Abstract
Plus-strand viral RNAs contain sequences and structural elements that allow cognate RNA-dependent RNA polymerases (RdRp) to correctly initiate and transcribe asymmetric levels of plus and minus strands during RNA replication. cis-acting sequences involved in minus-strand synthesis, including promoters, enhancers, and, recently, transcriptional repressors (J. Pogany, M. R. Fabian, K. A. White, and P. D. Nagy, EMBO J. 22:5602-5611, 2003), have been identified for many viruses. A second example of a transcriptional repressor has been discovered in satC, a replicon associated with turnip crinkle virus. satC hairpin 5 (H5), located proximal to the core hairpin promoter, contains a large symmetrical internal loop (LSL) with sequence complementary to 3′-terminal bases. Deletion of satC 3′-terminal bases or alteration of the putative interacting bases enhanced transcription in vitro, while compensatory exchanges between the LSL and 3′ end restored near-normal transcription. Solution structure analysis indicated that substantial alteration of the satC H5 region occurs when the three 3′-terminal cytidylates are deleted. These results indicate that H5 functions to suppress synthesis of minus strands by sequestering the 3′ terminus from the RdRp. Alteration of a second sequence strongly repressed transcription in vitro and accumulation in vivo, suggesting that this sequence may function as a derepressor to free the 3′ end from interaction with H5. Hairpins with similar sequence and/or structural features that contain sequence complementary to 3′-terminal bases, as well as sequences that could function as derepressors, are located in similar regions in other carmoviruses, suggesting a general mechanism for controlling minus-strand synthesis in the genus.
RNA genomes of positive-strand RNA viruses share similar replication strategies despite vast differences in genome organization, primary sequence, and secondary and tertiary structure. After release of the genomic RNA(s) from the viral capsid, translation proceeds using host ribosomes, leading to expression of the RNA-dependent RNA polymerase (RdRp). The RdRp, together with known or hypothetical host proteins and additional viral proteins, forms an active replicase that recognizes specific cis-acting elements, leading to initiation of minus-strand synthesis (1, 5). Minus strands, either still complexed with plus strands (26) or single stranded (2, 16), are transcribed into full-length plus strands such that an asymmetric ratio of plus and minus strands accumulates, ranging from 10:1 to 1,000:1 (5). In addition, minus strands can be templates for the synthesis of 3′ coterminal subgenomic RNAs (sgRNAs) using internal promoters (34).
Since the RdRp must be recruited to the 3′ ends of plus or minus strands to initiate synthesis of full-length complementary RNAs, cis-acting elements thought to recruit the RdRp are usually located within the 3′ noncoding regions (12). In addition, internal locations can also bind RdRp, with RNA bridges directing the RdRp to the 3′ end (28, 33). 3′-proximal sequences comprising diverse forms such as tRNA-like structures, poly(A) tails, pseudoknots, hairpins, and short primary sequences without high-order structures have been identified as core promoters for minus-strand synthesis (11). Involvement of 5′-proximal sequences and structural elements in minus-strand synthesis has also been found for an increasing number of viruses (4, 14, 23, 31, 61, 66, 69). In addition, since plus strands cannot be templates for further translation during active replication (15), viral RNAs must contain the necessary information to switch between replication and translation as well as to support asymmetric synthesis of plus and minus strands, requirements that may include transcriptional enhancers (36, 39, 42, 43) and repressors that function via RNA-RNA (39) or protein-RNA (11, 73) interactions.
Turnip crinkle virus (TCV), a member of the genus Carmovirus in the family Tombusviridae, contains a single-stranded, plus-sense RNA genome of 4,054 bases that encodes five proteins (22) (Fig. 1). p28 and p88 are translated from the genomic RNA and are required for replication in vivo. In contrast, only p88, which contains the canonical GDD active-site motif of RdRp, is required for transcription in cell-free (in vitro) assays (41). p8 and p9, which are required for virus movement, are translated from the larger of two sgRNAs. The multifunctional coat protein, which serves as the single capsid component and is also the viral suppressor of the host RNA silencing defense system (40, 60), is translated from the smaller sgRNA (22).
FIG. 1.
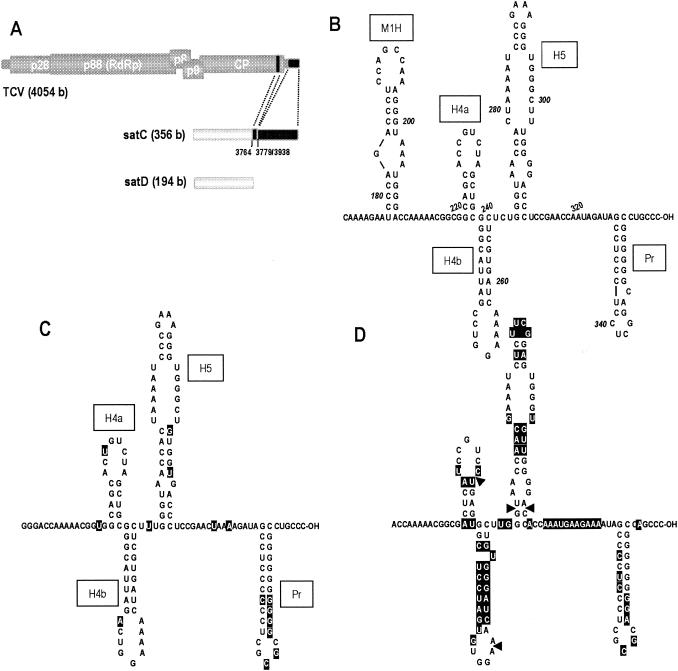
Sequence and structural relationships between satC, TCV, and the related carmovirus CCFV. (A) Schematic representation of TCV, satC, and satD. The five open reading frames of TCV, described in the text, are shown on the genomic RNA. satC is a chimeric molecule composed of satD sequence at the 5′ end and two regions of TCV at the 3′ end. Positions of TCV-related sequences in satC are indicated. (B) Computer-predicted lowest-free-energy structure for positions 170 to 356 of satC. Full-length satC was subjected to mfold analysis. Hairpin designations are boxed. Pr, core promoter on plus strands required for minus-strand synthesis of satC. M1H, replication enhancer on satC minus strands. M1H is also required as a hairpin on satC plus strands to bring together flanking single-stranded sequences that enhance helper virus movement by repressing virion formation (57, 71, 72). H4a, H4b, and H5 have not been reported previously. (C) Computer-predicted structure for the 3′ region of TCV. Bases that differ from those of satC are highlighted. (D) Analogous structure for the 3′ region of CCFV. Bases that differ from those of satC are highlighted. Arrowheads indicate locations of deleted bases.
By analyzing the replication requirements for small, noncoding replicons, sequences participating exclusively in RNA replication can be identified and their mechanistic roles can be determined. Subviral RNAs associated with TCV have proven particularly useful as model systems, allowing for the identification of cis-acting elements that function as core promoters and enhancers (46). Besides the genomic RNA, TCV is frequently associated with several small, dispensable nontranslated RNAs, such as defective interfering (DI) RNAs (30) and satellite RNAs (satRNAs) (47). satC (356 nucleotides) has been a particularly useful replicon for replication studies. This noncoding satRNA is a natural recombinant containing the nearly full-length sequence of a second satRNA (satD) at its 5′ end joined to two regions derived from TCV at its 3′ end, including 151 contiguous bases from the viral 3′ end (Fig. 1A). A number of cis-acting elements implicated in satC replication have been identified by whole-plant, protoplast, and in vitro assays. In vitro transcription assays, using purified or partially purified TCV RdRp, can accept exogenous plus and minus strands of satC as templates for de novo complementary-strand synthesis (41, 53).
satC plus strands contain a 3′ hairpin (Pr) flanked by a 3′-terminal motif (CCUGCCC-OH) that is conserved at the 3′ ends of all plus-strand RNAs replicated by the TCV RdRp (54). Several cis-acting elements that are important for replication in vivo or transcription in vitro have been identified on minus strands of satC. At the 3′ terminus of minus-strand satC is the carmovirus consensus sequence (CCS) [C2-3(A/U)3-7], which is conserved at the initiation sites for transcription of the genomic RNA and sgRNAs in all carmovirus minus-strand RNAs examined (17, 19). satC also contains a second CCS in an 11-base element (3′-proximal element) found near the 3′ ends of minus strands. This element is required for transcription in vitro in the absence of the 5′-proximal element described below,. The 3′-proximal element can also function independently as a promoter in vitro (17, 36). A hairpin (M1H) located in the central region of satC is a recombination hotspot and a transcriptional enhancer of plus-strand synthesis (8, 36). The 5′-proximal element, required for satC accumulation in vivo and the activity of minus-strand templates in vitro (in the absence of the 3′-proximal element), can also serve as a promoter in vitro (18, 19).
Deletion of the three 3′-terminal cytidylates (3′CCC) in full-length satC plus strands (producing construct CΔ3C) resulted both in a substantial increase in the transcription of full-length minus-strand products in vitro and in enhanced synthesis of internally initiated products (20). This result was surprising, because all templates transcribed by the TCV RdRp, including the genomic RNA, satRNAs, DI RNA, and sgRNAs, contain two or three 3′-terminal cytidylates. This sequence conservation had led to the assumption that 3′ cytidylates were essential for initiation of complementary-strand synthesis. Inoculation of turnip plants with CΔ3C led to recovery of wild-type (wt) satC at 2 weeks postinoculation, demonstrating that these residues could be repaired in vivo and were necessary for proper satC amplification. Sequencing analysis of products obtained by in vitro transcription of CΔ3C indicated that the RdRp was able to initiate de novo synthesis opposite the new 3′-terminal guanylate as well as to repair the 3-base deletion by polymerizing nontemplated nucleotides prior to initiating complementary-strand synthesis (20).
We now report several additional requirements for proper initiation of satC minus-strand synthesis in vitro. Hairpin 5 (H5), containing a large symmetrical internal loop (LSL) and located near the Pr core promoter, is required for correct initiation of minus-strand synthesis. In addition, H5 functions to repress synthesis of minus strands by binding to and sequestering the 3′ terminus from the RdRp. Repression of transcription can be derepressed in vitro by the availability of a second cis-acting sequence that is complementary to the 3′-terminal bases. Furthermore, the satC 5′ end affects transcription in vitro, possibly by stabilizing 3′-end structures. Hairpins with similar sequence and/or structural features that could function as repressors are located in identical positions in the genomic RNAs of TCV and other carmoviruses. This suggests a general mechanism for controlling minus-strand synthesis in this genus.
MATERIALS AND METHODS
Construction of satC mutants.
satC, either wt or containing progressive 5′-end deletions, was prepared by PCR amplification using Vent DNA polymerase (New England Biolabs) and pT7C+, which contains a T7 RNA polymerase promoter upstream of the full-length satC sequence (53). The 5′ primers, which also contained a T7 RNA polymerase promoter, were T7C5′ (5′-GTAATACGACTCACTATAGGGATAACTAAGGG-3′), T7C5′Δ2G (5′-GTAATACGACTCACTATAGATAACTAAGGGTTTC-3′), T7C5′Δ207 (5′-GTAATACGACTCACTATAGACCAAAAACGGCGGC-3′), T7C5′Δ218 (5′-GTAATACGACTCACTATAGCGGCAGCAGCACCG-3′), T7C5′Δ241 (5′-GTAATACGACTCACTATAGCATTAGCCTGGA-3′), and T7C5′Δ252 (5′-GTAATACGACTCACTATAGAAAACTAGTGCTCTC-3′). The 3′ primers were either oligo7 (5′-GGGCAGGCCCCCCGTCCGA-3′) or C3′ΔC (5′-CAGGCCCCCCGTCCGAGGA-3′), for production of wt or Δ3C ends, respectively.
satC containing G304C was generated by PCR using pT7C+, primer T7C5′, and a second oligonucleotide complementary to positions 287 to 356 at the 3′ end of satC (G304C). The latter oligonucleotide contained a C at position 304. H5RL and C300G were generated in a similar fashion except that the second oligonucleotide was complementary to positions 276 to 356 and had positions 296 through 302 containing 76% wt bases and 8% of each of the remaining three bases. To produce G353C and C300G/G353C, either pT7C+ or C300G was used for PCR, respectively. Primers were T7C5′ and a second oligonucleotide complementary to positions 337 to 356 at the 3′ end of satC, with an equal mixture of deoxynucleoside triphosphates at position 353. Deletion of the 3′CCC and/or the 5′GG (producing CΔ2G) was achieved by PCR using primers homologous to the 5′ end (T7C5′ or T7C5′Δ2G) and 3′ end (oligo7 or C3′ΔC). All mutants containing alterations in the LSL and 3′-terminal bases contained an additional alteration at position 176 (A to G), creating a BamHI site that permitted one to distinguish between satC containing reversions of the original mutations and possible contamination of wt satC. PCR products were treated with T4 polynucleotide kinase and the Klenow fragment and were ligated into the SmaI site of pUC19. To construct G218C, a primer complementary to positions 206 to 257 that contained equal amounts of C and G at positions 218 and 229 was used (position 229 was altered for a separate study). To construct C220G, a primer complementary to positions 208 to 257 that contained a G at position 220 was used. The second primer for these two constructs was T7C5′. PCR products were digested with SpeI and NcoI and ligated into similarly digested pT7C+, replacing the endogenous fragment. Deletion of the 3′CCC or 5′GG was performed as described above. After digestion with SmaI, wt and mutant satC's were synthesized by using T7 RNA polymerase, which generated transcripts with precise 5′ and 3′ ends.
RNA structure prediction and solution structure probing.
Secondary structures of satC and the 3′ ends of TCV and other members of the genus Carmovirus were predicted by mfold, version 3.1 (32, 74). Solution structure probing was performed as previously described (6, 63) with modifications. Briefly, purified satC transcripts (11 μg) synthesized by using T7 RNA polymerase and templates (wtC, CΔ2G, or CΔ3C) prepared by PCR as described above were mixed with 110 μg of yeast tRNA and 675 μl of modification buffer (70 mM HEPES [pH 7.5], 10 mM MgCl2, 0.1 mM EDTA, 100 mM KCl). The mixture was heated to 60°C, slowly cooled to 35°C, and incubated at 25°C for 20 min. Fifty-microliter samples of the RNA were added to an equal volume of modification buffer containing either no additional reagents (control) or one of the following: 10% (vol/vol) diethylpyrocarbonate (DEPC; Sigma), 1% (vol/vol) dimethylsulfate (DMS; Sigma), 0.05 U of RNase T1, or 0.03 U of RNase V1 (Ambion). Primer extension reactions were performed using 1 pmol of various oligonucleotides complementary to positions 339 to 356, 250 to 269, and 126 to 142 and [α-35S]-radiolabeled dATP. To determine the chemical and enzymatic reactivity of bases at the 3′ end of satC, satC transcripts that contained 220 vector-derived bases at the 3′ end were prepared from pT7C+.
In vitro RdRp assay.
The TCV p88 expression plasmid was a generous gift from P. D. Nagy (University of Kentucky). The expression and purification of p88 were carried out as described previously (41). TCV RdRp was also alternatively prepared from Arabidopsis thaliana protoplasts inoculated with TCV-CPmT, a TCV mutant containing altered bases in the initiation codon and an alternative initiation codon in the coat protein open reading frame to eliminate translation of the coat protein (64). RdRp was extracted at 40 h post-inoculation (hpi) of protoplasts (5 × 107) by using the same method as that for extraction from infected turnip leaves (53). In vitro RdRp assays using either of the two RdRp preparations were performed as previously described (19, 53). Briefly, 1 μg of the purified RNA template was added to a 25-μl reaction mixture containing 50 mM Tris-HCl (pH 8.2), 100 mM potassium glutamate, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM each ATP, CTP, and GTP, 0.01 mM UTP, 10 μCi of [α-32P]UTP (Amersham), and 12.5 μl of plant-derived RdRp (from ∼4.2 × 104 protoplasts)or 2 μg of Escherichia coli-derived TCV p88. After a 90-min incubation at 20°C, 1 μg of yeast tRNA was added, and the mixture was subjected to phenol-chloroform extraction and ammonium acetate-isopropanol precipitation. Radiolabeled products were analyzed by denaturing 5% polyacrylamide-8 M urea gel electrophoresis followed by autoradiography. Full-length and aberrant products were analyzed separately by densitometry (with a Bio-Rad imaging densitometer). Values were normalized for the amount of incorporated radioactive UTP possible in the full-length templates.
Protoplast inoculations, RNA extraction, and Northern blotting.
A. thaliana (ecotype Col-0) protoplasts were prepared from callus cultures derived from seeds as previously described (29). Protoplasts (5 × 106) were inoculated with 20 μg of TCV genomic RNA transcripts (38) and 2 μg of satC transcripts as previously described (29). Equal amounts of total RNA extracted from protoplasts at 40 hpi were subjected to electrophoresis through a 1.5% nondenaturing agarose gel followed by Northern hybridization analysis (62). After blotting, plus-strand TCV and satC RNAs were probed with a [γ-32P]dATP-labeled oligonucleotide complementary to positions 3950 to 3970 of TCV genomic RNA and 250 to 269 of satC.
RESULTS
Structure of the 3′ region of satC.
As described in the introduction, deletion of the satC 3′CCC results in enhanced transcription of full-length and aberrantly initiated complementary strands. One explanation for enhanced synthesis of products in the absence of 3′-terminal bases may be that the satC 3′ terminus is sequestered from the RdRp by base pairing with upstream complementary sequences. Deletion of the 3′CCC would enhance transcription by liberating the 3′ end and making it available to the RdRp. To determine the validity of this hypothesis and locate possible upstream sequence partners for the 3′CCC, full-length satC was subjected to computational structure prediction using mfold, version 3.1 (32, 74). The structure shown in Fig. 1B was predicted by mfold to be the most stable conformation of the 3′ region of satC (bases 170 to 356). The structure included five hairpins (M1H, H4a, H4b, H5, and Pr) and four single-stranded regions of 4 bases or more. The region of TCV that was the progenitor of the 3′ portion of satC also was predicted to fold into a similar structure, with base differences not affecting the overall structure (Fig. 1C). These five hairpins were also predicted to form in the lowest-free-energy structure when full-length TCV genomic RNA was subjected to mfold analysis (data not shown). Since phylogenetic comparisons can help reveal the existence of structural elements necessary for basic amplification of the genera, the analogous 3′ region in the most closely related carmovirus, Cardamine chlorotic fleck virus (CCFV) (38, 52), was also subjected to computational analysis. Using both the 3′ untranslated region (3′ UTR) and full-length CCFV genomic RNA, mfold predicted the existence of H5 and Pr only in the 10 lowest-free-energy structures. However, the CCFV sequence just upstream of H5 can fold into hairpins very similar to H4a and H4b, both in overall structure and in location, despite the fact that it shows only 35% sequence identity with the same region in satC (Fig. 1D).
Effects of deletions on RdRp-mediated transcription in vitro by using satC templates with and without 3′CCC.
Our original hypothesis suggested that synthesis of satC minus strands is repressed because of base pairing between the 3′CCC and upstream guanylate residues. Progressively deleting upstream sequences may thus identify these guanylates, since such deletions should abolish transcriptional enhancement when the 3′CCC is also deleted. To test this possibility, deletions of 2 to 252 bases were generated from the 5′ end of satC (Fig. 2A). Each satC deletion mutant was constructed with wt and CΔ3C 3′ ends. RNA transcripts synthesized from these constructs were assayed in an in vitro RdRp reaction programmed with radioactive UTP and partially purified RdRp extracted from TCV-infected Arabidopsis protoplasts (see Materials and Methods). Following gel electrophoresis, complementary minus strands were quantified by densitometer scanning of template-sized products as well as of products migrating both more slowly and faster than the template-sized products. It has previously been shown that some of the faster-migrating products are due to aberrant de novo initiation by the RdRp at internal locations using various initiating nucleotides (20). Other products migrating either more slowly or faster than full-length products have previously been determined to be foldback products synthesized by extension from the 3′-terminal hydroxyl of the template opposite either 3′-end sequences or internal positions (55). These foldback products can migrate either faster or more slowly than the full-length product depending on their level of denaturation, which is temperature dependent and can vary in individual gels.
FIG. 2.
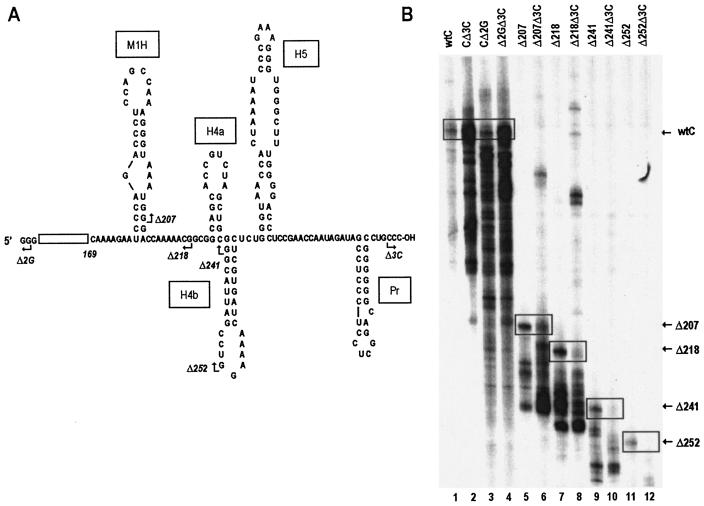
Deletion analysis to uncover sequences involved in enhancing minus-strand synthesis in the absence of the 3′CCC. (A) Schematic representation of the deletion mutants. Open box represents sequence that is not shown. Nucleotides are numbered from the 5′ end. Deletion end points are indicated by arrows, and the number of bases deleted is indicated in the adjacent names. Hairpin designations are from Fig. 1B. (B) Analysis of template activities of deletion mutants in an in vitro TCV RdRp assay. Partially purified TCV RdRp was isolated from Arabidopsis protoplasts. The radiolabeled minus-strand products were separated on denaturing polyacrylamide gels. Template-sized products are boxed, and construct designations are given on the right. Templates that contain a deletion of the 3′-terminal CCC (Δ3C) and/or a deletion of the 5′-terminal GG (Δ2G) are indicated.
Transcription of CΔ3C by TCV RdRp in vitro enhanced the synthesis of full-length products 3.5-fold and that of total products 8-fold over transcription of wt satC (Fig. 2B, lanes 1 and 2), as previously found (20). To determine if the 5′-terminal GGG could be the complementary partner of the 3′CCC, CΔ2G was constructed. Transcription of CΔ2G resulted in a 10-fold increase in overall template activity relative to activity with wt satC (Fig. 2B, lane 3), with less than a 2-fold increase in full-length satRNA synthesis. Transcription of satC containing both Δ2G and Δ3C resulted in an additive effect on the products of transcription, with a 12-fold enhancement in levels of both template-length and faster-migrating products (Fig. 2B, lane 4). These results were interpreted as indicating that transcription of CΔ2G was not precisely equivalent to that of CΔ3C. Therefore, while the results did not contradict possible 3′- and 5′-end interaction, other possible interactions could not be ruled out.
To further investigate possible interactions between the 3′-end and internal sequences, templates containing additional 5′-end deletions were constructed. Transcription of satC containing a deletion extending to position 207 (Δ207) near the base of M1H showed a 5-fold enhancement of full-length and aberrant-sized products, with levels increasing to 13-fold above wt when the template also included Δ3C (Fig. 2B, lanes 5 and 6). Transcription of satC with a deletion extending to near the base of H4a (Δ218) was enhanced 13-fold over wt (Fig. 2B, lane 7). However, addition of Δ3C (Fig. 2B, lane 8) did not further increase synthesis of aberrant products, while synthesis of full-length complementary strands was reduced threefold. Transcription of templates containing further deletions to the base (Δ241) and center (Δ252) of H4b resulted in fivefold or no enhancement, respectively, over transcription of wt satC. Inclusion of Δ3C with these templates suppressed transcription, with a 6.8- or 34-fold reduction in synthesis of full-length products, respectively. These results suggested that the 3′CCC is involved in suppressing full-length and aberrant minus-strand synthesis while the 5′GG mainly suppresses aberrant minus-strand synthesis in vitro. In addition, lack of enhanced synthesis of full-length products in the Δ218Δ3C, Δ241Δ3C, and Δ252Δ3C constructs suggested that the truncated 3′ end is no longer becoming more accessible to the RdRp. This could be due to (i) the involvement of sequence between positions 207 and 218 in initiation of minus-strand synthesis and/or (ii) alterations in downstream structures that are important for 3′-end interactions.
Deletion of 3′CCC alters the structure of H5.
While the deletion study did not identify internal base-pairing partners for 3′CCC, it did suggest that these residues reside in the 3′ 149 bases of satC (3′ of position 207). This region contains six locations with three or more consecutive guanylates. Since deletion of 3′CCC might result in structural changes to its partner sequences, wt satC, CΔ3C, and CΔ2G transcripts were subjected to in vitro chemical and enzymatic mapping. Bases residing in a single stranded conformation that were accessible to DMS methylation (N1 and N3 positions of adenosines and cytidines, respectively), DEPC carboxylation (N7 position of adenosines and guanosines), or RNase T1 cleavage after the 3′ phosphate of guanosines were detected following primer extension with reverse transcriptase, using oligonucleotides complementary to sequences throughout satC. RNase V1 was similarly used to detect bases that were present in a double-stranded or stacked helical conformation.
To determine the structure of the 3′-terminal 22 bases in wt satC, a satC template that contained an additional 220 vector-derived bases at the 3′ end (satC+220) was used. In vitro transcription of satC+220 by the TCV RdRp gave rise to wt satC-length products, indicating that satC+220 has the same structural recognition features for minus-strand synthesis as wt satC (53). All nucleotides upstream of position 334 (the first position detected by using the wt satC template) that were discernible by use of both the wt satC and the satC+220 template were identically susceptible to chemical and enzymatic reagents (data not shown).
A summary of the data obtained from these experiments is shown in Fig. 3. With the exception of H5, there was good to excellent correlation between the hairpins found in the most stable wt satC structure predicted by mfold and the results obtained by solution structure analysis. M1H, an element that was previously shown by a similar analysis to be present on minus strands (6), was strongly predicted by the present analysis to be present in its plus-strand complement. The 3′-terminal Pr hairpin differed slightly at the base from the computer structural prediction. G328 at the 5′ base of the hairpin was susceptible to DEPC treatment, suggesting that breathing by the terminal base pair may be occurring. In addition, C351, flanking the 3′ side of Pr, was susceptible to RNase V1 and may be base paired in this or an alternative 3′-end structure.
FIG. 3.
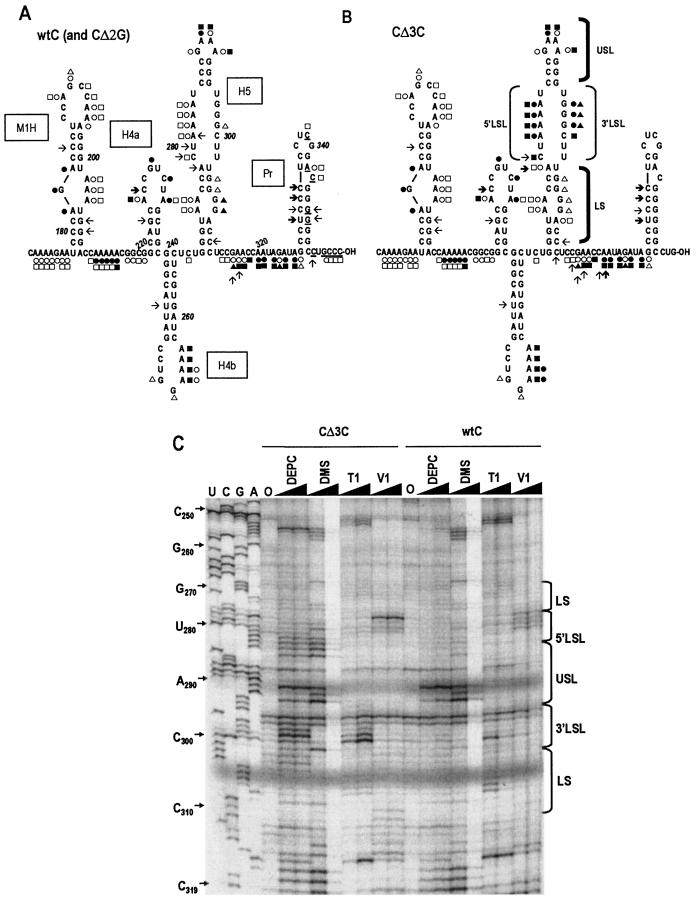
Chemical and enzymatic probing of the 3′ regions of wt satC, CΔ3C, and CΔ2G. Sample transcripts were treated for 10 or 20 min with DEPC and DMS, and for 5 or 10 min with RNase T1 and RNase V1. Untreated, modified, or cleaved RNAs were subjected to primer extension analysis using primers throughout satC. The products of the primer extension reactions were subjected to electrophoresis through 8% acrylamide-50% urea gels. Sequencing ladders were generated by using the same primers and pT7C+ as a template. (A) Summary of solution structure probing of wt satC and CΔ2G, which gave identical results at all positions throughout the satRNA. (B) Solution structure probing of CΔ3C. The initial residues at the 3′ ends of CΔ3C and CΔ2G that were discernible (by using an oligonucleotide complementary to the 3′-terminal sequence) were at position 334. For wt satC, an additional template that contained 220 bases of vector-derived sequence at the 3′ end was used to ascertain thestructure of the satC 3′-terminal nucleotides (underlined) (see Materials and Methods). Low or high sensitivity to reagents is indicated by open or solid symbols, respectively, or by light or bold arrows, respectively. Circles, DEPC; squares, DMS; triangles, RNase T1; arrows, RNase V1. Regions of H5 indicated in panel C are shown. (C) Sample gel showing H5 and surrounding sequence. The regions corresponding to the lower stem (LS), 3′ side of the LSL, 5′ side of the LSL, and upper stem and tetraloop (USL) are indicated. The sequencing ladder is shown in the first four lanes, and bases corresponding to specific nucleotides are indicated at the left. “0” indicates sample that was not treated with reagents prior to primer extension. Solid triangles above lanes indicate increasing incubation time. The strong DEPC signal at position C232 was found in multiple experiments.
The most obvious divergences between the computer-predicted structure and the bases accessible to chemical and enzymatic treatment were found for H5. Although the GAAA tetraloop was supported by the solution structure analysis, the LSL region was not. The LSL 5′-side adenylates and cytidylate were reactive with DMS and DEPC; however, only G299 on the LSL 3′ side was weakly digested by RNase T1. Furthermore, the three lower bases (bases 279 to 281) on the LSL 5′ side, including the DMS- and DEPC-reactive A281, were susceptible to RNase V1 cleavage. Such signal overlap again suggests plasticity in the structure in this region of the satRNA.
No differences were detected between wt satC and positions 1 to 334 of CΔ2G, the region assayed for CΔ2G (data not shown). With the exception of the adenylate in position 4 that was susceptible to RNase V1 in wt satC but not in CΔ3C, no changes were detected between wt satC and CΔ3C from the 5′ end to position 253 (Fig. 3C and data not shown). Positions 254 and 255 differed slightly between wt satC and CΔ3C in that both adenylates were weakly reactive with DEPC in wt satC and strongly reactive in CΔ3C. In both transcripts, these two adenylates reacted strongly with DMS. Identical reactivity of wt satC and CΔ3C to chemicals and enzymes was also evident for bases in positions 321 to 334.
In contrast, substantial variation was found between wt satC and CΔ3C in the lower stem and LSL of H5 and the 3′ sequence flanking H5 (Fig. 3). In the absence of the 3′CCC, all bases in the LSL that could react with DMS, DEPC, or RNase T1 became highly sensitive, including the three consecutive guanylates on the 3′ side of the LSL. In addition, only two of the four residues on the LSL 5′ side remained consistently reactive with RNase V1, with A278 becoming strongly reactive. In the lower stem, the two guanylates in the small symmetrical loop (positions 306 and 307) became less reactive with RNase T1 in the absence of the 3′CCC, while sensitivity to DMS and DEPC developed for the adjacent A308. Differences were also found in the 3′ sequences flanking H5, with four residues gaining sensitivity to RNase V1 in CΔ3C, including C311 at the base of H5. These results suggest major structural changes to the LSL of H5 and the 3′ region flanking H5 when the 3′CCC is deleted, without discernible disruption of the remaining satC structure outside of the minor change in the loop of H4b.
Compensatory changes between H5 LSL and a 3′-terminal base support an interaction between these sequences.
The simplest explanation for enhanced accessibility of the 3′ side of the LSL to single-stranded-specific reagents when the 3′CCC is deleted is that the 3′ terminal GCCC-OH may base pair with the LSL GGGC (5′ to 3′) in wt satC. To determine if an interaction exists between the 3′-terminal bases and the 3′ side of the LSL, three constructs were generated in satC that would disrupt and reestablish putative base pairing between the two sequences (Fig. 4A). The new satC constructs contained a cytidylate-to-guanylate change at position 300 in the LSL (C300G), a guanylate-to-cytidylate change at position 353 (G353C), and both alterations (C300G/G353C), which would reestablish the putative base pairing (Fig. 4A). If our hypothesis regarding sequestration of the 3′ end was correct, then disruption of base pairing between the two sequences should enhance synthesis of complementary strands by making the 3′ end more accessible to the RdRp.
FIG. 4.
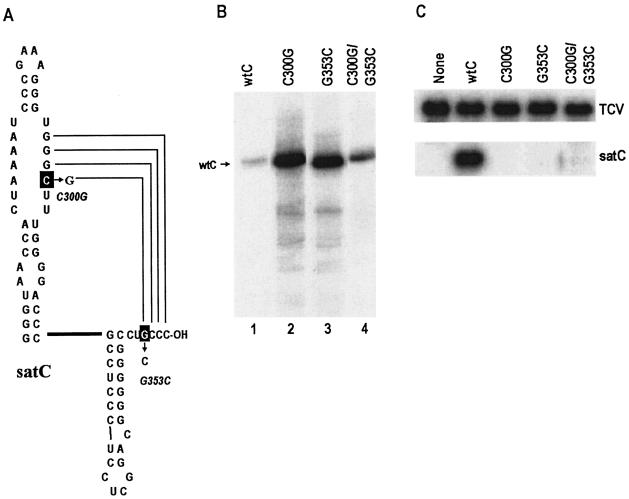
Compensatory exchanges between satC LSL and a base near the 3′ end. (A) Locations of the point mutations generated in satC. Bases that were altered are boxed; arrows point to the new bases. Names of the altered constructs are shown nearby. Possible base pairing between the 3′-terminal bases and the 3′ side of the LSL is shown. (B) In vitro transcription of satC containing the mutations diagrammed in panel A by using purified TCV RdRp expressed in E. coli. wtC, wt satC; C300G/G353C, satC containing both mutations. (C) Northern blot of mutant satC and helper virus (TCV) plus strands. Total RNA was extracted from protoplasts at 40 hpi and probed with an oligonucleotide complementary to both TCV and satC.
Mutant and wt satC transcripts were subjected to in vitro transcription by using RdRp expressed in, and purified from, E. coli. This newly available RdRp preparation gave results identical to those of the partially purified RdRp from infected plants in tests of several constructs (41) (data not shown). C300G and G353C transcripts increased the synthesis of full-length complementary strands relative to that with wt satC by similar amounts (10- and 11-fold, respectively) while also increasing the levels of aberrantly initiated products. The compensatory construct (C300G/G353C) produced much lower levels of full-length products than did transcripts containing only G353C or C300G. These results strongly support the existence of an interaction between the 3′CCC and the H5 LSL that represses minus-strand synthesis.
To determine the effect of these mutations on satC accumulation in vivo, transcripts were coinoculated with wt TCV onto Arabidopsis protoplasts, and satRNA levels were examined at 40 hpi by Northern blot analysis. None of the satC constructs accumulated to detectable levels (Fig. 4C). This suggests that disrupting the base-pairing interaction strongly inhibits satC accumulation. Furthermore, since the compensatory exchange did not restore satC viability, either C300G or G353C is likely essential for additional functions, and thus, the compensatory construct is still nonviable. Alternatively, the 3.7-fold elevation in in vitro transcription of minus strands by use of C300G/G353C compared with wt satC is sufficient to disrupt satC accumulation in vivo.
The H5 structure and satC 5′ end are also important for minus-strand synthesis in vitro.
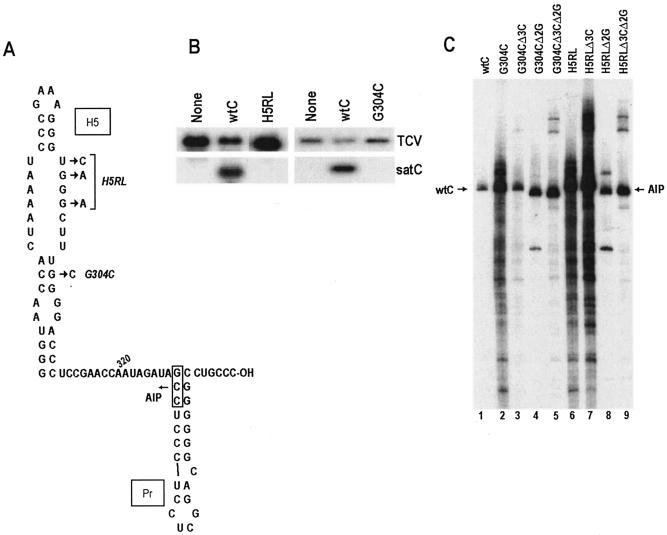
The covariation analysis of the LSL and the 3′ end of satC, coupled with the structural changes evident in H5 when the 3′CCC is deleted, suggests that the H5 LSL represses minus-strand synthesis by interacting with the 3′ end and sequestering the 3′ terminus from the RdRp. In addition, a role for the 5′GG in minus-strand synthesis was suggested by the finding that transcription of CΔ2G also leads to enhanced synthesis of products (Fig. 2B, lane 3). To further explore requirements for minus-strand synthesis, three positions (U296C, G297A, and G299A) were altered in the satC H5 LSL (generating construct H5RL); these alterations should disrupt base pairing with the 3′ end. To explore the consequences of disrupting H5 structure without changing the LSL, a single base was separately altered at position 304 (G304C) in the middle of the 3-base lower stem. This alteration was predicted by mfold to eliminate the H5 structure (Fig. 5A). As expected, H5RL and G304C did not accumulate to detectable levels in Arabidopsis protoplasts (Fig. 5B).
FIG. 5.
Effects of mutations in the LSL and lower stem of H5 on satC minus-strand synthesis in vitro and in vivo. (A) Alterations in H5 are shown. Names of the constructs are given on the right. Boxed sequence with an arrow at the base of Pr marks location of the internal initiation site for the major faster-migrating product (AIP) shown in panel C. (B) Northern blot of mutant satC and helper virus (TCV) plus strands. Total RNA was extracted from protoplasts at 40 hpi and probed with an oligonucleotide complementary to both TCV and satC. (C) In vitro transcription of complementary strands using TCV RdRp purified from E. coli. Positions of satC full-length (wtC) and alternative (AIP) products are indicated. Templates that also contain a deletion of the 3′CCC (Δ3C) and/or deletion of the 5′GG (Δ2G) are indicated.
Transcripts of the mutants with and without 3′CCC and 5′GG were assayed for complementary-strand synthesis in vitro by using TCV RdRp from E. coli. Both H5RL and G304C transcripts produced enhanced synthesis of full-length and aberrant products (Fig. 5C, lanes 2 and 6), as did other constructs that disrupted the 3′-end-LSL interaction (CΔ3C [Fig. 2B, lane 2], C300G [Fig. 4B, lane 2], and G353C [Fig. 4B, lane 3]). This result supports the hypothesis that either mutating the sequence proposed to interact with the 3′ end in the LSL (H5RL) or abolishing the hairpin structure (G304C) releases the 3′-terminal bases. Unexpectedly, the two mutants gave very different results when 3′CCC was also deleted (H5RLΔ3C and G304CΔ3C). Transcription of H5RLΔ3C produced products similar to those produced by H5RL (Fig. 5C; compare lanes 6 and 7), indicating that the 3′-end deletion did not further affect RdRp access to the 3′ end (or internal initiation sites). However, transcription of G304CΔ3C resulted in the loss of nearly all aberrant initiation products and no additional synthesis of full-length complementary strands (Fig. 5C, lane 3). Since the major difference between the two H5 mutants is that H5RL is predicted to retain the H5 structure while G304C does not, one possible explanation is that H5 participates in transcription beyond simple sequestration of the 3′ end.
In contrast, the two mutants gave identical but unexpected results in the absence of the 5′GG that were independent of the presence or absence of the 3′CCC. Transcription of H5RLΔ2G, H5RLΔ3CΔ2G, G304CΔ2G, or G304CΔ3CΔ2G resulted in one main product that migrated slightly faster than full-length complementary products. This product was designated the alternative initiation product (AIP). To determine the approximate initiation site for the AIP, AIPs synthesized from H5RLΔ2G and G304CΔ2G were isolated and subjected to 5′ RACE, using dCTP or dATP to tail the 3′ end of the cDNA. Despite multiple attempts, we were unable to successfully add poly(A) to the 3′ end of the AIP. However, poly(C) was successfully used to clone the AIP from both samples. Sequencing of the resultant clones indicated that transcription initiated between positions 328 and 330 at the 5′ base of Pr (Fig. 5A). Our inability to precisely identify the AIP 3′-end nucleotide was due to the presence of cytidylates at positions 329 and 330 that could have been derived from 3′ end tailing with dCTP. It has previously been demonstrated that although all natural TCV templates initiate opposite multiple cytidylates, the TCV RdRp is capable of initiating internally opposite guanylates as well (20).
Since transcription of CΔ2G did not result in the near-exclusive synthesis of the AIP (Fig. 2B, lane 3), a functional H5 that can base pair with the 3′GCCC must repress AIP synthesis. A model for how this may occur and for how the 5′GG may be involved in transcription is presented in the Discussion.
Conservation of H5 and Pr among carmoviruses.
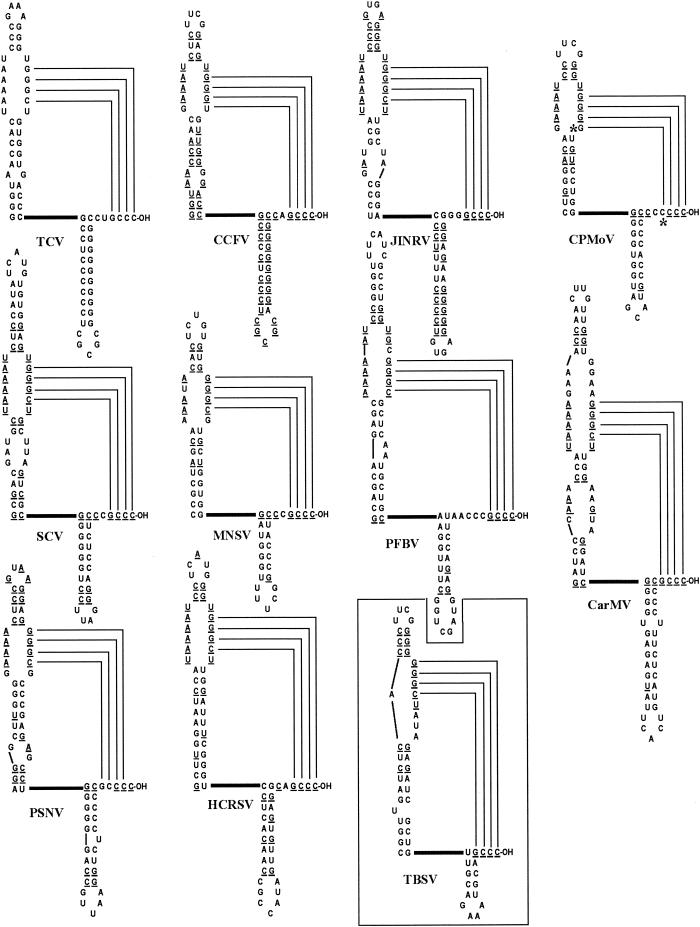
To determine if other carmoviruses have similar minus-strand initiation elements, the 3′ UTR of each known carmovirus was subjected to mfold analysis. With the exception of Galingosa mosaic virus (10), which contains no structures equivalent to either Pr or H5, all carmoviruses have a 3′-terminal hairpin structurally similar to the satC and TCV Pr (Fig. 6). In addition, all these carmoviruses have a second hairpin located 16 to 27 bases upstream of Pr that shares primary and secondary structural features with H5. When viewed as a whole, the defining feature of the carmovirus H5 is the LSL in the upper portion of the stem, which is symmetrical in 8 of the 10 viruses. The two exceptions, Carnation mottle virus (CarMV) and Pelargonium flower break virus, contain 1 additional base on the 3′ side of the loop. The LSL ranges from 10 bases for CCFV to 17 bases for CarMV. Four carmoviruses (TCV, Japanese iris necrotic ring virus, Saguaro cactus virus, and Hibiscus chlorotic ringspot virus) contain identical sequence in the LSL (5′-UAAAAU-3′ and 5′-UGGGCU-3′), while most others differ symmetrically from this sequence. The hairpin stems are poorly conserved, with upper stems ranging from 2 to 7 bp capped by either the main classes of stable tetraloops (GNRA, UNCG, or CUYG) (35), pentaloops, or hexaloops. The four 3′-terminal bases of all these carmoviruses could form similar base-pairing interactions with their H5 LSL. Most notably, Cowpea mottle virus (CPMoV), the only carmovirus that terminates in four cytidylates, is also the only carmovirus with four consecutive guanylates in the LSL. This phylogenetic analysis supports a role for H5 in repressing minus-strand synthesis in the genus.
FIG. 6.
Conserved structures in the 3′ ends of carmoviruses. 3′ UTRs of the following carmoviruses were subjected to computer structural predictions using mfold: TCV, Hibiscus chlorotic ringspot virus (HCRSV) (24), Japanese iris necrotic ring virus (JINRV) (59), Saguaro cactus virus (SCV) (65), CCFV (52), CarMV (21), CPMoV (70), Maize necrotic streak virus (MNSV) (44), Pea stem necrosis virus (PSNV) (58), and Pelargonium flower break virus (PFBV; GenBank accession number AJ514833). Bases identical to those in TCV are underlined. Putative interacting bases are indicated. Asterisks indicate covariation in the LSL-3′-end interaction in CPMoV. The boxed structure is the replication silencer and 3′-terminal hairpin of the tombusvirus Tomato bushy stunt virus (TBSV) (13, 39).
Involvement of additional internal sequences with initiation of minus-strand synthesis.
Based on the studies described above, the carmovirus H5 LSL likely serves as a repressor of minus-strand synthesis by binding to and sequestering the 3′ end from the RdRp. If this model is correct, then a missing component would be a derepressor that serves to derepress minus-strand synthesis by releasing the 3′ end from interaction with the LSL. Such a derepressor could be one or more sequences that disrupt LSL-3′-end base pairing by interacting with either the LSL, the 3′-terminal sequence, or both. One possibility for such a sequence was provided by the results of Fig. 2. Transcripts containing deletions extending from the 5′ end to position 218 and beyond showed decreased transcription of full-length products when the 3′CCC was also deleted (the opposite result from wt satC). Since transcripts containing deletions extending to position 207 still showed enhanced transcription in the absence of the 3′CCC, the region between positions 207 and at least 218 could play a role in transcription initiation in vitro. One possible interpretation of these results is that sequence in the vicinity of positions 207 to 218 may disrupt the interaction of the LSL with the 3′ end.
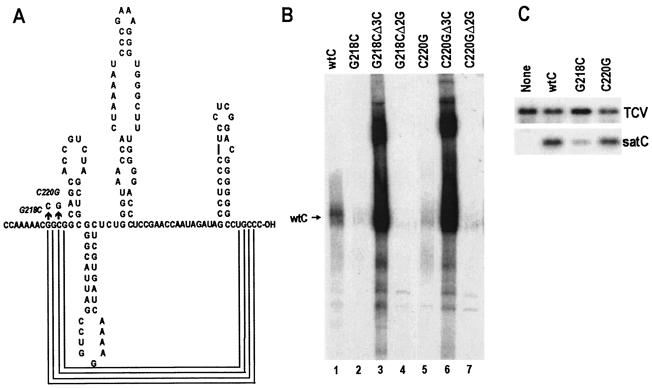
mfold computational analysis, which is not designed to detect an interaction between the 3′ end and the LSL of H5, predicts that the UGCCC-OH at the 3′ end of satC base pairs with positions 218 to 221 (5′ GGCG). To test if positions 218 to 221 function as a derepressor, two mutations were individually generated in satC at positions 218 (G218C) and 220 (C220G), and mutants were tested for transcription in vitro in the presence or absence of the 3′CCC and 5′GG (Fig. 7). If our hypothesis is correct, then mutating the putative derepressor sequence should strongly suppress minus-strand synthesis. Furthermore, mutations in the derepressor sequence should have no (additional) effect on the transcription of templates that contain Δ3C. Transcripts of the mutants showed either 11-fold (C220G) or 30-fold (G218C) suppression of full-length minus-strand synthesis in the presence of the 3′CCC (Fig. 7B; compare lane 1 with lanes 2 and 5). Deletion of the 3′CCC in combination with the G218C or C220G mutation resulted in enhancement of both full-length and aberrant transcription (Fig. 7B, lanes 3 and 6), results similar to those obtained with CΔ3C. In contrast, deletion of 5′GG had no effect on transcription of either mutant (Fig. 7B, lanes 4 and 7). These results are consistent with a role for positions 218 to 221 as a derepressor of minus-strand synthesis in vitro.
FIG. 7.
Effects of mutations in a possible derepressor sequence on minus-strand synthesis in vitro and in vivo. (A) Alterations of bases and possible interactions with the 3′-terminal sequences are shown. (B) In vitro transcription of complementary strands by using purified TCV RdRp expressed in E. coli. The position of the full-length satC product (wtC) is shown. Templates that also contain a deletion of the 3′CCC (Δ3C) or of the 5′GG (Δ2G) are indicated. (C) Northern blot of mutant satC and helper virus (TCV) plus strands. Total RNA was extracted from protoplasts at 40 hpi and probed with an oligonucleotide complementary to both TCV and satC.
To determine the effects of mutations at positions 220 and 218 on the accumulation of satC in protoplasts, wt satC, C220G, and G218G were individually inoculated with TCV onto Arabidopsis protoplasts, and total RNA was extracted at 40 hpi. Accumulation of G218C was 3.3-fold less than that of wt satC, supporting a role for position 218 in satC replication. However, the alteration in C220G had a more modest effect, with C220G accumulating to 80% of wt satC levels.
DISCUSSION
Repression and derepression of minus-strand synthesis.
A proposal has been made that the three 3′-terminal bases of plus-strand RNA viruses are the core element required for minus-strand synthesis, while structural elements in the region serve both to direct the 3′ end to the RdRp and to sequester alternative initiation sites (11). This hypothesis is based on in vitro transcription studies using RdRp from bacteriophage Qβ, Turnip yellow mosaic virus, and TCV (67, 68). Studies on flaviviruses and bromoviruses have also indicated that some or all of the 3′-terminal nucleotides are critical for replication in vivo and transcription in vitro (3, 25, 37, 49-51), although these nucleotides may not be important for stable interaction between the RdRp and the template (9). Despite the ability of TCV RdRp to initiate opposite the penultimate C in a sequence containing CCA repeats (68), the 3′CCC on the natural satC template is dispensable for basic transcription in vitro (20). In the present study, we have extended this analysis and propose additional functions for the 3′-terminal bases of carmoviruses based on studying subviral satC associated with TCV: the 3′-terminal bases are necessary both to repress minus-strand synthesis and, in conjunction with H5, to direct the RdRp to initiate uniformly at the 3′ terminus. Repression is achieved when the GCCC-OH at the 3′ end binds to the complementary GGGC (5′ to 3′) in the H5 LSL, thereby fully pairing all 3′-terminal bases, including the ultimate cytidylate, and restricting de novo initiation by the RdRp, which likely requires a free 3′-terminal base (27, 45, 48, 51). Deleting the 3′-terminal cytidylates (CΔ3C), eliminating the H5 structure (G304C), or mutating the LSL (H5RL and C300G) or 3′ interacting sequence (G353C) relieves both the repression and the near-exclusive direction of the RdRp to the 3′ terminus. This allows the RdRp to synthesize high levels of full-length minus strands as well as aberrant internally initiated and primer extension products.
If minus-strand synthesis is repressed by the 3′ end base pairing with the LSL, an important question is how this repression is overcome so that the RdRp can initiate transcription de novo opposite the terminal cytidylate. Our model, presented in Fig. 8, suggests that an additional sequence in positions 218 to 221, termed a derepressor, acts to disengage the 3′ end from its LSL complement. This sequence (GGCG) can potentially base pair with UGCCC-OH, leaving the terminal cytidylate unpaired and available for de novo initiation by the RdRp. We have chosen not to term this sequence an “activator,” since it is not required for transcriptional activation and is not an enhancer of transcription in the absence of repression. The existence of the derepressor is based on two pieces of evidence presented in this report: (i) the fact that transcription is no longer enhanced in the absence of the 3′CCC when the region of the derepressor is deleted (Fig. 2) and (ii) the fact that mutations in this sequence (G218C and C220G) strongly repress transcription in vitro in the presence of the 3′CCC (Fig. 7). It should be noted, however, that only the G218C alteration significantly reduced satC levels in protoplasts. At present, it is not clear why C220G had only a modest effect on the accumulation of satC in vivo. One possibility is that C220G, which converts the GGCG motif to GGGG, still permits base pairing with the 3′-terminal CCC. In G218C, the motif is altered to CGCG, which should substantially disrupt base pairing with the 3′ end. It is also possible that the reconstituted in vitro assay amplifies the consequence of the C220G alteration beyond a more limited effect in vivo.
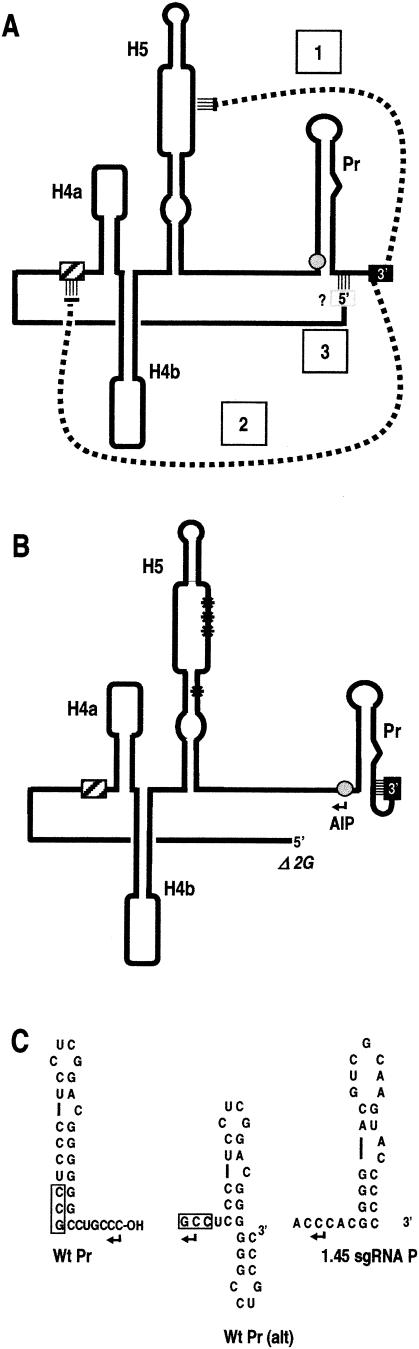
FIG. 8.
Model for the initiation of full-length satC minus strands and the AIP. (A) Diagram showing interactions supported by the present data. Solid box, 3′-terminal bases; shaded box, 5′-terminal bases. Solid line, satC sequence; dashed lines, long-distance base-pairing interactions. In step 1, interaction between the 3′ end and the LSL suppresses minus-strand synthesis by sequestering the 3′ end from the RdRp. In step 2, derepression of minus-strand synthesis is achieved when the 3′ end is released from the LSL by base pairing with the derepressor (hatched box). In step 3, base pairing between the 5′-terminal bases and sequence in the vicinity of Pr stabilizes the structureof Pr, keeping the AIP initiating bases (shaded circle) unavailable within the stem of Pr. The exact location of sequences that interact with the 5′ end is not known. (B) Synthesis of the AIP. Δ2G in combination with loss of H5 structure (G304C) (single asterisk in the H5 stem) or 3′-end-interacting sequences in the LSL (H5RL) (triple asterisk in LSL) leads to rearrangement of Pr and exposure of the AIP initiating sequence. (C) Possible Pr structural rearrangements that lead to base pairing of the 3′CCC internally within Pr. (Left) wt Pr; (center) alternative Pr structure; (right) core promoter for the 1.45-kb sgRNA, showing features similar to the alternative Pr structure.
Both TCV and CCFV contain identical or similar sequence (GGCG and GGUG, respectively) to that of satC just upstream from H4a that is complementary to 3′-end sequences. In addition, in vivo genetic selection of satC, which involved generating satC transcripts with random bases in positions 209 to 220 and then selecting for functional satRNA in plants, indicated that positions 217 to 220 were highly conserved, with all functional satC containing the sequence (U/C)GG(U/C) in that location (X. Sun and A. E. Simon, unpublished data). The presence of a uridylate or cytidylate at position 220 suggests that this base may pair with a guanylate, as proposed in our model.
5′-terminal bases of satC are involved in minus-strand initiation in vitro.
Our present results also suggest the involvement of the satC 5′GG in initiation of minus-strand synthesis in vitro. Published studies using a variety of viral genomic RNA and subviral RNA replicons also support a role for the 5′ end in minus-strand synthesis, through either protein-protein bridges, direct RNA-RNA interactions, or proposed 5′ stabilization of 3′ elements (4, 14, 23, 31, 61, 66, 69). Deletion of the satC 5′GG resulted in the synthesis of many additional products in the absence of any other alterations. Since the presence or absence of the 5′GG had no effect on enhanced transcriptional repression mediated by the mutated derepressor sequence (G218C, C220G, G218CΔ2G, and C220GΔ2G [see Fig. 7]), it seems unlikely that the 5′ end is required for the interaction between the 3′CCC and the LSL. However, in the absence of an interaction between the 3′ end and the LSL (e.g., H5RL and G304C [Fig. 5]), deletion of 5′GG substantially altered the transcription initiation site, leading to near-exclusive synthesis of a faster-migrating product (AIP), the result of internal initiation opposite nucleotides normally positioned within the lower stem of Pr.
One model that can explain the role of the 5′ end of satC in minus-strand synthesis is presented in Fig. 8. We propose that the 5′ end interacts with sequence in or near Pr, stabilizing the structure of the promoter and permitting the RdRp to properly recognize the 3′ end when the derepressor relieves repression. In this wt scenario, the initiation sequence for the AIP remains buried within Pr (Fig. 8A and 8C, left), supporting the proposal that one role for RNA structures in the 3′ region is to sequester alternative initiation sequences from the RdRp (11). When the 3′ end cannot base pair with the LSL, and without the 5′ end to stabilize the Pr structure, we propose that the 3′ end assumes an alternative conformation (Fig. 8B and 8C, center), with the 3′CCC base paired to nearby sequence and sequestered from the RdRp. The AIP initiation sequence is no longer part of the stem of Pr and is available to the RdRp for internal initiation. The proposed alternative structure for Pr resembles the core promoter for the 1.45-kb sgRNA, which contains a series of C:G base pairs upstream of a single-stranded sequence consisting of multiple cytidylates (62) and directs initiation to a similar downstream location (Fig. 8C, right). The alternative Pr structure is not as stable as the wt satC mfold-predicted structure (ΔG, −11.3 versus −15.7 kcal/mol, respectively). However, others have shown that the stability of a hairpin RdRp promoter does not necessarily correlate with its efficiency at promoting complementary-strand synthesis (51). Furthermore, by mutagenesis and in vivo genetic selection approaches, hairpins with many different structures and sequences were able to correctly promote de novo initiation in vitro and accumulation in vivo (7, 56). Since there was no discernible difference between the solution structures of satC in the presence or absence of the 5′GG, it is not currently understood how the 5′ end might stabilize the 3′-terminal structure. It should be noted, however, that we would be unable to detect any alterations that were present 3′ of base 334 by our experimental procedure.
Several earlier in vitro and in vivo studies support a role for 5′-terminal sequence in the synthesis of satC minus strands. Deletion of the seven 3′-terminal bases in otherwise full-length satC minus strands did not affect plus-strand synthesis in vitro, while deletion of an additional five bases reduced, but did not abolish, transcription (19). However, changing the guanylates in positions 2 and 3 individually to uridylate or cytidylate substantially reduced or eliminated detectable accumulation in protoplasts (17). In addition, in vivo genetic selection of satC containing randomized sequence in the 21 5′-terminal bases of the plus strand resulted in the recovery of 2 or 3 guanylates at the 5′ end in all sequenced satRNAs (17). Since these guanylates were not necessary for plus-strand synthesis in vitro, this suggests involvement of the 5′ guanylates in minus-strand synthesis.
Additional H5 properties.
It is not known at present if the RdRp interacts directly with H5 or if H5 functions in some capacity besides that of repressor. Interestingly, H5RL, which contains an altered 3′-end-interacting sequence within the LSL, and G304C, which should not contain an H5 structure, generated very different products in the absence of the 3′CCC when transcribed in vitro (Fig. 5). Transcription of G304CΔ3C produced a single full-length product, while H5RLΔ3C transcripts produced both full-length and aberrantly initiated products, like H5RL and G304C. One possible explanation for this result is that the RdRp might interact with H5 prior to interaction with Pr and, in the absence of the 3′CCC, would be unable to interact correctly with Pr and target a single initiation site. In the absence of the H5 structure, the RdRp might interact solely with Pr and thus be in the vicinity of the 3′ terminus, which might be necessary for exclusive 3′-end transcription in the absence of the 3′CCC.
A similar replication silencer is found in tombusviruses.
A hairpin very similar to H5 was recently found to be a strong suppressor of minus-strand synthesis in the Tombusvirus genus (39). The hairpin, labeled a replication silencer, is located in an upstream position similar to that of H5 relative to a 3′-terminal hairpin (13) (Fig. 6). Five potential base pairs are possible between the 3′-terminal AGCCC of tombusviruses (and viruses in related genera) and the asymmetrical loop sequence, which was proposed to sequester the 3′ terminus from the RdRp by pairing up the 3′-terminal nucleotide. Elements that repress minus-strand synthesis may thus be a general feature of RNA virus regulation of minus-strand synthesis.
Acknowledgments
We are very grateful to P. Nagy for providing the p88 expression plasmid. We also thank C. Song for help with the solution structure studies.
This work was supported by Public Health Service grant GM61515-01 and National Science Foundation grant MCB-0086952 to A.E.S.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Axelrod, V. D., E. Brown, C. Priano, and D. R. Mills. 1991. Coliphage Qβ RNA replication: RNA catalytic for single-strand release. Virology 184:595-608. [DOI] [PubMed] [Google Scholar]
- 3.Ball, L. A. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. USA 91:12443-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter, C. D., J. W. Oh, C. Zhang, and A. E. Simon. 1995. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J. Mol. Biol. 245:608-622. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter, C. D., and A. E. Simon. 1998. Analysis of sequences and predicted structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 26:2426-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascone, P. J., T. Haydar, and A. E. Simon. 1993. Sequences and structures required for RNA recombination between virus-associated RNAs. Science 260:801-805. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, M. R., and C. C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffreda, P., L. Rubino, and M. Russo. 1998. Molecular cloning and complete nucleotide sequence of Galinsoga mosaic virus genomic RNA. Arch. Virol. 143:173-180. [DOI] [PubMed] [Google Scholar]
- 11.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 12.Duggal, R., F. Lahser, and T. Hall. 1994. cis-acting sequences in the replication of plant viruses with plus-sense RNA genomes. Annu. Rev. Phytopathol. 32:287-309. [Google Scholar]
- 13.Fabian, M. R., H. Na, D. Ray, and K. A. White. 2003. 3′-terminal RNA secondary structures are important for accumulation of tomato bushy stunt virus DI RNAs. Virology 313:567-580. [DOI] [PubMed] [Google Scholar]
- 14.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, M., R. Mamoun, and J. M. Bove. 1980. TYMV RNA replication in vivo: replicative intermediate is mainly single stranded. Virology 104:357-374. [DOI] [PubMed] [Google Scholar]
- 17.Guan, H., C. D. Carpenter, and A. E. Simon. 2000. Analysis of cis-acting sequences involved in plus-strand synthesis of a TCV-associated satellite RNA identifies a new carmovirus replication element. Virology 268:345-354. [DOI] [PubMed] [Google Scholar]
- 18.Guan, H., C. D. Carpenter, and A. E. Simon. 2000. Requirement of a 5′-proximal linear sequence on minus-strands for plus-strand synthesis of a satellite RNA associated with TCV. Virology 268:355-363. [DOI] [PubMed] [Google Scholar]
- 19.Guan, H., and A. E. Simon. 1997. RNA promoters located on (−)-strands of a subviral RNA associated with turnip crinkle virus. RNA 3:1401-1412. [PMC free article] [PubMed] [Google Scholar]
- 20.Guan, H., and A. E. Simon. 2000. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc. Natl. Acad. Sci. USA 97:12451-12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guilley, H., J. C. Carrington, E. Balazs, G. Jonard, K. Richards, and T. J. Morris. 1985. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 13:6663-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacker, D. L., I. T. D. Petty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, M., D. C. Koh, L. J. Weng, M. L. Chang, Y. K. Yap, L. Zhang, and S. M. Wong. 2000. Complete nucleotide sequence and genome organization of hibiscus chlorotic ringspot virus, a new member of the genus Carmovirus: evidence for the presence and expression of two novel open reading frames. J. Virol. 74:3149-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khromykh, A. A., N. Kondratieva, J.-Y. Sgro, A. Palmenberg, and E. G. Westaway. 2003. Significance in replication of the terminal nucleotides of the Flavivirus genome. J. Virol. 77:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, C. H., C. C. Kao, and I. Tinoco. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7:415-423. [DOI] [PubMed] [Google Scholar]
- 28.Klovins, J., and J. van Duin. 1999. A long-range pseudoknot in Qβ is essential for replication. J. Mol. Biol. 294:875-884. [DOI] [PubMed] [Google Scholar]
- 29.Kong, Q., J. Wang, and A. E. Simon. 1997. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell 9:2051-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X. H., L. A. Heaton, T. J. Morris, and A. E. Simon. 1989. Turnip crinkle virus defective interfering RNAs intensify viral symptoms and are generated de novo. Proc. Natl. Acad. Sci. USA 86:9173-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyons, T., K. E. Murray, A. W. Roberts, and E. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, F., H. Weber, and C. Weissmann. 1981. Interactions of Qβ replicase with Qβ RNA. J. Mol. Biol. 153:631-660. [DOI] [PubMed] [Google Scholar]
- 34.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Moore, P. B. 1999. Structural motifs in RNA. Annu. Rev. Biochem. 68:287-300. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, P. E., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nomaguchi, M., M. Ackermann, C. Yon, S. You, and B. Padmanabhan. 2003. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J. Virol. 77:8831-8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh, J.-W., Q. Kong, C. Song, C. D. Carpenter, and A. E. Simon. 1995. Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant-Microbe Interact. 8:979-987. [DOI] [PubMed] [Google Scholar]
- 39.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of Turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranjith-Kumar, C. T., X. Zhang, and C. C. Kao. 2003. Enhancer-like activity of a Brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray, D., and K. A. White. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riviere, C. J., and D. M. Rochon. 1990. Nucleotide sequence and genomic organization of melon necrotic spot virus. J. Gen. Virol. 71:1887-1896. [DOI] [PubMed] [Google Scholar]
- 45.Schuppli, D., G. Miranda, H. C. Tsui, M. E. Winker, J. M. Sogo, and H. Weber. 1997. Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc. Natl. Acad. Sci. USA 94:10239-10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, A. E. 1999. Replication, recombination, and symptom-modulation properties of the satellite RNAs of turnip crinkle virus. Curr. Top. Microbiol. Immunol. 239:19-36. [DOI] [PubMed] [Google Scholar]
- 47.Simon, A. E., and S. H. Howell. 1986. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′-end of the helper virus genome. EMBO J. 5:3423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh, R. N., and T. W. Dreher. 1998. Specific site selection in RNA resulting from a combination of non-specific secondary structure and -CCR-boxes: initiation of minus-strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA 4:1083-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivakumaran, K., Y. Bao, M. J. Roossinck, and C. C. Kao. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 74:10323-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sivakumaran, K., and C. C. Kao. 1999. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J. Virol. 73:6415-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sivakumaran, K., C. H. Kim, R. Tayon, Jr., and C. C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skotnicki, M. L., A. M. Mackenzie, M. Torronen, and A. J. Gibbs. 1993. The genomic sequence of cardamine chlorotic fleck carmovirus. J. Gen. Virol. 74:1933-1937. [DOI] [PubMed] [Google Scholar]
- 53.Song, C., and A. E. Simon. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+) and (−)-strands of virus-associated RNAs. Proc. Natl. Acad. Sci. USA 91:8792-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:4-6. [DOI] [PubMed] [Google Scholar]
- 55.Song, C., and A. E. Simon. 1995. Synthesis of novel products in vitro by an RNA-dependent RNA polymerase. J. Virol. 69:4020-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stupina, V., and A. E. Simon. 1997. Analysis in vivo of turnip crinkle virus satellite RNA C variants with mutations in the 3′-terminal minus-strand promoter. Virology 238:470-477. [DOI] [PubMed] [Google Scholar]
- 57.Sun, X., and A. E. Simon. 2003. Fitness of a turnip crinkle virus satellite RNA correlates with a sequence-nonspecific hairpin and flanking sequences that enhance replication and repress the accumulation of virions. J. Virol. 77:7880-7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki, S., S. Hase, H. Takahashi, and M. Ikegami. 2002. The genome organization of pea stem necrosis virus and its assignment to the genus Carmovirus. Intervirology 45:160-163. [DOI] [PubMed] [Google Scholar]
- 59.Takemoto, Y., T. Kanehira, M. Shinohara, S. Yamashita, and T. Hibi. 2000. The nucleotide sequence and genome organization of Japanese iris necrotic ring virus, a new species in the genus Carmovirus. Arch. Virol. 145:651-657. [DOI] [PubMed] [Google Scholar]
- 60.Thomas, C. L., V. Leh, C. Lederer, and A. J. Maule. 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33-41. [DOI] [PubMed] [Google Scholar]
- 61.Vlot, A. C., and J. F. Bol. 2003. The 5′ untranslated region of alfalfa mosaic virus RNA1 is involved in negative-strand RNA synthesis. J. Virol. 77:11284-11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, J., and A. E. Simon. 1997. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology 232:174-186. [DOI] [PubMed] [Google Scholar]
- 63.Wang, J., and A. E. Simon. 1999. Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology 253:327-336. [DOI] [PubMed] [Google Scholar]
- 64.Wang, J., and A. E. Simon. 1999. Symptom attenuation by a satellite RNA in vivo is dependent on reduced levels of virus coat protein. Virology 259:234-245. [DOI] [PubMed] [Google Scholar]
- 65.Weng, Z. M., and Z. G. Xiong. 1997. Genome organization and gene expression of saguaro cactus carmovirus. J. Gen. Virol. 78:525-534. [DOI] [PubMed] [Google Scholar]
- 66.Wu, B., W. B. Vanti, and K. A. White. 2001. An RNA domain within the 5′ untranslated region of tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol. 305:741-756. [DOI] [PubMed] [Google Scholar]
- 67.Yoshinari, S., and T. W. Dreher. 2000. Internal and 3′ RNA initiation by Qβ replicase directed by CCA boxes. Virology 271:363-370. [DOI] [PubMed] [Google Scholar]
- 68.Yoshinari, S., P. D. Nagy, A. E. Simon, and T. W. Dreher. 2000. CCA initiation boxes without unique promoter elements support in vitro transcription by three RNA-dependent RNA polymerases. RNA 6:698-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 70.You, X. J., J. W. Kim, G. W. Stuart, and R. F. Bozarth. 1995. The nucleotide sequence of Cowpea mottle virus and its assignment to the genus Carmovirus. J. Gen. Virol. 76:2841-2845. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, F., and A. E. Simon. 2003. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 312:8-13. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, G., and A. E. Simon. 2003. A multifunctional turnip crinkle virus replication enhancer revealed by in vivo functional SELEX. J. Mol. Biol. 326:35-48. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, X., C. H. Kim, and C. C. Kao. 2003. Stable RNA structure can repress RNA synthesis in vitro by the brome mosaic virus replicase. RNA 9:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]