Abstract
To fuel unregulated proliferation, cancer cells alter metabolism to support macromolecule biosynthesis. Cell culture studies have revealed how different oncogenic mutations and nutrients impact metabolism. Glucose and glutamine are the primary fuels used in vitro; however, recent studies have suggested that utilization of other amino acids as well as lipids and protein can also be important to cancer cells. Early investigations of tumor metabolism are translating these findings to the biology of whole tumors and suggest that additional complexity exists beyond nutrient availability alone in vivo. Whole body metabolism and tumor heterogeneity also influence the metabolism of tumor cells, and successful targeting of metabolism for cancer therapy will require an understanding of tumor metabolism in vivo.
Keywords: cancer metabolism, tumor metabolism, cell proliferation, nutrient availability
Studies to understand cancer metabolism
Investigation into the mechanisms governing metabolic adaptations in cancer cells has undergone a dramatic expansion in recent years. Studies using in vitro culture systems have led to important insights regarding nutrient utilization and the regulation of metabolic pathways by describing how cancer cells exploit existing metabolic programs to fuel proliferation and survival. Examining tumor metabolism in vivo introduces new complexities, but taking this step is critical to gain a deeper understanding of how whole animal physiology impacts nutrient availability, as well as to appreciate the role of tumor heterogeneity and interactions between different cell types in tissues. Gaining this insight will be critical to developing new therapies that exploit metabolic pathways and improve patient therapies. In this review, we first discuss the current understanding of cancer cell metabolism gained primarily from cell culture studies, and then focus on emerging insights arising from experiment using patients and mouse models with the intent of highlighting the strengths and limitations of each experimental context and calling attention to key unanswered questions.
Defining proliferative metabolism using cell culture
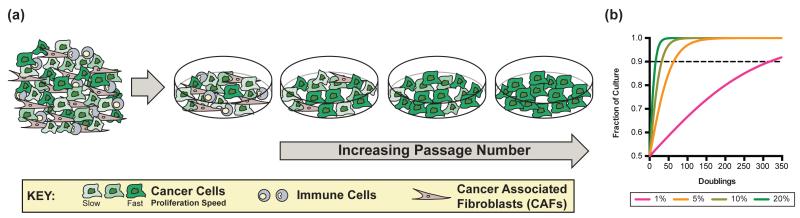
Cancer is defined by unconstrained proliferation of transformed cells. Establishing cell lines in culture selects for the fastest growing malignant clones from the tumor with concomitant loss of non-dividing and slowly proliferating cancer cells as well as any other cell types that were part of the original tumor tissue (Figure 1A). To illustrate, one clone with a slight 5% proliferation advantage will almost completely eliminate a second clone in fewer than 65 generations (Figure 1B). Thus by its nature, cell culture selects for a relatively homogeneous population of cancer cells, generating clean systems with which to investigate the contributions of specific oncogenic mutations to metabolic programs and the underlying metabolic requirements of cell proliferation. The common oncogenic drivers Ras and Myc both promote cell-autonomous metabolic changes associated with malignant transformation, namely the diversion of metabolic substrates into anabolic (see Glossary) pathways. Oncogenic Ras increases glucose and glutamine consumption [1,2], while Myc enhances glutamine metabolism through a transcriptional program that increases expression of genes involved in glutamine uptake and catabolism (see Glossary) [3,4]. Myc also ties increased glutaminolysis to changes in glucose metabolism [5], and can directly control expression of genes involved in aerobic glycolysis [6]. Mutations in other key cancer genes also influence metabolism. Loss of p53 promotes glucose uptake and metabolism [7,8], and can impact how glucose is used by cells [9]. In all cases, genetic alterations associated with cancer are accompanied by metabolic alterations that favor anabolism, enabling the acquisition and utilization of nutrients to satisfy increased ATP (see Glossary) demands and produce the nucleotides, lipids and proteins needed for rapid cell division [10].
Figure 1.
Establishing tumor-derived cell lines in culture selects for the fastest proliferating clones in the population, and non-dividing and less proliferative cells are lost upon serial passaging. This inevitable consequence of cell culture is illustrated graphically in (A), using the example of cell line generation from a tumor. Panel (B) shows a model demonstrating how many cell doublings are required for a clone to take over the culture population if that clone has the proliferation advantage indicated (key; proliferation advantage indicated as % faster than control doubling time). The model assumes competition between two distinct clones plated at equal density with one clone having a fixed advantage that is invariant over time. We further define one clone representing greater than 90% of the cultured population as having taken over the culture. This threshold is reached after 317 doublings with a 1% proliferation advantage, 64 with 5%, 32 with 10%, and only 16 with 20%. Additional details of the model are included as supplemental material.
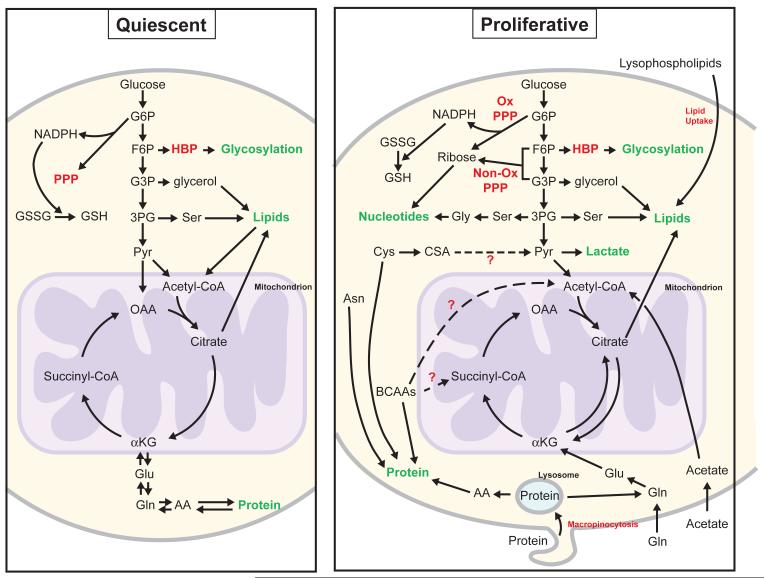
The metabolic differences between proliferating and non-proliferating cells have received less scrutiny. Studies utilizing mammalian primary fibroblasts and lymphocytes where culture conditions are manipulated to promote quiescent (see Glossary) or proliferative states have demonstrated that in contrast to proliferating cells, quiescent cells favor catabolic metabolism [5,11] (Figure 2). Maintaining homeostasis requires nutrient breakdown to generate ATP, as well as NADPH production to cope with redox stress [11,12]. Quiescent cells also strive to balance fatty acid and protein degradation with synthesis [11], a finding consistent with the absence of an increase in cell mass in these non-proliferating cells. Nevertheless, these cultured cells still rely on glucose and glutamine, while many differentiated mammalian tissues use other nutrients [13]. For example, the heart can consume fatty acids, glucose, ketones, or amino acids to support the large amount of ATP required for electrical activity and continuous mechanical contraction [14], while the brain relies almost exclusively on glucose metabolism, only switching to ketones when glucose is not available [15]. Thus, caution is needed when generalizing studies of specialized quiescent cell systems in culture to diverse cell types in intact tissues in an organism.
Figure 2.
/B> Differences between quiescent and proliferative metabolism. In quiescent metabolism (left), cells balance catabolic and anabolic process such as lipogenesis and β-oxidation, or protein synthesis and degradation. In proliferative metabolism (right), anabolic processes are favored to allow the net production of biomass. Red labels indicate metabolic pathways not shown in detail. Green labels indicate macromolecules (or protein modifications such as glycosylation) that represent the starting points of catabolic pathways and/or the end points of anabolic pathways. Broken arrows represent potential fates of the identified nutrients that have not been extensively investigated. The mitochondrion is represented in purple to illustrate compartmentalized reactions. Abbreviations: AA = amino acids, Asn = Asparagine, αKG = α-ketoglutarate, BCAAs = branched chain amino acids, CSA = cysteine sulfinic acid, Cys = cysteine, F6P = fructose-6-phosphate, Gln = glutamine, Glu = glutamate, Gly = glycine, GSH = reduced glutathione, GSSG = oxidized glutathione, G3P = glyceraldehyde-3-phosphate, G6P = glucose-6-phosphate, HBP = hexosamine biosynthetic pathway, NADPH = reduced nicotinaminde adenine dinucleotide phosphate, OAA = oxaloacetate, ox = oxidative, PPP = pentose phosphate pathway, Pyr = pyruvate, Ser = serine, 3PG = 3-phosphoglycerate.
Nevertheless, the different metabolic phenotypes of proliferating and non-proliferating cells in culture illustrate that these states have different metabolic requirements. At a first approximation, proliferating cells favor biomass production while non-proliferating cells favor biomass maintenance. Relevant to understanding tumor metabolism, not all cancer cells actively proliferate in many solid tumors [16], and the mechanics of serially passaging cancer cell lines selects against quiescent or more slowly proliferating cancer cells (Figure 1B), limiting the study of these tumor cell populations to date. It is important to recognize that studies of cancer cell metabolism in culture fail to capture the metabolic phenotype of less proliferative tumor cells.
Glucose and glutamine: primary substrates of proliferative metabolism in vitro
Glucose
In addition to taking up more glucose, proliferating cancer cells in culture metabolize glucose differently than non-proliferating cells, converting most of the pyruvate derived from glucose to lactate, rather than oxidizing it via the tricarboxylic acid (TCA) cycle. For most normal cells, increased conversion of glucose to lactate is favored in oxygen-limited conditions, while cancer cells exhibit this phenotype even when oxygen is abundant, an observation first described by Otto Warburg [17]. Termed aerobic glycolysis or the Warburg effect, this metabolic phenotype is a well described feature of cancer cells that has been extensively studied [10].
Increased glucose flux (see Glossary) through glycolysis is thought to promote shunting of metabolites into branch pathways for the synthesis of macromolecules (see Glossary) [10]. For instance, metabolism of glucose-6-phosphate via the oxidative arm of the Pentose Phosphate Pathway (PPP) produces NADPH and ribose-5-phosphate, two critical components for new cell generation. NADPH is critical for managing redox stress and for reductive biosynthetic reactions, while ribose-5-phosphate is a required precursor for de novo nucleotide synthesis [18]. Additionally, some cancer cells depend on flux of downstream glycolytic intermediates into the non-oxidative arm of the PPP for ribose-5-phosphate production (Figure 2) [19,20]. Glucose metabolism also contributes to the generation of nucleotide bases, and slowing this production can limit proliferation in some situations [21].
Diverting fructose-6-phosphate, another product of glycolysis, into the hexosamine biosynthetic pathway (HBP) provides the necessary substrates for glycosylation of proteins and lipids, an abundant modification that has been implicated in several aspects of tumor progression [22]. Both oncogenic K-ras and phosphoinositide 3-kinase (PI3K)/Akt pathway activation have been implicated as important drivers of glucose flux through the HBP [20,23], linking these signaling pathways directly to this aspect of glucose metabolism. Thus, the HBP serves as an integration hub, ensuring that continued mitogenic signaling and proliferation occur only under favorable conditions.
Several branch points of glucose metabolism provide components for de novo lipogenesis to generate new lipid membranes in dividing cells. Oxidation of glucose carbon to citrate can yield cytosolic acetyl-CoA, which is used for the synthesis of fatty acids and sterols. Glycolytic products contribute to two different lipid head groups as well. Glyceraldehyde-3-phosphate can be further metabolized to glycerol-3-phosphate to form the glycerol component of triacylglycerides and membrane phospholipids [18]. Synthesis of ceramides and other related structural and signaling lipids requires serine, one source of which is 3-phosphoglycerate from glycolysis [18]. Interestingly, amplification of the gene encoding phosphoglycerate dehydrogenase (PHGDH), the enzyme catalyzing the initial step of the serine biosynthesis pathway from glucose, is found in some cancers [24,25]. While knockdown of PHGDH in amplified cell lines impairs proliferation, exogenous serine is unable to rescue this effect, indicating the flux through this pathway contributes to proliferation in a manner beyond serine production alone.
Glutamine
After glucose, glutamine represents the second most consumed nutrient in cell culture [26]. Glutamine is the most abundant amino acid in both plasma [27] and culture media, and its consumption in large excess relative to demand for nucleotide and protein synthesis is well described [28,29]. With little glucose-derived carbon entering the TCA cycle, glutamine oxidation allows cells to replenish TCA cycle intermediates for use in biosynthetic reactions [1,29]. Like their glycolytic counterparts, TCA metabolites are required for anabolic processes to sustain cell proliferation [18]. For example, several non-essential amino acids necessary for nucleotide and protein synthesis, such as aspartate and glutamate, can be derived from TCA cycle intermediates. Glutamine also serves as a primary source of nitrogen for proliferating cells in culture, which is likewise critical for de novo nucleotide and amino acid synthesis [29]. Oncogenic mutations driving enhanced uptake and utilization of glutamine ensures a continuous and abundant supply of nitrogen for transformed cells. The exact mechanism by which this nitrogen is extracted and glutamine carbon is further metabolized can differ, as cells lines with increased Myc favor glutaminolysis (see Glossary) with nitrogen released as ammonia release while mutant K-ras can favor transamination of the nitrogen to α-ketoacids [3,4,30].
Other amino acids as nutrients for cancer cells
Although much of the investigation of nutrient utilization in cell culture has focused on the fates of glucose and glutamine, cancer cells in vivo have access to other nutrients including amino acids that could support proliferation. For instance, although many cancer cells synthesize serine and glycine de novo from glucose, recent studies have suggested uptake of these nutrients from media can also be important. Profiling of spent media from the NCI-60 panel of cell lines found that glycine levels correlated with proliferation rate [26]. This finding might be reflective of serine depletion and amino acid use during proliferation [31] since serine uptake is dramatically increased in the fastest proliferating cells to a rate almost 8-fold that of glycine [32]. Although serine and glycine can be interconverted by serine hydroxymethyl transferases (SHMT), the directionality of this interconversion can have a profound impact on proliferating cells. In particular, serine can supply the necessary intracellular glycine and one-carbon units for nucleotide biosynthesis in the absence of glycine, while glycine cannot compensate for serine-free conditions in all cells [31].
When serine levels are low, excess glycine depletes one-carbon units and blocks de novo nucleotide synthesis, halting proliferation [31]. The mitochondrial isoform of SHMT (SHMT2) likely plays a critical role in this block, as it generates folates from serine for nucleotide biosynthesis in cell lines [33,34], and coordination of this enzyme with activity of the glycine cleavage system can impact proliferation of poorly vascularized tumors [35]. Like glutamine, cells also take up serine in excess of that needed for protein synthesis. This is not always true for glycine, a finding consistent with serine generating some glycine used to make proteins [36]. However, rates of DNA synthesis correlate directly with glycine uptake [36] in agreement with studies showing direct incorporation of 13C-glycine from the culture media into the purine backbone of nucleotides [26]. Because glucose metabolism is also important for nucleotide base synthesis [21], understanding the use of glucose-derived serine and glycine relative to exogenous amino acids will be important. Glycine also contributes to glutathione synthesis, which is critical in p53 null cells [32]. Thus, while these studies highlight distinct contributions of available extracellular and newly synthesized serine and glycine, they also emphasize how metabolic context can influence pathway fluxes in cells with concomitant effects on proliferation and growth. Determining what regulates serine and glycine interconversion, and how the use of these amino acids in different metabolic processes is regulated, represent important open question for the field.
Catabolism of amino acids other than glutamine may contribute to tumor progression as well. For example, expression of branched chain amino acid transaminase 1 (BCAT1) is necessary for the growth and progression of isocitrate dehydrogenase (IDH) wild-type gliomas [37]. These wild-type IDH gliomas represent a distinct subset of brain tumors with worse clinical outcomes than those with gain-of-function mutations in IDH [38]. As branch chain amino acids (BCAAs) are abundant amino acids in serum [27] (Table 1), BCAT1 expression may allow BCAAs to serve as an additional carbon source for the TCA cycle [37]. Nevertheless, the contribution of BCAA breakdown to cancer cell metabolism in this subset of gliomas, as well as in other cancers, requires further characterization.
Table 1.
The nutrient levels in plasma/serum that are also found in standard tissue culture media as reported across several studies. The concentrations of the same nutrients in different standard media formulations are also shown for comparison. All concentrations are in μM.
| PLASMA/SERUM | MEDIA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | BME | MEM | DMEM | RPMI | ||||||||
| Blau et al.a [100] |
Albrittonb [101] |
McMenamy et al.c [102] |
Newgard et al.d [103] |
Pitkanen et al.e [104] | Stein and Mooref [27] |
Wuu et al.g [105] |
Albrittonh [101] |
||||||
| Males 40- 59yrs |
Females 40-59yrs |
||||||||||||
| Alanine | 146-494 | 449 | 320 | 367.3 | 406 | 347 | 383 | 243.7 | 662 | ||||
| Arginine | 28-96 | 132.0 | 68 | 115.3 | 128 | 99 | 86.6 | 25.5 | 55.7 | 100 | 600 | 400 | 1149 |
| Asparagine | 32-92 | 16.5 | 65 | 58 | 43.9 | 77 | 379 | ||||||
| Aspartate | 2-9 | 17 | 18 | 22.5 | 9.1 | 150 | |||||||
| Cystine | 24-54 | 58.3 | 49.1 | 23.2 | 33.0 | 50 | 100 | 200 | 208 | ||||
| Glutamate | 6-62 | 54.4 | <20 | 81.2 | 66 | 45 | 47.6 | 19.5 | 224.3 | 136 | |||
| Glutamine | 466-798 | 450 | 561 | 556 | 568 | 261.8 | 1000 | 2000 | 4000 | 2055 | |||
| Glycine | 147-299 | 240 | 235 | 328.4 | 262 | 315 | 205 | 168.7 | 253 | 400 | 1334 | ||
| Histidine | 72-108 | 90.2 | 88 | 81.9 | 107 | 96 | 74.1 | 65.5 | 103.1 | 50 | 200 | 200 | 97 |
| Isoleucine | 46-90 | 122.0 | 57 | 149 | 74 | 58 | 67.9 | 95.1 | 114.4 | 200 | 400 | 800 | 382 |
| Leucine | 113-205 | 145 | 79 | 151 | 121 | 129 | 121.8 | 183 | 200 | 400 | 800 | 382 | |
| Lysine | 135-243 | 205 | 143 | 232 | 203 | 186 | 182.9 | 438 | 200 | 400 | 800 | 219 | |
| Methionine | 13-37 | 33.5 | 28 | 27.6 | 33 | 26 | 25.5 | 17.1 | 67.0 | 50 | 100 | 200 | 101 |
| Phenylalanine | 46-74 | 84.8 | 56 | 61.6 | 67 | 60 | 50.9 | 55.5 | 145.3 | 100 | 200 | 400 | 91 |
| Proline | 97-297 | 142 | 158.1 | 205 | 131.1 | 156 | 174 | ||||||
| Serine | 89-165 | 85 | 116.7 | 132 | 155 | 107 | 117.9 | 400 | 285 | ||||
| Threonine | 92-180 | 168 | 135 | 148 | 142 | 117 | 97.1 | 294 | 200 | 400 | 800 | 168 | |
| Tryptophan | 25-65 | 53.9 | 21 | 54 | 51 | 54.4 | 126.4 | 102.8 | 20 | 50 | 80 | 24 | |
| Tyrosine | 37-77 | 82.8 | 70 | 67.1 | 80 | 62 | 56.8 | 54.2 | 138.0 | 100 | 200 | 400 | 110 |
| Valine | 179-335 | 239 | 174 | 235.3 | 267 | 204 | 246 | 176.4 | 367 | 200 | 400 | 800 | 171 |
| Glucose | 4996i | 5578i | 5500 | 5500 | 25000 | 11100 | |||||||
| Pyruvate | 27-160 | 136 | 90 | 1000 | |||||||||
plasma, males, mean ± 2SD
plasma
plasma, mean
serum, median; lean subjects
serum, mean
plasma, mean
serum, mean
plasma
fasted
Examination of metabolites from patient-derived glioma samples suggested that partial cysteine catabolism and accumulation of the cysteine sulfinic acid (CSA) correlates with increasing tumor grade [39]. Although most cancer research involving cysteine metabolism has focused on its contribution to glutathione [40,41], blocking cysteine catabolism to CSA independently inhibits tumor growth [39]. The ultimate fate of CSA in these tumors remains to be determined.
The catabolism of additional amino acids to support proliferation is a subject of interest; however, existing data suggests that most amino acids are used primarily for protein synthesis in dividing cells [36,42]. Breakdown of amino acids might be more important to maintain ATP and cellular redox state to support cell survival when other nutrients are limiting. Consistent with this assertion, MYC induces a switch from proline catabolism to anabolism to support proliferation [43].
Scavenging macromolecules, reductive TCA flux, and acetate
In addition to the metabolism of glucose and free amino acids, cancer cells can catabolize extracellular protein to supplement their nutritional requirements in some genetic and environmental contexts. Oncogenic Ras expression stimulates macropinocytosis, a process that enables clathrin-independent uptake of extracellular material [44]. Some of this extracellular material can then be trafficked to the lysosome where protein is degraded to provide amino acids that can enter central carbon metabolism [45]. This process allows tumor cells in culture to proliferate when glutamine or essential amino acids are limiting [45,46], and pharmacologic inhibition of macropinocytosis can slow growth of xenograft tumors [45]. Understanding the extent to which tumor cells rely on metabolizing extracellular material in their environment in vivo is an area of active study.
Extracellular lipids can also be an important fuel for cancer cells. In cell culture, glucose contributes substantial amounts of carbon for the de novo synthesis of lipids. In the setting of both oncogenic Ras expression or hypoxia, however, uptake of lysophospholipids can also contribute to fatty acid pools [47]. This uptake compensates for the decreased flux of glucose carbon through pyruvate dehydrogenase (PDH) observed in both conditions [1,48]. Hypoxia also alters the fate of glutamine carbon entering the TCA cycle by favoring reductive rather than oxidative metabolism to serve as a source of acetyl-CoA for fatty acid synthesis [48-50], in part to compensate for decreased contribution of glucose to citrate [51].
Other sources of carbon can also be used for lipid synthesis. Recent data suggest that despite being present at relatively low levels in cell culture media and serum, acetate can also contribute to fatty acid synthesis and TCA cycle metabolism [52-54]. Acetate is converted to acetyl-CoA by the action of acetyl-CoA synthetases (ACSS), and ACSS2 is critical for growth of at least some tumor types in vivo [55]..
Location, location, location
The many studies showing cancer cells can utilize a variety of fuels in culture suggests metabolic flexibility exists despite genetic and environmental contexts favoring the use of certain substrates. This argues that the metabolism of cancer cells in tumors will be influenced heavily by nutrient availability, and uncovers a potential limitation of cell culture in that it does not necessarily mimic nutrient conditions in tissues. Changing culture conditions to maintain constant nutrient levels is one approach that has been used to generate insight [56], however standard cell culture media contains many-fold excess the nutrient levels in plasma and the ratios of different nutrients are also different from those found in vivo (Box 1 and Table 1). There are other nutrients, including acetate, which are found in blood but not added to standard media (Table 2). Culturing cells in hypoxia or low levels of specific nutrients maybe be informative, but in most instances these perturbations are accompanied by superphysiologic levels of other nutrients in the media, failing to mimic the reality of hypoperfusion. As the best approaches to model the tumor environment remains unclear, efforts to study the metabolism of tumors in vivo will be critical to take the next step in understanding cancer metabolism.
Box 1. Development of cell culture media.
Early cell culture media required supplementation with large amounts of serum such that detailed investigation of growth requirements in vitro remained impractical. In 1955-56, Harry Eagle and colleagues outlined, for the first time, the specific amino acids vitamins, salts and other nutrients that enabled the continued culture of two cell lines, adult mouse fibroblasts and HeLa cells, with minimal added serum [28,108,109]. These studies formed the basis for the original standardized media formulation that became known as Basal Medium Eagle (BME) (Table 1). Two observations stand out from these studies. The first is that glutamine was considered separate from other amino acids and equivalent to glucose as a requirement for proliferation in culture. The second is that although the optimal concentrations of amino acids to support growth were determined empirically, concentrations were deliberately kept close to the physiologic ranges reported by Albritton [101,108].
Additional work over the next several years by Eagle and colleagues led to the development of minimal essential media (MEM) [110]. This media, which allowed for the culture of additional cell lines, contained two-fold or greater increases in the amino acids included in BME (Table 1). These changes were intended to more closely conform to the amino acid composition of total protein in cultured human cells [110]. In 1960, Marguerite Vogt and Renato Dulbecco, in their studies of the interactions between the polyoma virus and embryonic cells, further modified MEM to include four-fold the concentration of amino acids and vitamins described by Eagle [111]. This media, now known as DMEM (Dulbecco’s Modified Eagle Medium), which also contains glucose at five-times the concentration that is normal in blood (Table 1), has become a standard culture media used today.
In a manner analogous to Eagle’s studies of the previous decade, George Moore and colleagues at the Roswell Park Memorial Institute (RPMI) undertook a similar effort with the primary focus on the development of culture medias for both normal and transformed blood cells. This work spawned a large series of similar medias, including RPMI 1640 [112], which was originally designed for the culture of normal primary leukocytes and has become another workhorse of modern tissue culture. RPMI differs mainly from DMEM in the number of added amino acids (Table 1).
Since these early studies, there have been many additional modifications to generate specialized media, although in large part these are all based on the pioneering and painstaking work of Eagle and Moore, with the major differences in most cases being the addition of new growth factors and hormones. It is now possible to grow some cells in completely defined (serum-free) media, however the nutrient composition is still based on the classic formulations.
Table 2.
Plasma/serum levels of metabolites not added to standard tissue culture media that could be a source of nutrients for tissues in vivo. All metabolites listed were detectable in at least 90% of control patients, and all concentrations are in μM. All values are from Hoffman et al [106], except where indicated.
| Nutrient | Average | Rangea |
|---|---|---|
| Lactate | 1700 | 700–3300 |
| Glycolate | 27 | 9–42 |
| 2-Hydroxybutyrate | 54 | 8–80 |
| 3-Hydroxybutyrateb | 180 | 22–170 |
| 3-Hydroxyisobutryrate | 20 | 4–48 |
| Glycerate | 10 | n.d.–24 |
| Pyroglutamate | 51 | 13–161 |
| Acetoacetateb | 21 | n.d.–86 |
| 2-oxo-3-methyl-n-valerate | 18 | 8–31 |
| 2-oxoisocaproate | 28 | n.d.–58 |
| Laurate | 12 | 2–37 |
| Isocitrate | 6 | n.d.–10 |
| Citrate | 190 | 30–400 |
| Acetate [107] | 51 | 2–99 |
| Triglycerides | 450–1710 |
n.d. = not detected
ketone bodies
Investigating the metabolic requirements of cells in vivo adds many layers of biological and experimental complexity. Nutrient availability in vivo can vary between cell types, both within and across organs. For example, the architecture of the liver results in gradients of oxygen and nutrients across zones of hepatocytes [57]. Tumors also have gradients of glucose and oxygen availability caused by disrupted microvasculature, leading to variable nutrient uptake across tumors and in comparison with adjacent normal tissues [58-61]. Furthermore, homeostatic regulation of metabolites and the involvement of multiple tissues and cell types in determining nutrient levels in vivo complicates the manipulation of nutrient levels for experimental purposes [32].
Differences in nutrient availability among normal tissues may dictate the metabolic programs utilized by those tissues. Interestingly, it appears these programs constrain the metabolic repertoire available to tumor cells in vivo. Expression analysis of metabolic genes from 22 human tumor types and normal tissue demonstrated that the expression pattern of tumors more closely resembled those of their tissue of origin than they did tumors from other tissues [62]. Tissue-of-origin even constrains the effects of the same oncogenic drivers in different tissues, with Myc-driven liver and lung cancers exhibiting different phenotypes related to glutamine metabolism [63]. Further complicating matters, tumors driven by different oncogenes that arise in the same tissue can also exhibit different metabolic programs [63]. Thus, while upregulation of some central metabolic pathways such as nucleotide biosynthesis occurs irrespective of tissue type and driver mutation, the use of other pathways is heterogeneous, possibly reflecting both the tissue of cancer origin and adaptations to available nutrients [62].
Tumor cells also have metabolic interactions with normal host tissues that can have a profound effect on cancer pathogenesis. For example, whole body metabolic alterations such as obesity and diabetes influence the development and progression of many cancers, including pancreatic cancer [64-67]. Pancreatic cancer can in turn alter whole body metabolism causing new onset diabetes and cachexia in many patients and tumors can secrete factors that can alter whole body metabolic rate [68-72],. Given the vagaries of nutrient supply in vivo, these whole-body metabolic interactions suggest tumors may actively strive to manipulate their environment to induce conditions favorable conditions for their growth [73]. To this end, early pancreatic cancers can drive increased whole-body protein turnover with concomitant amino acid release many years prior to diagnosis [74].
Metabolic heterogeneity of tumors
Another essential difference between tumor cells in vitro and in vivo is that while every cell in vitro must proliferate or be lost at passaging, in vivo only a fraction of tumor cells appear to be actively dividing at any given time [16]. Dramatic cell-to-cell variation in the expression profiles of cell cycle markers in glioblastoma cells has been observed and is consistent with variable proliferation throughout the tumor [75]. This same study also found hypoxia-related gene expression associated with intratumor O2 gradients, providing evidence for intratumor heterogeneity of metabolic programs [75]. Differences in pyruvate kinase activity requirements also suggest that proliferating and non-proliferating tumor cells use glucose differently, and that how glucose is metabolized can impact the ability of cells to proliferate in vivo [16].
Spatial heterogeneity of nutrient availability and metabolic phenotypes allows for metabolic cooperation between different populations of cells within tumors. For example, lactate released by hypoxic cancer cells can serve as oxidative fuel for the growth of neighboring normoxic cells [76,77]. It has also been proposed that tumor cells might induce a similar metabolic phenotype in cancer associated fibroblasts (CAFs), inducing them to release lactate, dipeptides and ketone bodies to fuel oxidative tumor metabolism [78-80]. Metabolic interactions with non-malignant tumor stromal cells can also directly influence disease progression, metastasis and redox status. Ovarian tumors, which frequently metastasize to the omentum, are drawn there in part by release of adipokines from omental adipocytes [81]. Once there, direct transfer of lipids from adipocytes to tumor cells supports cancer cell proliferation as adipocytes upregulate lipolysis in concordance with an increase in β-oxidation in tumor cells [81]. In chronic lymphocytic leukemia, metabolism of cystine by bone marrow stromal cells generates cysteine to support glutathione synthesis in the tumor cells [82]. New methods to use mass spectrometry-based assessment of the spatial distribution of metabolites in tissue are likely to lend further insight [83,84], although additional new approaches will need to be developed to elucidate the complex interactions between different cells types in tumor tissue.
Measurement of tumor metabolism in vivo
Studies of cancer metabolism in vivo have found that, consistent with cell culture models, most tumors increase nutrient uptake relative to surrounding normal tissue. Physicians leverage this to image tumors using [18F]-2-deoxyglucose positron emission tomography (FDG-PET), an approach that is useful for cancer staging and as a measure of therapeutic response [85,86]. [18F]-glutamate and glutamine analogs are also being studied as tumor imaging agents [87,88], as is [11C]-acetate [89], and these agents may help track FDG-PET negative tumors [88]. To date, PET tracers (see Glossary) have been limited in providing insight beyond nutrient uptake because distinguishing labeled species in downstream metabolism is not feasible via this imaging approach. Nevertheless, glutamine incorporation into protein and conversion to glutamate has been assessed by tissue sampling [88].
Both radioactive and stable isotopes have been used to study cancer metabolism phenotypes in vivo [90]. While earlier studies were limited to investigations of whole body metabolism, they confirmed increased glucose oxidation [91,92], protein turnover [93] and lipolysis [94] in tumor-bearing patients. More recent studies have coupled the use of stable isotope tracers to nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS) to determine the exact fates of different nutrients in tissues. In one example, bolus injections of [U-13C]glucose found that the Warburg effect in vivo may not be universal, as Myc-driven liver tumors showed increased glucose to lactate conversion while Met-driven ones did not [63]. Use of NMR-spectroscopy can also provide insight into dynamic changes in tumor metabolism in live animals [95], although even with hyperpolarization methods to improve sensitivity, the diversity of metabolites that have been tracked to date is limited [96].
To compare nutrient fates across tissues in vivo, methods to achieve steady state plasma enrichment of stable isotope tracers can be used [16,97]. Infusions of [U-13C]glucose into patients with FDG-PET positive intracranial tumors revealed that while glucose was metabolized to lactate as anticipated, a large amount of glucose was also oxidized via the TCA cycle [98]. Glucose was also found to contribute to de novo glutamine synthesis in the brain tumors studied [98]. Labeled glucose infusions into mice with orthotopic human glioblastoma transplants further confirmed a higher than expected flux from glucose into the TCA cycle as well as glutamine production from glucose [97]. While studies of additional in vivo models are needed to know whether brain cancers represent a unique case in either metabolic phenotype or the similarities between mouse and human tumors (Box 2), the results illustrate that nutrient use in vivo can be different from what is observed in most cell lines.
Box 2. Interspecies differences in metabolism.
More than a decade ago Rangarajan and Weinberg intoned, “mice are not small people,” when discussing the use of mice to model human cancer [113]. However, mice remain a powerful tool to study cancer with their ease of genetic manipulation, relatively short generation time and many physiologic similarities with humans. Nevertheless, there are important species-specific differences. Spontaneous cancers in mice have both a different natural history and tissue spectrum than humans [113]. Introduction of tissue-specific pro-neoplastic genetic alterations are powerful tools to model tumorigenesis in an organ, and while these often recapitulate many pathological features of the human disease there are also differences including far less genetic heterogeneity among tumor cells in mice. Additionally, the seven-fold higher metabolic rate in rodents [114] may play a role in how tissues respond to the genetic changes and the altered signaling pathways that are found in human cancers.
Nutrient handling can also differ dramatically across mammalian species. Although mice and rats are commonly used to study type-2 diabetes, rodents show differences from humans in glucose metabolism at every level from glucose-regulatory transcription factors to mechanisms of insulin secretion to the primary tissue of glucose disposal [115]. Unsurprisingly, amino acids also display species-specific differences in their requirement for protein synthesis and the relative catabolism of individual amino acids across tissues [116,117]. These differences in macronutrient (see Glossary) handling are likely a consequence of the fact that mammalian species consume diets with very different compositions. Failing to account for these metabolic differences could affect the translatability of findings when different species are used as a research tool to study metabolism and its impact on disease [115].
Despite these caveats, important insight can be gained from mouse models of human cancer and provide a powerful tool to study cancer metabolism in vivo. Both humans and mice have the same elevations in branched chain amino acids (BCAAs) initiated by early stage pancreatic cancers, and the ability to study this phenotype in mice uncovered that this change in amino acid levels reflects increased protein turnover [74]. Similarly, detailed studies of glucose metabolism of human glioblastomas in mice showed nearly identical phenotypes to what was observed in tumors of patients with glioblastomas [97,98]. Given the complexity of intra-operative studies to gain metabolic insight on human tumors in vivo, the concordance with findings in animal studies argues that true mechanistic insight into human disease can be gained from more tractable experiments using mouse models. Careful validation of phenotypic findings in each species enhances the likelihood that mechanistic dissection of phenotypes in mice will be informative for improved patient care.
Concluding remarks
The study of cancer metabolism in culture has advanced our understanding of the metabolic requirements for proliferation. In vitro cell culture will continue to allow investigation of the metabolic characteristics of single cell types and the effects of discreet genetic and environmental manipulations. Indeed, numerous examples of metabolic symbiosis between cell types have been defined using culture systems [77,79-82]. To continue to derive insight from culture systems, however, a greater appreciation for the limitations discussed above will be important (Box 3). The limited data from in vivo studies argues that cancer metabolism in whole animals can be different from that observed in culture. Whether this difference reflects the metabolic phenotype of less proliferative cancer cell populations that form the bulk of tumors remains to be determined, but studies in vivo will enable the study of these cells as well as cancer types that have not been amenable to cell culture. Studies in vivo will also be important to probe tumor-host tissue interactions that can have a major impact on patient outcomes [70]. Finally, the ability to understand the metabolic changes associated with tumor growth in different tissue contexts will also benefit from studies of cancer metabolism in vivo, and inform our understanding of metastasis [99]. Most importantly, in vivo studies will help guide more effective targeting of those metabolic changes that aid tumor progression in relevant clinical settings.
Box 3. Outstanding Questions.
How does prolonged culture shape the metabolic dependencies of cancer cells over time?
What is the importance of different metabolic substrates such as cysteine, acetate, BCAAs and protein relative to glucose and glutamine across a variety of cancer types?
How do the metabolic requirements of non-proliferating tumor cells differ from proliferating tumor cells and can conditions be developed to study these non-proliferating cells in culture?
What are the metabolic phenotypes of “unculturable” tumor cell populations?
What is the metabolic relationship between tumor cell and normal cell populations present within the tumor and in distant tissues, and can systems be developed to investigate these symbioses in vivo?
How does tissue of origin and tissue site of tumor growth affect the metabolic phenotype of cancer cells in vivo?
How faithfully do cancer models recapitulate the metabolic alterations observed in human cancers?
Supplementary Material
Highlights.
Quiescent metabolism is catabolic while proliferative metabolism requires anabolism.
Metabolic phenotypes in cell culture reflect proliferative metabolism.
Most cancer cells in vivo are not proliferating rapidly and may display distinct metabolism.
The in vivo context of a tumor cell, including hypoperfusion, can strongly influence metabolism.
Metabolic cooperation can exist among different cells within a tumor.
Acknowledgements
We thank all members of the Vander Heiden lab for their thoughtful discussion and feedback throughout the course of developing this manuscript, and specifically thank A.M. Hosios and D.Y. Gui for assistance with generating the model of proliferation in culture and S.M. Davidson for discussions regarding metabolism in vivo. We thank R.J. Deberardinis, A.M. Hosios and W.J. Israelsen for input into compiling nutrient values in plasma. We also thank the reviewers for their time and thoughtful critiques to improve our manuscript. For space limitations we regret being unable to cite many excellent papers. J.R.M. was supported by F30 CA183474. M.G.V.H. acknowledges support from the Burroughs Wellcome Fund, the Lustgarten Foundation, the American Association for Cancer Research and the NCI.
Glossary Box
- Anabolic:
Biochemical reactions requiring the input of energy for the synthesis of new macromolecules in cells.
- ATP:
Adenosine triphosphate. The primary energy currency of the cell.
- Catabolic:
Biochemical reactions that provide energy for use by cells via the oxidation of various nutrients.
- Flux:
Rate of metabolite flow per unit time through a metabolic pathway.
- Glutaminolysis:
Oxidative metabolism of glutamine by the TCA cycle.
- Macromolecules:
Large polymers that compose much of the structure of a cell including DNA, proteins and lipids
- Macronutrient:
General term describing nutrients required in large amounts such as carbohydrates, proteins, and fats.
- NADPH:
Reduced nicotinamide adenine dinucleotide phosphate. A co-factor for intracellular redox reactions that is used primarily as a source of electrons for reduction reactions. It is important for many anabolic reactions and for coping with reactive oxygen species.
- Quiescent:
Non-proliferating.
- Tracer:
A version of a metabolite in which one or more of the atoms has been replaced with a different isotope version of that atom such that the difference can be tracked using mass spectrometry or nuclear magnetic resonance spectroscopy. If the isotope used is radioactive, radioactivity detection methods can also be used.
Footnotes
Disclosures M.G.V.H. is a consultant and scientific advisory board member for Agios Pharmaceuticals, which seeks to target cancer metabolism for therapeutic benefit.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Molecular Systems Biology. 2011;7:523–523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vizan P. K-ras Codon-Specific Mutations Produce Distinctive Metabolic Phenotypes in Human Fibroblasts. Cancer Research. 2005;65:5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- 3.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R, et al. The Transcription Factor Myc Controls Metabolic Reprogramming upon T Lymphocyte Activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim H, et al. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proceedings of the …. 1997 doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartzenberg-Bar-Yoseph F, et al. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Research. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 8.Bensaad K, et al. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell. 2006 doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 9.Maddocks O, Vousden KH. Metabolic regulation by p53 - Springer. Journal of molecular medicine. 2011 doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns RA, et al. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 11.Lemons JMS, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khairallah M. Profiling substrate fluxes in the isolated working mouse heart using 13C-labeled substrates: focusing on the origin and fate of pyruvate and citrate carbons. AJP: Heart and Circulatory Physiology. 2003;286:H1461–H1470. doi: 10.1152/ajpheart.00942.2003. [DOI] [PubMed] [Google Scholar]
- 13.Metallo CM, Vander Heiden MG. Understanding Metabolic Regulation and Its Influence on Cell Physiology. Molecular cell. 2013;49:388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake KJ, et al. Amino acids as metabolic substrates during cardiac ischemia. Experimental …. 2012 doi: 10.1258/ebm.2012.012025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen OE, et al. Brain metabolism during fasting. J Clin Invest. 1967;46:1589–1595. doi: 10.1172/JCI105650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Israelsen WJ, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warburg O, Posener K. Ueber den Stoffwechsel der Carcinomzelle. 1924 Die Naturwissenschaften. [Google Scholar]
- 18.Lunt SY, Vander Heiden MG. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 19.Boros LG, et al. Transforming Growth Factor β2 Promotes Glucose Carbon Incorporation into Nucleic Acid Ribose through the Nonoxidative Pentose Cycle in Lung Epithelial Carcinoma Cells. Cancer Research. 2000 [PubMed] [Google Scholar]
- 20.Ying H, et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lunt SY, et al. Pyruvate Kinase Isoform Expression Alters Nucleotide Synthesis to Impact Cell Proliferation. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.10.027. DOI: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 23.Fang M, et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143:711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain M, et al. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein WH, Moore S. The free amino acids of human blood plasma. J Biol Chem. 1954;211:915. [PubMed] [Google Scholar]
- 28.Eagle H, et al. The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J Biol Chem. 1956;218:607–616. [PubMed] [Google Scholar]
- 29.Deberardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labuschagne CF, et al. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 32.Maddocks ODK, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2012;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis CA, et al. Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.05.008. DOI: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discovery. 2014 doi: 10.1158/2159-8290.CD-14-0250. DOI: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D. Nature. submitted. [Google Scholar]
- 36.Dolfi SC, et al. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metab. 2013;1:20. doi: 10.1186/2049-3002-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tönjes M, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nature Medicine. 2013;19:901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prabhu A, et al. Cysteine Catabolism: A Novel Metabolic Pathway Contributing to Glioblastoma Growth. Cancer Research. 2014;74:787–796. doi: 10.1158/0008-5472.CAN-13-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung WJ, et al. Inhibition of cystine uptake disrupts the growth of primary brain tumors. J Neurosci. 2005;25:7101–7110. doi: 10.1523/JNEUROSCI.5258-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogunrinu TA, Sontheimer H. Hypoxia increases the dependence of glioma cells on glutathione. J Biol Chem. 2010;285:37716–37724. doi: 10.1074/jbc.M110.161190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, et al. Asparagine Plays a Critical Role in Regulating Cellular Adaptation to Glutamine Depletion. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.08.018. DOI: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. 2012;109:8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 45.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamphorst JJ. Cancer Research. in press. [Google Scholar]
- 47.Kamphorst JJ, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011 doi: 10.1038/nature10602. DOI: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011 doi: 10.1038/nature10642. DOI: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. 2011. [DOI] [PMC free article] [PubMed]
- 51.Fendt S-M, et al. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat Comms. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamphorst JJ, et al. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer & …. 2014 doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mashimo T, et al. Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schug ZT. Cancer Cell [Google Scholar]
- 55.Comerford SA, et al. Acetate Dependence of Tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birsoy K, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puchowicz MA, et al. Zonation of acetate labeling across the liver: implications for studies of lipogenesis by MIDA. Am J Physiol. 1999;277:E1022–7. doi: 10.1152/ajpendo.1999.277.6.E1022. [DOI] [PubMed] [Google Scholar]
- 58.Hirayama A, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Research. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 59.Tatum JL, et al. Hypoxia: importance in tumor biology, noninvasive measurement by imaging, and value of its measurement in the management of cancer therapy. Int. J. Radiat. Biol. 2006;82:699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]
- 60.Helmlinger G, et al. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nature Medicine. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 61.Vaupel P, et al. Blood Flow, Oxygen and Nutrient Supply, and Metabolic Microenvironment of Human Tumors: A Review. Cancer Research. 1989 [PubMed] [Google Scholar]
- 62.Hu J, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature biotechnology. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuneva MO, et al. The Metabolic Profile of Tumors Depends on Both the Responsible Genetic Lesion and Tissue Type. Cell Metabolism. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Everhart J, Wright D. Diabetes mellitus as a risk factor for pancreatic cancer. A meta-analysis. Jama. 1995;273:1605–1609. [PubMed] [Google Scholar]
- 65.Wang F, et al. The relationship between diabetes and pancreatic cancer. Mol. Cancer. 2003;2:1–5. doi: 10.1186/1476-4598-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khasawneh J, et al. Inflammation and mitochondrial fatty acid β-oxidation link obesity to early tumor promotion. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3354. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zyromski NJ, et al. Obesity potentiates the growth and dissemination of pancreatic cancer. Surgery. 2009;146:258–263. doi: 10.1016/j.surg.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 68.Pannala R, et al. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10:88–95. doi: 10.1016/S1470-2045(08)70337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pannala R, et al. Temporal association of changes in fasting blood glucose and body mass index with diagnosis of pancreatic cancer. Am. J. Gastroenterol. 2009;104:2318–2325. doi: 10.1038/ajg.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dewys WD, et al. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am. J. Med. 1980;69:491–497. doi: 10.1016/s0149-2918(05)80001-3. [DOI] [PubMed] [Google Scholar]
- 71.Wigmore SJ, et al. Changes in nutritional status associated with unresectable pancreatic cancer. Br J Cancer. 1997;75:106–109. doi: 10.1038/bjc.1997.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kir S, et al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature. 2014;513:100–104. doi: 10.1038/nature13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shukla SK, et al. Metabolic reprogramming induced by ketone bodies diminishes pancreatic cancer cachexia. Cancer Metab. 2014;2:18. doi: 10.1186/2049-3002-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nature Medicine. 2014 doi: 10.1038/nm.3686. DOI: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sonveaux P, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guillaumond F, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 79.Pavlides S, et al. Warburg Meets Autophagy: Cancer-Associated Fibroblasts Accelerate Tumor Growth and Metastasis viaOxidative Stress, Mitophagy, and Aerobic Glycolysis. Antioxidants & Redox Signaling. 2012;16:1264–1284. doi: 10.1089/ars.2011.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaudhri VK, et al. Metabolic Alterations in Lung Cancer-Associated Fibroblasts Correlated with Increased Glycolytic Metabolism of the Tumor. Molecular Cancer Research. 2013;11:579–592. doi: 10.1158/1541-7786.MCR-12-0437-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nieman KM, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature Medicine. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang W, et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol. 2012;14:276–286. doi: 10.1038/ncb2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDonnell LA, Heeren R. Imaging mass spectrometry - McDonnell - 2007 - Mass Spectrometry Reviews - Wiley Online Library. Mass spectrometry reviews. 2007 doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 84.Steinhauser ML, et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012 doi: 10.1038/nature10734. DOI: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hawkins RA, Phelps ME. PET in clinical oncology. Cancer Metastasis Rev. 1988;7:119–142. doi: 10.1007/BF00046482. [DOI] [PubMed] [Google Scholar]
- 86.Ben-Haim S, Ell P. 18F-FDG PET and PET/CT in the evaluation of cancer treatment response. J. Nucl. Med. 2009;50:88–99. doi: 10.2967/jnumed.108.054205. [DOI] [PubMed] [Google Scholar]
- 87.Baek S, et al. Exploratory Clinical Trial of (4S)-4-(3-[18F]fluoropropyl)-l-glutamate for Imaging xC– Transporter Using Positron Emission Tomography in Patients with Non–Small Cell Lung or Breast Cancer. Cancer Research. 2012 doi: 10.1158/1078-0432.CCR-12-0214. [DOI] [PubMed] [Google Scholar]
- 88.Lieberman BP, et al. PET Imaging of Glutaminolysis in Tumors by 18F-(2S,4R)4-Fluoroglutamine. J. Nucl. Med. 2011;52:1947–1955. doi: 10.2967/jnumed.111.093815. [DOI] [PubMed] [Google Scholar]
- 89.Ho C-L, et al. 11C-acetate PET imaging in hepatocellular carcinoma and other liver masses. J. Nucl. Med. 2003;44:213–221. [PubMed] [Google Scholar]
- 90.Sakurai Y, Klein S. Metabolic alteration in patients with cancer: nutritional implications. Surg. Today. 1998;28:247–257. doi: 10.1007/s005950050116. [DOI] [PubMed] [Google Scholar]
- 91.Holroyde CP, et al. Altered glucose metabolism in metastatic carcinoma. Cancer Research. 1975;35:3710–3714. [PubMed] [Google Scholar]
- 92.Shaw JH, Wolfe RR. Glucose and urea kinetics in patients with early and advanced gastrointestinal cancer: the response to glucose infusion, parenteral feeding, and surgical resection. Surgery. 1987;101:181–191. [PubMed] [Google Scholar]
- 93.Inculet RI, et al. Altered leucine metabolism in noncachectic sarcoma patients. Cancer Research. 1987;47:4746–4749. [PubMed] [Google Scholar]
- 94.Shaw JH, Wolfe RR. Fatty acid and glycerol kinetics in septic patients and in patients with gastrointestinal cancer. The response to glucose infusion and parenteral feeding. Ann. Surg. 1987 doi: 10.1097/00000658-198704000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodrigues TB, et al. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nature Medicine. 2014;20:93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilson DM, Kurhanewicz J. Hyperpolarized 13C MR for Molecular Imaging of Prostate Cancer. J. Nucl. Med. 2014;55:1567–1572. doi: 10.2967/jnumed.114.141705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metabolism. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maher EA, et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 2012;25:1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang L, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Molecular Systems Biology. 2014;10:728–728. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blau N. Physician’s guide to the laboratory diagnosis of metabolic diseases. 2003.
- 101.Albritton EC. Standard Values in Blood. American Institute of Biological Sciences. 1952 [Google Scholar]
- 102.McMenamy RH, et al. Unbound Amino Acid Concentrations in Human Blood Plasmas. J Clin Invest. 1957;36:1672–1679. doi: 10.1172/JCI103568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Newgard CB, et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pitkänen HT, et al. Serum amino acid concentrations in aging men and women. Amino Acids. 2003;24:413–421. doi: 10.1007/s00726-002-0338-0. [DOI] [PubMed] [Google Scholar]
- 105.Wuu JA, et al. Amino acid concentrations in serum and aqueous humor from subjects with extreme myopia or senile cataract. Clin. Chem. 1988;34:1610–1613. [PubMed] [Google Scholar]
- 106.Hoffmann GF, et al. Physiology and pathophysiology of organic acids in cerebrospinal fluid. J. Inherit. Metab. Dis. 1993;16:648–669. doi: 10.1007/BF00711898. [DOI] [PubMed] [Google Scholar]
- 107.Tollinger CD, et al. Measurement of acetate in human blood by gas chromatography: effects of sample preparation, feeding, and various diseases. Clin. Chem. 1979;25:1787–1790. [PubMed] [Google Scholar]
- 108.Eagle H. The Specific Amino Acid Requirements of a Human Carcinoma Cell (Strain HeLa) in Tissue Culture. Journal of Experimental Medicine. 1955;102:37–48. doi: 10.1084/jem.102.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eagle H. The specific amino acid requirements of a mammalian cell (strain L) in tissue culture. J Biol Chem. 1955;214:839–852. [PubMed] [Google Scholar]
- 110.Eagle H. Amino acid metabolism in mammalian cell cultures. Science. 1959;130:432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- 111.Vogt M, Dulbecco R. Virus-Cell Interaction with a Tumor-Producing Virus. Proc. Natl. Acad. Sci. U.S.A. 1960;46:365–370. doi: 10.1073/pnas.46.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moore GE. Culture of Normal Human Leukocytes. Jama. 1967;199:519. [PubMed] [Google Scholar]
- 113.Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 114.Ames BN, et al. Oxidants, antioxidants, and the degenerative diseases of aging. Proceedings of the …. 1993 doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chandrasekera PC, Pippin JJ. Of rodents and men: species-specific glucose regulation and type 2 diabetes research. ALTEX. 2014;31:157–176. doi: 10.14573/altex.1309231. [DOI] [PubMed] [Google Scholar]
- 116.Harper A, et al. Branched-chain amino acid metabolism. Annual review of nutrition. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 117.Harper AE. Some concluding comments on emerging aspects of amino acid metabolism. J. Nutr. 1994;124:1529S–1532S. doi: 10.1093/jn/124.suppl_8.1529S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.