiated at DR2 in the α element mutants could be due to an elongation defect rather than a defect in plus-strand primer translocation and subsequent initiation of plus-strand DNA synthesis. If this were the case, then the mutants would synthesize short lengths of plus-strand DNA with 5′ termini located at DR2. To confirm the presence of this short plus-strand DNA initiated at DR2 in the α element mutants, we used an oligonucleotide (B primer) capable of annealing to plus-strand DNA at a site located 59 nt away from DR2 and upstream of the circularization point (Fig. 4C and D, B primer). Primer extension analysis with the B primer revealed that far fewer 5′ termini located at DR2 were detected in the R022 and R045 mutants in comparison to the wild type (Fig. 6A, lane 1 versus lanes 4 and 5). Based on this result, we concluded that the α element mutants are defective in primer translocation to DR2 or subsequent initiation from translocated DR2 rather than in elongation and/or circularization during plus-strand RC DNA synthesis.
FIG. 6.
Detection of plus-strand DNA before circularization. (A) The B primer was used to determine the level and position of the 5′ end of plus-strand DNA before circularization. The position of DR2 is indicated. A sequencing ladder made with the B primer is shown for comparison. (B) The amount of plus-strand DNA initiated at DR2 relative to the amount of minus-strand DNA initiated at DR1 was calculated for each mutant after normalization as described for Fig. 5C. For each mutant, the mean and standard deviation of the ratio of plus-strand DNA to minus-strand DNA from three independent transfections is shown. WT, wild type.
Identification of cis-acting elements γ and δ.
As stated above, we have identified two additional novel cis-acting elements that are involved in facilitating HBV plus-strand RC DNA synthesis. Southern blot analysis suggested that four deletion mutants (R060, R048, R027, and R053) led to a reduction in RC DNA synthesis (Fig. 3A, lanes 2, 3, 15, and 16). A region deleted in the R060 and R048 mutants, termed the γ element, is located near the 3′ end of the minus strand, whereas a region deleted in R027 and R053 mutants, termed the δ element, is located near DR2 on the minus-strand DNA. To further characterize these elements, we first examined minus-strand DNA synthesis. In the γ and δ element mutants, primer extension analysis with the M primer indicated that the amount of minus-strand DNA initiated at DR1* was at a level comparable to that of the wild type, suggesting that the γ and δ element mutants are not defective in minus-strand DNA synthesis (Fig. 5B, lanes 2, 3, 6, and 7). Next, we examined whether these mutants are defective in initiating plus-strand DNA synthesis. To determine the relative amount of 5′ termini at DR2 made from these mutants, primer extension analysis with the P primer was carried out (Fig. 5A). This analysis indicated that the amount of 5′ termini of plus-strand DNA at DR2 as well as DR1 was reduced in all four γ and δ element mutants relative to the wild type (Fig. 5A [lanes 2, 3, 6, and 7], C, and D). In other words, it appeared that both in situ priming and the initiation at DR2 following translocation were commonly suppressed in these mutants. Unlike the case for α element mutants described above, the reduction of plus-strand DNA initiated at DR2 in the γ and δ element mutants apparently did not result in a proportional increase in DNA synthesis initiated at DR1 (i.e., in situ priming) (Fig. 5A and D). We interpreted this finding as a possible indication that both the γ and δ elements act at a step prior to the commitment to initiate plus-strand DNA synthesis, since the initiation of plus-strand synthesis at DR2 as well as DR1 was reduced.
Alternatively, the reduction in the accumulation of plus-strand DNA initiated at DR2 in the γ and δ element mutants could be due to a defect in the template switch that circularizes the DNA genome (Fig. 1H). To examine this possibility, primer extension analysis was performed with the B primer that is designed to anneal to plus-strand DNA upstream of the circularization point (Fig. 4 and 6A). The analysis indicated that the amount of 5′ termini located at DR2 in the γ and δ element mutants was reduced approximately threefold relative to the wild type (Fig. 6A [lanes 2, 3, 6, and 7] and B), suggesting that these mutants are defective primarily in the RNA primer translocation to DR2. The observation that the magnitude of defect exhibited by these mutants in Fig. 5C is slightly greater than that measured in Fig. 6B suggested that the γ and δ element mutants are also partly defective in the template switching mediating circularization. This finding, together with data presented above, suggested that the γ and δ elements contribute to multiple steps during plus-strand RC DNA synthesis: (i) a step prior to the commitment to initiate plus-strand DNA synthesis, (ii) primer translocation to DR2, and (iii) the circularization step in some degree.
The γ and δ element mutants are competent for full-length minus-strand template synthesis.
The data presented above indicated that the γ and δ elements contribute to multiple steps during plus-strand DNA synthesis. However, the possibility that mutants lacking the γ or δ elements fail to synthesize full-length minus-strand DNA template is not excluded. In this case, the inability of the mutants to generate a capped RNA primer would lead to a defect in subsequent plus-strand DNA synthesis. This possibility was excluded by our ability to detect PCR products with a primer that is specific to the last 18 nt at the 3′ end of the minus-strand DNA (data not shown). Consequently, incomplete minus-strand DNA synthesis cannot account for the reduced accumulation of the RC DNA in the γ and δ element mutants. Therefore, we conclude that the γ and δ element mutants are defective in a step subsequent to the completion of minus-strand DNA synthesis. Though unlikely, it is formally possible that a few (<ca. 4) nucleotides at the very end of minus-strand DNA are still missing but nevertheless amplified by PCR with a limited base pairing.
In addition, previous studies have established that the hepadnavirus pgRNA contains a cis-acting element (posttranscriptional regulatory element) which facilitates the nuclear export of unspliced viral transcripts (2, 9, 24). Because one of the two functional stem-loop structures of the posttranscriptional regulatory element (i.e., HSLβ1) is disrupted by the δ element mutants (i.e., R027 and R053), these two mutants are possibly defective in RNA processing; however, this possibility was excluded, as the encapsidation efficiency measured by RNase protection analysis of the pgRNA derived from these mutants was comparable to that of the wild type (data not shown).
Further deletion analysis to delineate the boundary of the α region.
On the other hand, studies have shown that deletion mutants of the pre-S1 domain isolated in patients are capable of replication; in fact, some of these deletions overlap with the α element (19, 20). To determine the boundary of the α region, five smaller deletion mutants were made (Fig. 7A). As shown in Fig. 7B, for most mutants, transfection of each mutant resulted in comparable or slightly reduced accumulation of RC DNA relative to the wild type. In contrast, two mutants failed to produce detectable RC DNA (Fig. 7B, lanes 3 and 4), suggesting that the 88-nt region deleted by these two mutants (nt 2818 to 2905) represents the minimal α element that is essential for efficient RC DNA synthesis. Importantly, the observation that the accumulation of DL DNA by these two α region mutants appeared to be significantly increased (Fig. 7B, lanes 3 and 4) is consistent with the phenotype we observed above (Fig. 5A and D), in that the reduction in RC DNA synthesis is accompanied by a proportional increase of plus-strand DNA synthesis by in situ priming.
DISCUSSION
Here, we report three novel cis-acting elements that are required for plus-strand RC DNA synthesis: (i) the α element, located in the middle of the minus-strand DNA; (ii) the γ element, located near the 3′ end of the minus-strand DNA; and (iii) the δ element, located near DR2. Based on the magnitude of the defect exhibited by each deletion mutant, these cis-acting elements could be divided into two groups: (i) the α element, when deleted, led to a severe defect, and (ii) the γ and δ elements, when deleted, led to a modest defect, in the plus-strand RC DNA synthesis. Importantly, these two groups appear to be distinctive not only in terms of the magnitude of the defect exhibited by each mutant but also with respect to the step of plus-strand DNA synthesis at which these elements exert their effect. In other words, our data suggested that the α element contributes to the primer translocation or subsequent initiation during plus-strand DNA synthesis, whereas the γ and δ elements promote the plus-strand RC DNA synthesis at multiple steps: (i) a step earlier than commitment to plus-strand DNA synthesis, (ii) primer translocation, and (iii) circularization.
How does the α element contribute to plus-strand DNA synthesis? Intriguingly, the α element mutations that led to a failure to initiate plus-strand DNA synthesis at DR2 resulted in a concomitant increase in priming at DR1 (Fig. 5A). This finding was taken to imply that in situ priming is regulated through a mechanism by which the α element positively contributes to the initiation of plus-strand DNA synthesis at DR2. This observation contrasts with the situation for the 5E and M region of DHBV, in which the mechanism of in situ priming appears to be distinct from the mechanism by which 5E and M contribute to priming from DR2 (6). Being at least 0.8 or 1.7 knt away from either the donor (DR1) or acceptor site (DR2), respectively, it is not clear how this element contributes to the efficient translocation of the plus-strand primer to DR2. In the case of DHBV, however, a study has shown that the ends of the minus-strand template are juxtaposed via base pairing to facilitate the template switches during the plus-strand DNA synthesis; in fact, base pairing among 5E, M, and 3E was shown to be essential for their function (15). Similarly, we speculate that the α element in HBV might act to bring two ends of the minus-strand DNA template together to facilitate the template switch during plus-strand DNA synthesis. Our effort to find such a base-pairing involving the α element has revealed numerous potential base-pairings, including one with sequences within the γ element (J. Lee and W.-S. Ryu, unpublished observations). To substantiate such a global structure involving the α element, further analysis to determine the boundaries of theses cis-acting elements is warranted. Alternatively, the α element might be recognized by a protein, perhaps P protein, that facilitates the template switch for plus-strand DNA synthesis. As virion DNAs produced by the α element mutants were undetectable in culture medium (data not shown), the possibility that RC DNA was undetectable in cytoplasmic nucleocapsids as the virion secretion process was expedited by the α element mutants was excluded.
In addition, we identified two other novel elements, termed γ and δ, that are required for efficient plus-strand RC DNA synthesis. Unlike the α element, the γ and δ elements are not indispensable for the accumulation of the plus-strand RC DNA genome. Our data suggested that the γ and δ elements, though not indispensable, contribute to multiple steps of the plus-strand DNA synthesis. It is worth noting that the phenotypes of the γ and δ deletion mutants are indistinguishable, despite the fact that these two elements are distantly located on the minus-strand template. We suggest that these two cis-acting elements work together, possibly via base pairing, to contribute to multiple steps of plus-strand DNA synthesis. However, an attempt to find a base-pairing between the γ and δ elements revealed at best an 15-nt-long imperfect duplex having seven consecutive base pairs (Lee and Ryu, unpublished observations). However, the significance of such a base-pairing interaction for plus-strand RC DNA synthesis remains to be established. On the other hand, a formal possibility that the defects of the γ and δ deletion mutants are attributable to the size of the deletion as opposed to the sequences per se or a modified structure due to the deletions was not excluded.
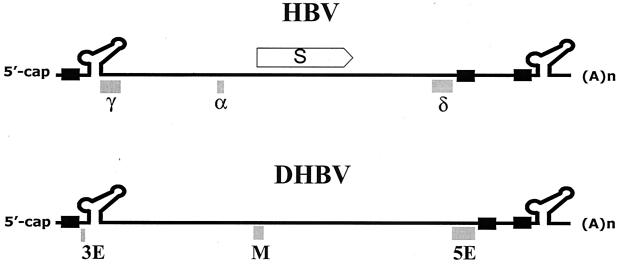
It should be informative to compare the three cis-acting elements identified in the present study to those found in the DHBV genome, i.e., 3E, M, and 5E (6, 7). Intriguingly, the positions of the three cis-acting elements (γ, α, and δ) of HBV are similar to those of the 3E, M, and 5E regions in the DHBV genome, respectively (Fig. 8). It is interesting that the α element, in particular, is analogous to the M region of DHBV with respect to its mutant phenotype as well as its position relative to other repeat elements in the genome. Since the phenotypes of the α element mutants are distinct from those of the γ and δ element mutants, our findings suggested that the α element of HBV does not work intimately with the γ and δ elements during plus-strand DNA synthesis. Thus, specific roles of these cis-acting elements appear to be distinct for HBV and DHBV. While this work was in preparation, mapping of the cis-acting elements in the HBV genome was reported (14); two cis-acting elements that those authors mapped in the region we examined overlap with the α and δ elements that we report here. On the other hand, Liu et al. suggested that the region (nt 1915 to 2110) that overlaps with the γ element described here might exert an inhibitory effect in RC DNA synthesis, as the relative proportion of RC DNA was enhanced when this region was deleted (14). Further studies are needed to clarify this discrepancy. Overall, multiple cis-acting elements located at distinct positions on the minus-strand template reflect the complex nature of hepadnavirus reverse transcription.
FIG. 8.
Comparison of cis-acting elements required for RC DNA synthesis by HBV with those required for RC DNA synthesis by DHBV. The direct repeats, DR1 and DR2, are not drawn to scale. The S (surface antigen) ORF is shown for comparison. The nucleotide sequence corresponding to 3E of DHBV, which is positioned between the 5′ copy of DR1 and 5′ ɛ, was not examined in this work.
Acknowledgments
This work was supported by a National Research Laboratory grant from the Ministry of Science and Technology of the Korean Government.
REFERENCES
- 1.Condreay, L. D., T. T. Wu, C. E. Aldrich, M. A. Delaney, J. Summers, C. Seeger, and W. S. Mason. 1992. Replication of DHBV genomes with mutations at the sites of initiation of minus- and plus-strand DNA synthesis. Virology 188:208-216. [DOI] [PubMed] [Google Scholar]
- 2.Donello, J. E., A. A. Beeche, G. J. Smith III, G. R. Lucero, and T. J. Hope. 1996. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J. Virol. 70:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 4.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 5.Goff, S. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In P. M. Hawley and D. M. Knipe (ed.), Field's virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 6.Havert, M. B., L. Ji, and D. D. Loeb. 2002. Analysis of duck hepatitis B virus reverse transcription indicates a common mechanism for the two template switches during plus-strand DNA synthesis. J. Virol. 76:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havert, M. B., and D. D. Loeb. 1997. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J. Virol. 71:5336-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsch, R. C., D. D. Loeb, J. R. Pollack, and D. Ganem. 1991. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J. Virol. 65:3309-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, Z. M., and T. S. Yen. 1995. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 15:3864-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong, J. K., G. S. Yoon, and W. S. Ryu. 2000. Evidence that the 5′-end cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J. Virol. 74:5502-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, J., H.-J. Lee, M.-K. Shin, and W.-S. Ryu. 2004. Versatile PCR-mediated insertion or deletion mutagenesis. BioTechniques 36:398-400. [DOI] [PubMed] [Google Scholar]
- 13.Lien, J. M., C. E. Aldrich, and W. S. Mason. 1986. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J. Virol. 57:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, N., L. Ji, M. L. Maguire, and D. D. Loeb. 2004. cis-acting sequences that contribute to the synthesis of relaxed-circular DNA of human hepatitis B virus. J. Virol. 78:642-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, N., R. Tian, and D. D. Loeb. 2003. Base pairing among three cis-acting sequences contributes to template switching during hepadnavirus reverse transcription. Proc. Natl. Acad. Sci. USA 100:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeb, D. D., K. J. Gulya, and R. Tian. 1997. Sequence identity of the terminal redundancies on the minus-strand DNA template is necessary but not sufficient for the template switch during hepadnavirus plus-strand DNA synthesis. J. Virol. 71:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeb, D. D., R. C. Hirsch, and D. Ganem. 1991. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 10:3533-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeb, D. D., and R. Tian. 1995. Transfer of the minus strand of DNA during hepadnavirus replication is not invariable but prefers a specific location. J. Virol. 69:6886-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melegari, M., S. Bruno, and J. R. Wands. 1994. Properties of hepatitis B virus pre-S1 deletion mutants. Virology 199:292-300. [DOI] [PubMed] [Google Scholar]
- 20.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1997. The small envelope protein is required for secretion of a naturally occurring hepatitis B virus mutant with pre-S1 deleted. J. Virol. 71:5449-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassal, M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeger, C., and J. Maragos. 1990. Identification and characterization of the woodchuck hepatitis virus origin of DNA replication. J. Virol. 64:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sells, M. A., M. L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 84:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, G. J., III, J. E. Donello, R. Luck, G. Steger, and T. J. Hope. 1998. The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function. Nucleic Acids Res. 26:4818-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavis, J. E., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]