Abstract
Cellular stress, induced by external or internal cues, activates several well-orchestrated processes aimed at either restoring cellular homeostasis or committing to cell death. Those processes include the unfolded protein response (UPR), autophagy, hypoxia, and mitochondrial function, which are part of the global ER stress (ERS) response. When one of the ERS elements is impaired, as often occurs under pathological conditions, overall cellular homeostasis may be perturbed. Further, activation of the UPR could trigger changes in mitochondrial function or autophagy, which could modulate the UPR, exemplifying cross-talk processes. Among the numerous factors that control the magnitude or duration of these processes are ubiquitin ligases, which govern overall cellular stress outcomes. Here we summarize crosstalk among fundamental processes governing ERS responses.
Keywords: ER stress, UPR, ubiquitin, hypoxia, autophagy, mitochondria
ER stress, an overview
The endoplasmic reticulum (ER) is a complex, dynamic organelle whose functions include protein folding, Ca2+ storage, and lipid and carbohydrate metabolism. Diverse cellular stresses such as perturbations in Ca2+ homeostasis, redox imbalance, altered protein glycosylation, or protein folding defects cause unfolded or misfolded proteins to accumulate in the ER lumen, a condition known as ER stress (ERS). To guard against or respond to ERS, cells have an integrated signaling system to restore homeostasis and normal ER function. Fundamental pathways that constitute integral parts of this response include the unfolded protein response (UPR), ER-associated degradation (ERAD), autophagy, hypoxic signaling and mitochondrial biogenesis. Concerted activity of all of these processes determines the extent of ERS and thus governs whether cells will re-establish homeostasis or activate cell death programs.
The UPR is a well-defined process that plays a critical role in restoring homeostasis following accumulation of potentially toxic misfolded proteins [1, 2]. The UPR is regulated by three ER membrane-embedded sensors—double-stranded RNA-activated protein kinase (PKR)-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1)—that are activated by perturbed ER homeostasis. All three activate specialized transcriptional programs mediated by distinct transducers—ATF4 (for PERK), cleaved ATF6 (for ATF6), and spliced XBP1 (sXBP1; for IRE1). These factors directly activate transcription of chaperones or proteins functioning in redox homeostasis, protein secretion, lipid biosynthesis or cell death programs. A description of the UPR-mediated processes is provided in Box 1.
Box 1. The UPR as part of the cellular ERS.
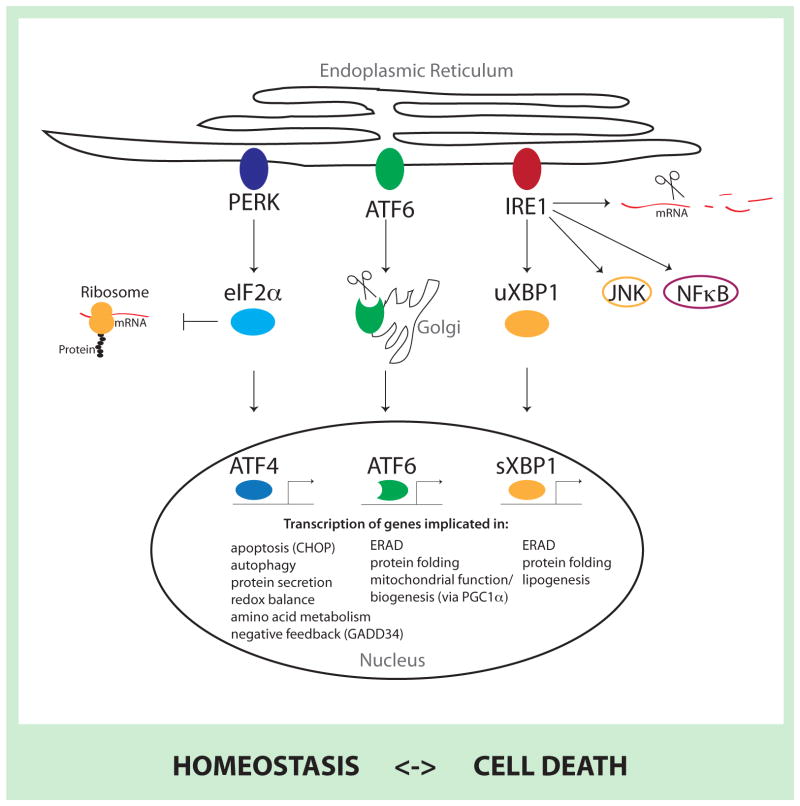
PERK, ATF6 and IRE1 have distinct cytosolic functions associated with the ability to activate respective transducers (ATF4, cleaved ATF6, and sXBP1, respectively) with a concomitant effect on protein translation, cellular metabolism and cell survival or cell death programs. For example, PERK inhibits general protein translation (via eIF2α phosphorylation) enabling dedicated translation of transcripts harboring an alternate open reading frame, including ATF4, a key transducer. ATF4 is also implicated in the induction of several ATG genes and, depending on the magnitude of stress stimuli, can activate cell death programs (in cooperation with CHOP) [1, 2]. IRE1 RNAse domain mediates splicing and activation of the transcription factor XBP-1, which induces expression of chaperones, ER-associated degradation (ERAD) components and proteins involved in lipogenesis. In addition, the IRE1 RNAse domain is also implicated in mRNA degradation by promoting mRNA decay and its kinase domain could be linked with other stress induced pathways such as JNK or NF-κB (CITE?). Upon endoplasmic reticulum stress (ERS), ATF6 translocates to the Golgi where it is cleaved by Site-1 protease (S1P) and Site-2 protease (S2P) to produce an active transcription factor that translocates to the nucleus and regulates genes such as CHOP, grp78 and ERAD components [59]. Furthermore, ATF6 activity stimulates mitochondrial biogenesis through its effect on PGC1α and related co-activators [42, 43]. Additionally, the UPR attenuates protein translocation to the ER (dependent on signal peptides) and enhances ERAD. Of note, some of these pathways can also be activated by other stress pathways independent of bona fide UPR components, including phosphorylation of eIF2α (by GCN2, PKR, or HRI) [60]. Also, non-canonical PERK and IRE signaling has been reported [61, 62]. Tissue- or cell type-dependent expression of UPR sensors also likely mediates specialized, lineage-dependent responses [63]. A simplified overview of molecular mechanisms of UPR signaling is provided in Figure I, and the reader is also directed to specific reviews in this area [1, 2].
UPR activity must be tightly regulated on several levels as current observations suggest that a high level or prolonged UPR signaling is associated with cell death, whereas more moderate or shorter UPR signaling enables restoration of homeostasis and cell survival (CITE?). For example, in yeast, under lower levels of stress, the ER chaperone grp78 reportedly regulates IRE1 activation–deactivation kinetics: IRE1 is only turned on once stress levels reach a certain threshold, and grp78–IRE1 interaction sensitizes IRE1 to those lower levels. Once stress is relieved, grp78–IRE1 binding enhances IRE1 deactivation by favoring its de-oligomerization [64]. IRE1 also auto-regulates its own activity by two mechanisms: Hrd1, a XBP-1 transcriptional target, ubiquitylates IRE1, promoting its degradation [47], and the IRE1 RNAse domain degrades its own mRNA [65]. Overall, the idea is emerging that crosstalk among UPR components—exemplified by the effect of IRE1 on PERK, or ATF6 modulation of IRE1 signaling [66]—amplifies the UPR signal, which ultimately regulates cellular responses to stress stimuli.
Figure I. Molecular mechanisms of UPR signaling.
The three ERS sensors PERK, ATF6 and IRE1 activate a complex transcriptional cascade with distinct cytosolic functions. PERK phosphorylates eIF2α to decrease overall translation while increasing specific translation of genes, including ATF4. Upon ERS, ATF6 is translocated to and is processed at the golgi apparatus to create a highly active transcription factor. IRE1 decreases overall protein flux to the ER by enhancing mRNA degradation, activates other cellular pathways (such as JNK or NFκB) to counteract ERS and leads to the activation of the XBP1 transcription factor by splicing the uXBP1 mRNA to create sXBP1 mRNA, which is more efficiently translated. All three transcription factors lead to the upregulation of chaperones, in addition to their respective specific targets (indicated in figure) to counteract ERS and restore homeostasis, or proceed to induce cell death pathways.
ATF: activating transcription factor; eIF2α: eukaryotic translation initiation factor 2 alpha; IRE1: inositol-requiring enzyme 1; JNK: c-Jun N-terminal kinase; NFκB: Nuclear Factor κB; PERK: double-stranded RNA-activated protein kinase (PKR)-like ER kinase; u/sXBP1: unspliced/spliced x-box binding protein 1.
The UPR is often associated with significant cellular threats stemming from altered environmental conditions (changes in nutrients or oxygen) or intracellular insult (carcinogens or oncogenic mutation), hence it is often closely linked to autophagy, hypoxia signaling, mitochondrial biogenesis or reactive oxygen species (ROS) responses. Given these interactions, it is not surprising that UPR dysfunction is implicated in many human diseases, including cancer, diabetes, neurodegeneration, ischemia, and infectious disease (reviewed in [3]). Accordingly, a better understanding of the complexity of the ERS response and the UPR could manifest novel therapeutic approaches.
Here, we review recent advances in our understanding of crosstalk between the UPR and mitochondrial function and autophagy, two fundamental processes associated with the cellular response to stress. In doing so, we emphasize the emerging roles played by E3 ubiquitin ligases in fine-tuning and regulating these processes.
The UPR and autophagy
Autophagy is a major catabolic process that delivers proteins, cytoplasmic components, and organelles to lysosomes for degradation and recycling. A well-orchestrated program including over 30 AuTophaGy-related (ATG) genes controls autophagy, which can be activated by nutrient starvation and subsequent inhibition of mechanistic target of rapamycin (mTOR) signaling [4], or by the UPR as aggregated misfolded proteins accumulate [5]. A link between autophagy and the UPR has been further substantiated by the demonstration that the PERK–eIF2α pathway is essential for autophagy induction after ERS (stimulated by tunicamycin treatment) [6]. Specifically, ATF4 and CHOP (C/EBP homologous protein, a transcription factor induced by ATF4, Box 1) were shown to transcriptionally regulate more than a dozen ATG genes [6]. In addition, IRE1 is also implicated in the activation of autophagy. TNF receptor-associated factor 2 (TRAF2)-dependent activation of IRE1 and c-Jun N-terminal kinase (JNK) reportedly results in Bcl-2 phosphorylation, enabling dissociation of Beclin-1 (an autophagy regulatory protein), activation of the Phosphoinositide-3-Kinase (PI3K) complex and autophagy [5]. Notably, it has been shown that following oxidative stress JNK contribution to the control of autophagy can be IRE1-independent [7].
Increased IRE1 activity and IRE1-dependent inflammation was observed in intestinal epithelial cells of ATG-knockout mice. The latter implies that dysregulated autophagy may also trigger the IRE1 activity with concomitant activation of the sXBP arm of the UPR, thereby pointing to a plausible feedback mechanism in the control of UPR signaling [5, 8]. As there have been recent reviews on some molecular mechanisms linking ERS and the UPR to autophagy [5], we focus here on the bi-directional interaction between autophagy and ERS (Figure 1).
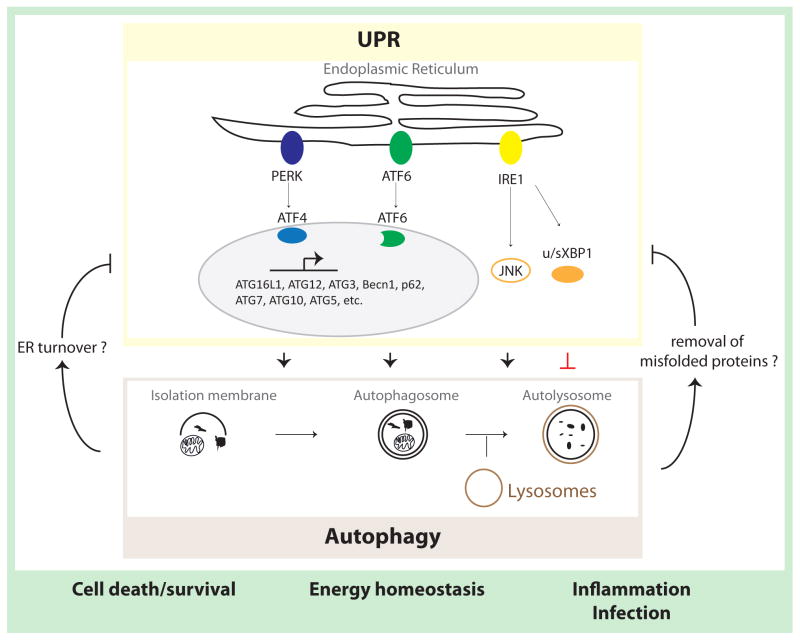
Figure 1. UPR crosstalk with autophagy.
Autophagy is initiated by encapsulation of cytoplasmic components (proteins and organelles) within isolation membranes to form autophagosomes. These structures eventually fuse with lysosomes, and the cargo is degraded. Activation of the PERK–eIF2α–ATF4 pathway upregulates expression of a large set of autophagy genes. While IRE1 signaling has been implicated in promoting autophagy (via JNK-mediated signaling), it was also shown to elicit negative regulation of autophagy. Functionally, autophagy promotes cell survival, increases energy supply and mediates innate immune responses. Loss of autophagy genes induces the UPR, indicative of a negative feedback mechanism. Autophagy may decrease cellular stress levels by removal of ER membranes, which contain UPR sensors, or decrease the amplitude of stress by clearing aberrant proteins from the ER.
ATF: activating transcription factor; ATG: autophagy-related gene; Becn1: Beclin 1; eIF2α: eukaryotic translation initiation factor 2; IRE1: inositol-requiring enzyme 1; JNK: c-Jun N-terminal kinase; NF-κB: Nuclear Factor kappa B; PERK: double-stranded RNA-activated protein kinase (PKR)-like ER kinase; UPR: unfolded protein response; u/sXBP1: unspliced/spliced x-box binding protein 1.
Autophagy and ERAD
In the canonical ERAD system, ubiquitylated un- or misfolded proteins are degraded by the ubiquitin-proteasome system (UPS) [9, 10]. However, activation of autophagy by UPR transducers also functions as a non-canonical ERAD pathway. Initially, autophagic degradation of unfolded proteins was viewed as a secondary response to ERS, engaged only when the canonical, proteasome-dependent pathway was overwhelmed by an excess of ubiquitylated proteins. Recent evidence suggests, however, that autophagy efficiently removes misfolded proteins that either form aggregates or, due to their structure, are not efficiently processed by the canonical ERAD machinery (such as mutant α1-antitrypsin or misfolded forms of the gonadotropin-releasing hormone receptor (GnRHR)) [11, 12]. Additionally, in select tissues, as shown for neuroblastoma cells, enhanced LC3 processing, a marker of autophagy, was observed, following UPR-inducing stimuli [13]. Likewise, phospho PERK-positive neurons derived from Alzheimer’s disease patients showed elevated LC3 processing [13]. Although the role of autophagy in the clearance of misfolded proteins remains to be established, the possibility that certain tissue types benefit from autophagy as a process that serves to maintain homeostasis is intriguing and merits further studies.
Autophagy serves as a pro-survival mechanism under ERS conditions
As the UPR induces not only survival but also cell death signals, understanding the nature of the switch between cellular outcomes is of great importance, and is likely mediated by the fine-tuning of UPR signals. Cell death is largely associated with increased ATF4 and CHOP levels, whereby autophagy (which can also be regulated transcriptionally by ATF4 and CHOP) is recognized as a major pro-survival mechanism that counteracts excessive UPR-signaling [14]. Although most studies indicate that autophagy has a pro-survival function following ERS, a heightened degree/duration of stress can induce autophagy-dependent cell death mechanisms [15–17].
The role of autophagy in regulating cell survival is best illustrated by the response of tumor cells to chemotherapies. In some cases, activation of autophagy has been shown to confer tumor cell resistance and inhibit cell death, thereby antagonizing the effect of therapy [15]. For example, although treatment of glioma cells with Cyclosporine A induces ERS, autophagy (in an IRE1- and PERK-dependent manner), and apoptosis, the extent of cell death increases as autophagy is inhibited [18]. Altered expression/activity of key UPR components (i.e., IRE1, PERK and related transducers) has been reported in tumor cells. Those may account for autophagy-mediated resistance phenotypes, as exemplified by a report of UPR upregulation in melanoma cell lines following BRAF inhibitor (BRAFi) treatment, an effect accompanied by a PERK-dependent induction of autophagy, and inhibition of autophagy sensitized melanoma cells to BRAFi-induced cell death [19]. Likewise, pharmacological inhibition of autophagy augmented BRAFi-induced antitumor activity in a xenograft model [19]. Notably, another study reported that induction of apoptosis following treatment with the BRAFi vemurafenib was associated with the ability of melanoma cells to induce ATF4 [20]. An emerging possibility, which remains to be confirmed, is that failure to activate the UPR is associated with resistance to therapy. This is attractive given the role of ATF4 in control of autophagy, thereby pointing to the link between UPR and autophagy as an underlying mechanism for tumor cell resistance mechanisms.
Autophagy and inflammation
The link between the UPR and autophagy can also explain protective mechanisms seen following inflammation of intestinal epithelial cells [8]. Loss of the IRE1 transducer, the transcription factor XBP-1, in cultured intestinal epithelial cells induces PERK- and p-eIF2α-dependent autophagy. This points to a plausible feedback mechanism and also indicates the need to balance the regulatory arms (sensors and their transducers) of the UPR. Consistent with this possibility is the observation that conditional knockout of XBP-1 in intestinal epithelial cells results in activation of autophagy in paneth cells, a secretory cell type important for the production of anti-microbial compounds, followed by enteritis, which is worsened when the ATG genes ATG7 or ATG16L1 are co-deleted. Both, XBP1−/ − ATG7−/ − and XBP1−/ − ATG16L1−/ − double-knockout mice exhibit a Crohn’s disease-like inflammatory phenotype driven by IRE1-dependent nuclear factor (NF)-κB activation. Furthermore, ATG16L1 conditional knockout mice exhibit increased GRP78 (a chaperone that is induced by the UPR and is also recognized as a regulator of UPR activation) as well as eIF2α phosphorylation and JNK activation, culminating in IRE1 expression and increased XBP-1 splicing in intestinal crypts. These changes substantiate a link between autophagy and select arms of the UPR, which is sufficient to increase epithelial cell death. These conditions enhance the response to pro-inflammatory agents that further challenge the UPR. The latter finding suggests a negative feedback mechanism in which ERS induces autophagy, which in turn negatively regulates the UPR
Importantly, the finding that XBP-1 loss induces autophagy indicates that the UPR can also limit the degree of autophagy. Besides activation of PERK/ATF4 [8], XBP-1 loss was reported to regulates autophagy via the Forkhead box protein O1 (FOXO1), a transcription factor known to induce autophagy [21]. Under glutamine-starvation conditions, it was shown that after phosphorylation by ERK1/2 (extracellular-signal regulated kinase1/2) unspliced XBP-1 (uXBP1) can bind to FOXO1 and mediate its proteasomal degradation [22]. Notably, in overexpression studies, spliced XBP-1 was also demonstrated to bind and elicit degradation of FOXO1 [23]. While pointing to XBP-1 as a regulator of autophagy, further studies are needed to shed light on the molecular mechanisms underlying XBP-1 ability to regulate autophagy under physiological-relevant ERS conditions.
Overall, the UPR link with autophagy constitutes an important regulatory pathway that mediates pro-survival signals in response to misfolded proteins, chemotherapy, and inflammation. The degree of initial insult, often regarded as the degree of ERS or UPR activation, can determine the balance between pro- and anti-survival signals, in which autophagy may serve to either promote or attenuate ERS and UPR signals. Bona fide autophagy genes might affect ERS and the UPR independent of their role in autophagy, as autophagy-independent processes are being described for autophagy-related genes. For example, the kinase ULK1 (unc-51 like autophagy activating kinase 1) was shown to affect microphtalmia-associated transcription factor (MITF) and melanogenesis independent of its role in autophagy and mTOR complex 1 (mTORC1) regulation [24]. Future studies are expected to illuminate this interesting regulatory cue.
A link between ERS, the UPR and autophagy may also explain how ER and possibly mitochondrial membranes serve as a source of autophagosomal membranes. Both omegasome sites in the ER and ER–Golgi contact areas provide a source for initiation of autophagosome membrane formation [25]. Furthermore, autophagosomes reportedly form at physical contact sites between the ER and the mitochondria (called mitochondrial associated membranes or MAM; Box 2). MAM disruption inhibits autophagosome formation [26]. Although the impact of these activities remains to be determined, they likely affect the rate of mitochondrial fusion and fission and ER signal integrity.
Box 2. ERS and mitochondria-associated ER membranes (MAMs).
MAMs are flexible, ER membrane-derived structures that form physical contact points between the ER and mitochondria. Mechanisms governing MAM formation are unclear, since most proteins enriched in MAMs (such as phosphatidylserine synthase, IP3R, VDAC1, calnexin, DRP1, ERO1, and PERK) appear not to serve a structural function but rather are associated with functions such as lipid biosynthesis, ER-mitochondria lipid or Ca2+ transfer, mitochondrial dynamics and bioenergetics, apoptosis, or autophagy [67, 68]. Most of these processes are also linked to ERS, and tethering of the two organelles is strengthened following ERS, supporting a key role for MAMs in ERS. MAM-mediated Ca2+ transfer from the ER to mitochondria may have either pro-survival or pro-death effects, perhaps contributing to enhanced mitochondrial ATP production to satisfy increased energy demands resulting from stress. Conversely, elevating mitochondrial Ca2+ may promote apoptosis. The ER is a source of autophagosome membrane components, and MAMs are directly implicated in autophagosome formation [68]. Autophagy may also control ER expansion through selective “ER-phagy”. Evidence suggests that MAMs serve as important hubs for mitochondrial fusion and fission. For example, ER-mitochondrial tethering contributes to the formation of constriction sites, a key step in Drp-1-mediated mitochondrial fission. Mfn-2 is an important regulator of mitochondrial fusion, and both Mfn-2 and PERK are critical to maintain the MAM. Thus, the fusion process is likely linked to the UPR [68].
The UPR link with mitochondrial function
Several regulatory components link the UPR with mitochondrial regulation and function. UPR transducers affect mitochondrial regulatory components, including ATF4, which was demonstrated to control expression of the ubiquitin ligase Parkin, a crucial regulator of mitochondria function and dynamics [27]. In turn, reciprocal activation is illustrated by the ability of Parkin to enhance select branches of the UPR signaling through the activation of the sXBP1 pathway [28]. ATF6 is also associated with the activation of PCG1α (peroxisome proliferator-activated receptor gamma, coactivator 1 alpha), a master regulator of mitochondrial biogenesis, thereby linking the UPR with metabolic gene programs [29].
Parkin exemplifies a regulatory node that underlies crosstalk between the UPR and mitochondria: while activated by ATF4, Parkin functions in mitochondrial dynamics [30, 31], bioenergetics [32], and mitophagy [33]. Furthermore, Parkin reportedly increases MAMs (see Box 2). By modulating the MAM, Parkin maintains ER-mitochondrial Ca2+ transfer and mitochondrial bioenergetics during ERS [32]. Although it is recognized that Parkin marks damaged or dysfunctional mitochondria for mitophagy, there is little evidence that this process is induced by ERS. Until recently, Parkin-mediated mitophagy upon accumulation of un- or misfolded proteins was reported only upon activation of the mitochondrial UPR [34] (a process that, like the endoplasmic UPR, communicates stress to the nucleus and increases expression of mitochondrial chaperones [35]). Nonetheless, ERS occurring as a result of cerebral ischemia has also been linked with mitophagy [36] and may protect cells from cell death signaling induced by ischemia/reperfusion-induced brain injury. Both mitophagy and cytoprotective effects are mediated by ATF4-dependent Parkin expression [36]. Consistent with these observations, a reduced level of UPR output under neuronal ischemic conditions was found to decrease infarct size and enhanced cell survival [37]. In these studies, reducing the expression of the Siah ubiquitin ligases, which are important in controlling the degree of UPR output (by stabilizing ATF4 levels) limited the degree of ATF4 and CHOP, and attenuated the induction of cell death programs under neuronal ischemic conditions [37] (see Box 3 for more details).
Box 3. UPR–HIF crosstalk.
Cells respond to decreased oxygen levels by activating adaptive pathways including induction or stabilization of the transcription factors HIF1 (which consists of the oxygen-regulated subunit HIF1α and the constitutively expressed subunit HIF1α) and HIF2 and their transcriptional targets, which promote vascularization, glycolysis and survival. Under severe hypoxia other mechanisms, such as the UPR, are activated to antagonize stress, although all of these pathways synergize to activate common downstream targets.
During development, nutrient deprivation, hypoxia, and increased protein secretion activate the UPR. One UPR target is VEGF, which is activated independent of HIF1α [69]. Notably, endothelial cells respond to VEGF by upregulating the UPR, which supports VEGF-mediated angiogenesis [70]. In addition, anti-angiogenic Type I interferon signaling is inactivated via both VEGF and UPR-induced degradation of the interferon receptor [71, 72]. HIF1α heterodimerization with sXBP1, which has been demonstrated in breast cancer cells, exemplifies cooperation between the UPR and hypoxia, an activity that was implicated in maintenance of triple-negative breast cancer (TNBC). Consistent with earlier findings that HIF1α is hyperactivated in TNBC, sXBP1 reportedly co-localizes with HIF1α in TNBC tumor cells at distinct regulatory elements, where both regulate established HIF targets and maintain cancer stem cell activity [73].
The ubiquitin ligases Siah1/2 function in both hypoxia and UPR signaling. Siah stabilizes HIF1α by proteasomal degradation of PHD proteins, which destabilize HIF [74]. In addition, under ischemia Siah1/2 are transcriptionally upregulated by ATF4 or sXBP1. Siah-dependent PHD down-regulation increases ATF4 levels and activity, which are required to activate cell death programs under ischemia [37]. Siah2 contribution to UPR and hypoxic conditions also impacts its role in the control of mitochondrial dynamics [45].
Additional mechanisms link the ERS, UPR and mitochondrial function. One is regulation of mitochondrial fusion and fission processes by cues initiated by the ERS and UPR. Genetic inactivation of mitofusin-2 (Mfn-2), a key component of the mitochondrial fusion process, promotes mitochondrial swelling, Ca2+-overload and reduced mitochondrial respiration [38]. Notably, PERK inhibition normalizes mitochondrial integrity in Mfn-2 mutant cells, identifying PERK as an important regulator of mitochondrial processes [38]. Accordingly, both Mfn-2 [39] and PERK [40] were shown to regulate MAM integrity. Although PERK function in control of mitochondrial dynamics may be independent of its function in the UPR (discussed in [41]), one cannot exclude UPR-dependent mechanisms. Consistent with the latter possibility is the observation that Mfn-2-deficient cells show hyperactivated UPR signaling and defective autophagy and apoptosis, which indicates a role of MAM integrity in UPR signaling [38].
The transcriptional co-regulator PGC1α, an important regulator of mitochondrial biogenesis and function, is also linked to the UPR in a tissue-specific manner. In skeletal muscle, PGC1α is upregulated upon ERS (induced by exercise) and then cooperates with ATF6 to mediate an adaptive response to stress [42]. In addition, in hepatoma cell lines ATF6 and PGC1α activity enhances expression of the estrogen-related receptor gamma (ERRγ), another regulator of mitochondrial function. In turn, ERRγ interacts with PGC1α to mediate transcription of ERR target genes. Interestingly, ATF6 has been identified as one ERRγ/PGC1α target, suggesting a feed forward mechanism whereby PGC1α mediates adaptation to stress [43].
An obvious function of UPR crosstalk with mitochondria is to control programmed cell death (reviewed in [14]). Small ubiquitin-like modifiers (SUMO) have been found to modulate ER–mitochondrial crosstalk and apoptosis through the deSUMOylating enzyme SENP3. ERS induces PERK- and cathepsin B-dependent degradation of SENP3, resulting in elevated levels of SUMOylated proteins. Among those is Drp-1, a key mediator of mitochondrial fission. DeSUMOylation of Drp-1 attenuates cytochrome c release and caspase-mediated cell death. Thus, Drp-1 SUMOylation constitutes an additional layer in the control of pro-survival signals from the ER to mitochondria [44]. Through their regulation of the adaptor protein AKAP121, which in turn controls the Drp1–Fis1 complex, the ubiquitin ligases Siah1/2 regulate mitochondrial fission under hypoxia conditions (known to induce ERS). The interplay between Siah1/2 control of fission complex and ATF4, which has also been associated with control of mitochondrial dynamics, is intriguing [45] (Box 3).
The UPR influences mitochondrial function on several levels: it induces mitophagy to clear stress-damaged mitochondria, regulates mitochondrial bioenergetics by influencing the MAM, and promotes loss of mitochondrial membrane potential. Parkin coordinates crosstalk between these activities; likewise, the ubiquitin ligases Siah1/2 function in the crosstalk between the UPR and mitochondrial functions under ischemic conditions [37].
Ubiquitin ligases fine-tune the ERS response
The observation that ubiquitin ligases regulate crosstalk between the UPR, autophagy and mitochondria suggests that they play a more dominant role within the ERS response than foreseen.
Ubiquitin ligases are central for the ERAD process (reviewed in [9, 10]) where they ubiquitylate misfolded proteins, tagging them for proteasomal degradation or removal by autophagy. However, emerging evidence suggests that E3 ubiquitin ligases contribute to ERS response through mechanisms beyond their role in ERAD. Exemplified by studies with Parkin and Siah1/2, ubiquitin ligases spatially and temporally regulate factors that fine-tune the magnitude and duration of UPR, autophagy or mitochondrial function, and thus, impact overall cellular stress outcome (Table 1). Mechanistically, SUMO and ubiquitin E3 ligases either regulate the abundance of major UPR proteins by targeting them for proteasomal degradation [44, 46–51], or modulate activity of UPR components by direct modification or modification of upstream regulators; for example, through modulation of XBP-1 transcriptional activity by SUMOylation [52, 53] or of ATF4 activity via prolyl hydroxylase 1/3 (PHD1/3)-mediated hydroxylation [37].
Table 1.
Ubiquitin ligases implicated in the ERS response
| E3 ligase | Target | Functions in | Ref |
|---|---|---|---|
| Hrd1 | Unfolded proteins IRE1, OASIS, BBF2H7, ATF6 |
ERAD IRE1-, ATF6 signaling |
[9, 10, 46, 47, 49] |
| gp78 | Unfolded proteins | ERAD | [9, 10] |
| Chip | Pael-R, CFTR | ERAD | [74, 75] |
| TMEM129 | MHC class I | ERAD | [76, 77] |
| RNF5 | CFTR, HERP, JAMP | ERAD | [78–80] |
| RNF185 | CFTR | ERAD | [81] |
| TRC8 | MHC class I, uXBP1 | ERAD, IRE1 signaling | [82, 83] |
| Parkin | Pael-R, | ERAD, mitophagy, ER- mitochondria tethering | [32, 34, 36, 84, 85] |
| TRIM13 | CD3d, p62, caspase-8 | ERAD, autophagy, apoptosis | [17, 86, 87] |
| cIAP | CHOP | apoptosis | [51] |
| P300 | CHOP | apoptosis | [50] |
| RNF186 | BNip1 | apoptosis | [88] |
| Siah1/2 | PHD1/3 | ATF4-signaling, apoptosis | [37] |
| Smurf1 | WFS1 | ATF6 signaling | [48] |
Other E3 ligases that function in ERS or autophagy (such as RNF5 [54]) or are relevant to mitochondrial function (like MITOL [55]) could also function in crosstalk between these processes. The recent finding that ubiquitin itself can be phosphorylated [56–58] serves to link two important signaling pathways and adds another level of complexity to mechanisms governing various cellular processes, including the ERS response.
Concluding remarks
Under normal growth conditions, concerted action of ERS components is required to maintain cellular homeostasis following external stimuli. Imbalance in these regulatory components, as often seen in pathological conditions, presents a challenge to develop therapies to restore such homeostasis.
Here, we point to the link between UPR-activating conditions, (i.e. hypoxia, ischemia, inflammation, and disruption of the ER-mitochondria contact sites) and the fine balance of UPR regulatory arms in defining the cellular outcomes including autophagy and cell death. We point to the importance of transcriptional and post-transcriptional programs, which dictate the duration and magnitude of signals, and thereby fine-tune the UPR.
From a therapeutic perspective, it will be of great importance to understand how the UPR could be pharmacologically manipulated to favor pro-death or pro-survival signaling. Understanding crosstalk among the different elements of the UPR as well as how all these activities are linked with mitochondrial function, hypoxia and autophagy should provide new treatment options for a variety of pathologies, including neurodegenerative disorders, heart disease, diabetes, inflammatory diseases and cancer.
Highlights.
Autophagy and UPR interaction underlies ER stress response outcomes
ER stress modulates mitochondrial biogenesis and function
Ubiquitin ligases fine-tune the ER stress response
Acknowledgments
We thank Sergey Y. Fuchs and Eric Lau for critical reading of this manuscript. Support by NCI grant (CA128814) to ZR is gratefully acknowledged. DS is supported by a Dr. Mildred Scheel Postdoctoral Fellowship from the German Cancer Aid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 2.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Deegan S, et al. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci. 2013;70:2425–2441. doi: 10.1007/s00018-012-1173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.B’Chir W, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haberzettl P, Hill BG. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol. 2013;1:56–64. doi: 10.1016/j.redox.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adolph TE, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruggiano A, et al. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279:G961–974. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- 12.Houck SA, et al. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell. 2014;54:166–179. doi: 10.1016/j.molcel.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nijholt DA, et al. Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: implications for Alzheimer’s disease. Cell Death Differ. 2011;18:1071–1081. doi: 10.1038/cdd.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubiolo JA, et al. Yessotoxin induces ER-stress followed by autophagic cell death in glioma cells mediated by mTOR and BNIP3. Cell Signal. 2014;26:419–432. [PubMed] [Google Scholar]
- 17.Tomar D, et al. TRIM13 regulates caspase-8 ubiquitination, translocation to autophagosomes and activation during ER stress induced cell death. Biochim Biophys Acta. 2013;1833:3134–3144. doi: 10.1016/j.bbamcr.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Ciechomska IA, et al. Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine a-induced cell death. Oncogene. 2013;32:1518–1529. doi: 10.1038/onc.2012.174. [DOI] [PubMed] [Google Scholar]
- 19.Ma XH, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck D, et al. Vemurafenib potently induces endoplasmic reticulum stress-mediated apoptosis in BRAFV600E melanoma cells. Sci Signal. 2013;6:ra7. doi: 10.1126/scisignal.2003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal RL, et al. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum Mol Genet. 2012;21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, et al. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 2013;23:491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalie E, et al. ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. PLoS One. 2013;8:e75313. doi: 10.1371/journal.pone.0075313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb CA, et al. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 26.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 27.Bouman L, et al. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress. Cell Death Differ. 2011;18:769–782. doi: 10.1038/cdd.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duplan E, et al. ER-stress-associated functional link between Parkin and DJ-1 via a transcriptional cascade involving the tumor suppressor p53 and the spliced X-box binding protein XBP-1. J Cell Sci. 2013;126:2124–2133. doi: 10.1242/jcs.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arensdorf AM, et al. Temporal clustering of gene expression links the metabolic transcription factor HNF4alpha to the ER stress-dependent gene regulatory network. Front Genet. 2013;4:188. doi: 10.3389/fgene.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng H, et al. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cali T, et al. Enhanced parkin levels favor ER-mitochondria crosstalk and guarantee Ca(2+) transfer to sustain cell bioenergetics. Biochim Biophys Acta. 2013;1832:495–508. doi: 10.1016/j.bbadis.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runkel ED, et al. Mitochondrial stress: Balancing friend and foe. Exp Gerontol. 2014;56C:194–201. doi: 10.1016/j.exger.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Endoplasmic reticulum stress induced by tunicamycin and thapsigargin protects against transient ischemic brain injury: Involvement of PARK2-dependent mitophagy. Autophagy. 2014;10:1801–1813. doi: 10.4161/auto.32136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scortegagna M, et al. Fine tuning of the UPR by the ubiquitin ligases Siah1/2. PLoS Genet. 2014;10:e1004348. doi: 10.1371/journal.pgen.1004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz JP, et al. Mfn2 modulates the UPR and mitochondrial function via repression of PERK. Embo J. 2013;32:2348–2361. doi: 10.1038/emboj.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 40.Verfaillie T, et al. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rainbolt TK, et al. Stress-responsive regulation of mitochondria through the ER unfolded protein response. Trends Endocrinol Metab. 2014;25:528–537. doi: 10.1016/j.tem.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13:160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misra J, et al. Transcriptional cross talk between orphan nuclear receptor ERRgamma and transmembrane transcription factor ATF6alpha coordinates endoplasmic reticulum stress response. Nucleic Acids Res. 2013;41:6960–6974. doi: 10.1093/nar/gkt429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C, et al. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. Embo J. 2013;32:1514–1528. doi: 10.1038/emboj.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H, et al. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44:532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fonseca SG, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120:744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao B, et al. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 2008;9:480–485. doi: 10.1038/embor.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X, et al. The E3 ligase Smurf1 regulates Wolfram syndrome protein stability at the endoplasmic reticulum. J Biol Chem. 2011;286:18037–18047. doi: 10.1074/jbc.M111.225615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kondo S, et al. Activation of OASIS family, ER stress transducers, is dependent on its stabilization. Cell Death Differ. 2012;19:1939–1949. doi: 10.1038/cdd.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeong K, et al. Cyclophilin B is involved in p300-mediated degradation of CHOP in tumor cell adaptation to hypoxia. Cell Death Differ. 2014;21:438–450. doi: 10.1038/cdd.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi Y, Xia P. Cellular inhibitor of apoptosis protein-1 (cIAP1) plays a critical role in beta-cell survival under endoplasmic reticulum stress: promoting ubiquitination and degradation of C/EBP homologous protein (CHOP) J Biol Chem. 2012;287:32236–32245. doi: 10.1074/jbc.M112.362160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, Qi L. SUMO modification regulates the transcriptional activity of XBP1. Biochem J. 2010;429:95–102. doi: 10.1042/BJ20100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Z, et al. SENP1 deficiency promotes ER stress-induced apoptosis by increasing XBP1 SUMOylation. Cell Cycle. 2012;11:1118–1122. doi: 10.4161/cc.11.6.19529. [DOI] [PubMed] [Google Scholar]
- 54.Kuang E, et al. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem Sci. 2013;38:453–460. doi: 10.1016/j.tibs.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugiura A, et al. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 56.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 57.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kazlauskaite A, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maurel M, et al. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 60.Donnelly N, et al. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coelho DS, et al. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep. 2013;5:791–801. doi: 10.1016/j.celrep.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Del Vecchio CA, et al. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol. 2014;12:e1001945. doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asada R, et al. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem. 2011;149:507–518. doi: 10.1093/jb/mvr041. [DOI] [PubMed] [Google Scholar]
- 64.Pincus D, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tirasophon W, et al. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida H, et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 67.Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM) Biochim Biophys Acta. 2013;1833:213–224. doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 68.van Vliet AR, et al. New functions of mitochondria associated membranes in cellular signaling. Biochim Biophys Acta. 2014;1843:2253–2262. doi: 10.1016/j.bbamcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Ghosh R, et al. Transcriptional regulation of VEGF-A by the unfolded protein response pathway. PLoS One. 2010;5:e9575. doi: 10.1371/journal.pone.0009575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karali E, et al. VEGF Signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol Cell. 2014;54:559–572. doi: 10.1016/j.molcel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 71.Bhattacharya S, et al. Anti-tumorigenic effects of Type 1 interferon are subdued by integrated stress responses. Oncogene. 2013;32:4214–4221. doi: 10.1038/onc.2012.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng H, et al. Vascular endothelial growth factor-induced elimination of the type 1 interferon receptor is required for efficient angiogenesis. Blood. 2011;118:4003–4006. doi: 10.1182/blood-2011-06-359745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakayama K, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 75.Imai Y, et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- 76.Meacham GC, et al. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 77.van den Boomen DJ, et al. TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1409099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van de Weijer ML, et al. A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat Commun. 2014;5:3832. doi: 10.1038/ncomms4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernasconi R, et al. Autoadaptive ER-associated degradation defines a preemptive unfolded protein response pathway. Mol Cell. 2013;52:783–793. doi: 10.1016/j.molcel.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 80.Morito D, et al. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol Biol Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tcherpakov M, et al. Regulation of endoplasmic reticulum-associated degradation by RNF5-dependent ubiquitination of JNK-associated membrane protein (JAMP) J Biol Chem. 2009;284:12099–12109. doi: 10.1074/jbc.M808222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.El Khouri E, et al. RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR) J Biol Chem. 2013;288:31177–31191. doi: 10.1074/jbc.M113.470500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen CY, et al. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. Embo J. 2014;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stagg HR, et al. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol. 2009;186:685–692. doi: 10.1083/jcb.200906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imai Y, et al. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 86.Xiong H, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lerner M, et al. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18:1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tomar D, et al. TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim Biophys Acta. 2012;1823:316–326. doi: 10.1016/j.bbamcr.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 89.Wang P, et al. A novel RING finger E3 ligase RNF186 regulate ER stress-mediated apoptosis through interaction with BNip1. Cell Signal. 2013;25:2320–2333. doi: 10.1016/j.cellsig.2013.07.016. [DOI] [PubMed] [Google Scholar]