Abstract
The natural evolution of human immunodeficiency virus type 1 infection often includes a switch in coreceptor preference late in infection from CCR5 to CXCR4, a change associated with expanded target cell range and worsened clinical prognosis. Why coreceptor switching takes so long is puzzling, since it requires as few as one to two mutations. Here we report three obstacles that impede the CCR5-to-CXCR4 switch. Coreceptor switch variants were selected by target cell replacement in vitro. Most switch variants showed diminished replication compared to their parental R5 isolate. Transitional intermediates were more sensitive to both CCR5 and CXCR4 inhibitors than either the parental R5 virus or the final R5X4 (or rare X4) variant. The small number of mutations in viruses selected for CXCR4 use were distinctly nonrandom, with a dominance of charged amino acid substitutions encoded by G-to-A transitions, changes in N-linked glycosylation sites, and isolate-specific mutation patterns. Diminished replication fitness, less-efficient coreceptor use, and unique mutational pathways may explain the long delay from primary infection until the emergence of CXCR4-using viruses.
The entry of human immunodeficiency virus type 1 (HIV-1) into target cells requires binding of the viral envelope glycoprotein gp120 to CD4 and one of two chemokine receptors, CCR5 or CXCR4 (1, 9, 15, 17, 19). Although other chemokine receptors have been identified as potential entry factors, only CCR5 and CXCR4 appear to be important for infection by clinical isolates of HIV-1 (53, 54). This observation is reflected in the current nomenclature for HIV-1 coreceptor use (E. A. Berger, R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss, Letter, Nature 391:240, 1998): R5 for viruses that use only CCR5, X4 for viruses that use only CXCR4, and R5X4 for viruses that can use both receptors. R5 virus isolates are equivalent to macrophage-tropic, non-syncytium-inducing (NSI) viruses, and R5X4 or X4 isolates are T-cell-tropic, syncytium-inducing viruses (43, 44, 47), with only a few interesting exceptions (51, 52). R5 HIV-1 accounts for the vast majority of primary infections regardless of the route of transmission (12, 43). During the evolution of virus populations (quasispecies) within an infected individual, coreceptor switching from R5 to X4 is common in clade B HIV-1 infection (12, 43) and less common in clade C infection (29, 40). The R5-to-X4 coreceptor switch is a harbinger of accelerated clinical disease progression and typically occurs after 8 to 10 years of infection (6, 11). The issue of coreceptor switching has become relevant to drug resistance as coreceptor inhibitors enter clinical trials (18, 37, 48, 49). Coreceptor switching is one route to resistance to these compounds (37, 49).
Coreceptor use maps to the variable V3 and V2 loops of gp120, and the R5-to-X4 switch is often accompanied by an increase in charged residues in the V3 loop (7, 8, 20, 25, 27, 42, 46). As few as one or two amino acid changes in V3 suffice for coreceptor switching (5, 8, 14, 24, 37). Given this minimum requirement for coreceptor switching and the high rate of mutation and recombination in the HIV-1 genome (10, 32, 35), evolution of X4 variants would be expected to be rapid and frequent. Since this is not observed in infected patients, there must be negative selective pressure against X4 variants or unexpected obstacles to the R5-to-X4 switch. Selective pressure influencing the emergence of coreceptor switch variants could be generated by host factors (such as target cell prevalence, chemokine levels, susceptibility to neutralizing antibody or CTL killing [e.g., see reference 22]) and/or intrinsic properties of the virus. We have used in vitro selection to investigate intrinsic obstacles to coreceptor switching.
In these experiments, we have replaced CCR5-bearing target cells with CXCR4-bearing target cells to select for virus variants capable of switching coreceptor use and examined the changes in viral properties during the transition from CCR5 to CXCR4 use. The transition was difficult despite the short distance (few mutations) in sequence space that separate R5 from R5X4 variants. Increased susceptibility to coreceptor inhibitors during the transition implies that coreceptor use is suboptimal. Diminished replication compared to parental R5 isolates implies a loss of competitive fitness. Additional constraints on the transition are imposed by the requirements of inserting charged amino acids at specific locations and by the strong bias in favor of G-to-A substitutions rather than random mutation. Silent mutations are exceedingly rare, implying strong selection for mutations conferring CXCR4 use. These three obstacles, loss of entry efficiency, replication fitness, and limited transitional pathways, may explain the long delay before coreceptor switching in infected patients.
MATERIALS AND METHODS
Cell lines.
U87.CD4.R5 and U87.CD4.X4 cells (4) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1 μg of puromycin/ml, and 300 μg of G418/ml. MT2 and MT2-R5 (39) cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Virus infection.
JR-CSF has been described previously (7). 242 and 230 are molecular clones derived by insertion of mutated V3 regions into NL4-3 (8). The 242 isolate used here has an extra mutation in V3, an R to H at position 21. To generate ADA and BaL infectious viruses, 293T cells were transfected with the pNL4-ADA plasmid (generated by inserting the ADA envelope glycoprotein sequences into the pNL4-3 plasmid; kindly provided by J. Sodroski, Dana-Farber Cancer Institute, Boston, Mass.) or with the pR8-BaL plasmid (generated by inserting the BaL envelope sequences into the pR8 plasmid; kindly provided by C. Aiken, Vanderbilt University, Nashville, Tenn.). All parental viruses were propagated in peripheral blood mononuclear cells (PBMC) cultured for 2 days in 2 μg of phytohemagglutinin/ml and two additional days in 20 U of interleukin 2/ml. The U87.CD4.R5 or X4 cell line was infected with viruses overnight and then washed with phosphate-buffered saline and incubated in culture medium for 4 to 8 days. When PSC-RANTES or AMD3100 entry inhibitor was used, the cells were preincubated with the inhibitors for 2 h before infection. Replication of HIV-1 was determined in the culture supernatant by p24 capsid antigen enzyme-linked immunosorbent assay (Perkin-Elmer, Boston, Mass.).
Generation of coreceptor switch variants.
All R5 viruses were first grown in U87.CD4.R5 cells for 4 days, and then the supernatant was collected and transferred to cultures containing 90% U87.CD4.X4 cells and 10% U87.CD4.R5 cells for another 4 days. This 10-fold dilution of R5 target cells was repeated twice more at 4-day intervals. For HIV-1 isolates ADA and BaL (see Fig. 1), we used a modified selection system in which the viruses were cultured for 7 days in mixtures of U87.CD4.X4 cells and 10, 5, 2, 1, and finally 0.1% U87.CD4.R5 cells. To determine if the viruses had switched to CXCR4 usage, they were tested for growth on U87.CD4.X4 cells after each passage. When viral replication on U87.CD4.X4 cells was detected by p24 enzyme-linked immunosorbent assay and syncytium formation, coreceptor switch variants were grown for two more passages in U87.CD4.X4 cells and then sequenced and tested for growth on MT-2 cells. Several virus isolates were initially unable to grow on MT-2 cells, and further passage was required to isolate viral descendants capable of growth on MT-2 cells. We therefore grew these transitional isolates for 6 days in MT2-R5low cells, which express both CXCR4 and CCR5, and then started adding MT2 cells every 6 days for 4 weeks. The resulting viruses, ADA-1, ADA-3, BaL-1A, BaL-1B, BaL-2A, and BaL-2B, were able to grow on MT2 cells and also on U87.CD4.X4 cells.
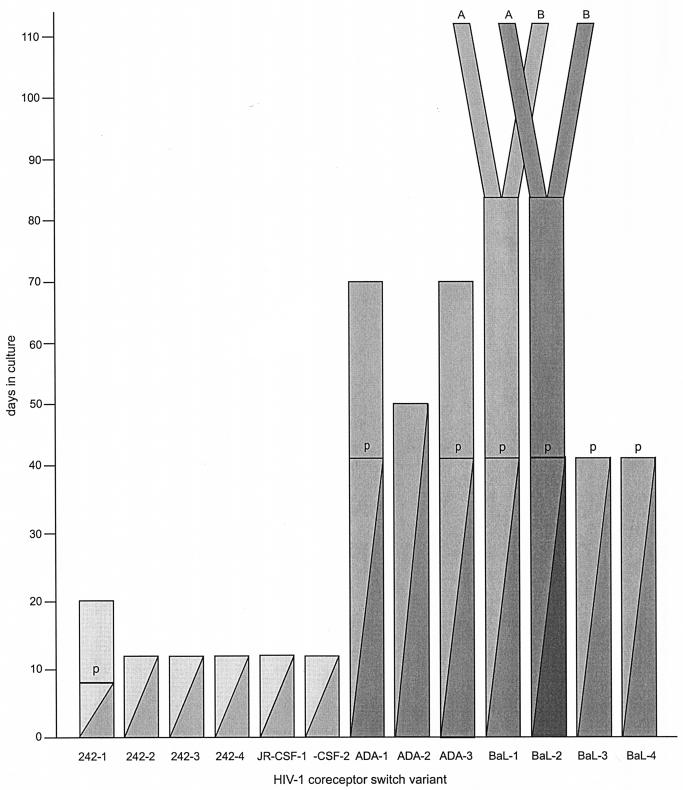
FIG. 1.
Timing of selection and emergence of coreceptor switch mutants. Initially, CCR5 target cells were diluted 10-fold into CXCR4 target cells every 4 days, as indicated by the declining value for the dark filled column. After each dilution, virus was isolated and tested for growth on pure CXCR4 target cells. R5X4 mutants were obtained after 8 to 12 days with starting HIV-1 isolates 242 and JR-CSF. Transitional mutants that replicated in U87-CD4-CXCR4 cells but not MT-2 cells are indicated by p (provisional). These were cultured with MT-2 cells until viruses capable of growth emerged (solid bars). HIV-1 isolates ADA and BaL did not produce coreceptor switch variants under these conditions, so the addition of new CXCR4 targets was performed every 7 days. Transitional (p) or full coreceptor switch mutants were isolated after 42 to 112 days of selection. Solid bars again indicate selection on CXCR4-bearing targets only. Two clonal derivatives of BaL-1p and BaL-2p were isolated. These are designated A and B, and their common origin is indicated by diverging columns. Each separately numbered isolate was generated in an independent experiment.
Virus sequences.
DNA was extracted from infected cells using the QIAamp DNA Mini kit (QIAGEN, Valencia, Calif.). The entire HIV envelope was amplified from cellular DNA by four overlapping PCRs with the following sets of primers: Lgp120S (5′-GAGAGAGAGCAGAAGACAGT-3′) and Lgp120AS (5′-ACTGCCATTTAACAGCAGTTGAGT-3′), outerV3S (5′-CCAATTCCCATACATTATTG-3′) and Mgp120AS (5′-ATCTCTTGTTAATAGCAGCCCAGT-3′), Jgp120S (5′-TGGAGGGGAATTTTTCTACTG-3′) and Jgp120AS antisense (ACCTACCAAGCCTCCTACTATC-3′), and K-end sense (5′-GGGTTGCTCTGGAAAACTCATT-3′) and K-end antisense (5′-GCTGGCTCAGCTCGTCTCCAT-3′). The PCR products were gel purified with the QIAquick gel extraction kit (QIAGEN) and sequenced directly in both directions with the same primers used for the PCR.
Entry inhibitors.
Parental viruses and coreceptor switch variants were evaluated for their sensitivity to the CCR5 inhibitor PSC-RANTES (39) (kindly provided by O. Hartley and R. Offord, University of Geneva) and the CXCR4 inhibitor AMD3100 (16) (kindly provided by G. Henson, AnorMed, Vancouver, British Columbia, Canada) using either U87.CD4.R5 or U87.CD4.X4 target cells. Fifty-percent inhibitory concentrations (IC50s) were calculated from a sigmoidal curve fitting program after log conversion of the inhibitor concentration (Prism 4; GraphPad Software, San Diego, Calif.).
Nucleotide sequence accession numbers.
All nucleotide sequences determined in this study have been deposited in GenBank (accession numbers AY426102 to AY426127).
RESULTS
Generation of HIV-1 coreceptor switch variants in vitro.
CCR5-using (R5) viruses derived from molecular clones were induced to switch coreceptor use by sequential passage into target cell mixtures containing a rapidly escalating percentage of CXCR4-bearing target cells (Fig. 1 and Materials and Methods). This method allowed the isolation of coreceptor switch variants from R5 isolates 242 and JR-CSF within 8 to 20 days. Identical experiments were repeated four times for virus isolate 242 and twice for JR-CSF to ensure the reproducibility of switching, to isolate independent switch mutants, and to avoid potential cross-contamination of PCR-amplified env genes. Switch mutants were isolated by growth on U87.CD4.X4 cells, and use of CXCR4 was confirmed by growth on MT-2 cells and PBMC from a CCR5 Δ32/Δ32 donor. One variant isolate (242-1p) was able to infect U87.CD4.X4 cells but not MT2 cells or CCR5-deficient PBMC. We designate this phenotype a provisional (p) switch variant. After three additional passages in U87.CD4.X4 targets, a secondary switch variant capable of infecting MT2 cells was isolated. As demonstrated below, these provisional switch variants are extremely poor at using CXCR4 for entry and appear to survive on U87.CD4.X4 cells because of the unusually high levels of CXCR4.
Two other R5 viruses (ADA and BaL) did not switch coreceptors using the initial rapid target cell replacement model, and a slower 7-day passage with less-rapid dilution of CCR5-target cells was adopted (Fig. 1 and Materials and Methods). This more sustained selection system allowed us to isolate three ADA switch variants and four BaL switch variants in seven independent experiments. Many of the early isolates displayed the provisional switch phenotype, and additional culture on CXCR4-expressing cells was required to isolate secondary switch variants capable of infecting MT2 cells (see Materials and Methods). None of these provisional switch variants were capable of replication on activated T cells from a CCR5 Δ32/Δ32 donor. In the 13 experiments conducted, coreceptor switch variants were isolated after 8 to 112 days of selection. HIV-1 isolates 242 and JR-CSF generated switch variants more rapidly and transitional variants less frequently than HIV-1 isolates ADA and BaL (Fig. 1). We were thus able to compare the sequence and biologic properties of two classes of coreceptor switch variants, rapid (242 and JR-CSF) and slow (ADA and BaL).
Identification of envelope gene mutations in coreceptor switch variants.
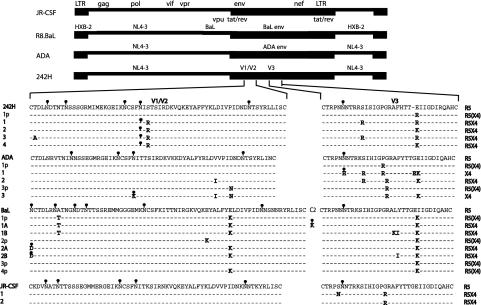
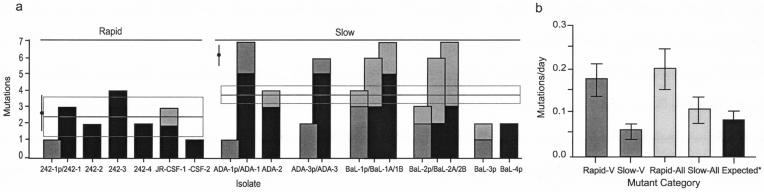
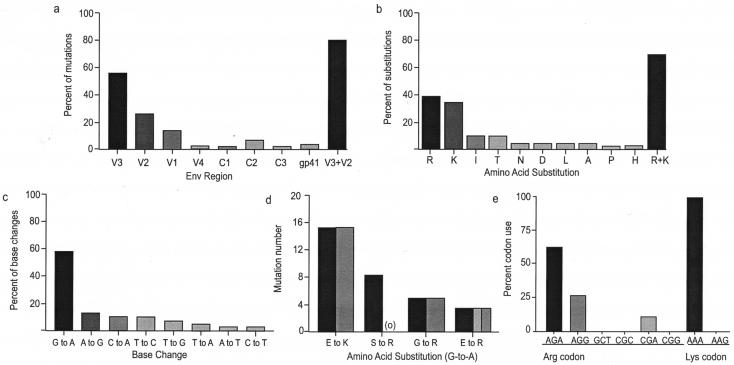
We sequenced the entire envelope region (∼2,500 bp) from starting isolates and their provisional and complete coreceptor switch variants. The V1/V2 and V3 translated amino acid sequences are shown (Fig. 2) because the majority of mutations occurred in these regions (Fig. 3a and 4a). Several features of the mutations associated with coreceptor switching are obvious (Fig. 2 to 4). (i) Substitutions that generated the charged amino acids Arg or Lys were heavily favored, and these were concentrated in V3 and to a lesser extent V2 (Fig. 2 and 4a). By far the most common substitution was from glutamic acid (E) to the charged amino acids lysine (K) or arginine (R) at position 25 of V3 (Fig. 2 and 4b), a hot spot for coreceptor switching previously noted (25). (ii) Several mutations abolished N-linked glycosylation sites, most commonly in V1/V2, as has been observed in other studies (38). In the 242 and JR-CSF switch variants that were generated under fast switch conditions (Fig. 3a), mutations were almost entirely confined to V1/V2 and V3, and the mean number of substitutions required for coreceptor switching was 2.33 ± 1.03 (mean ± standard deviation). By contrast, the ADA and BaL switch variants that were generated under slower switch conditions had more V1/V2 and V3 mutations (3.74 ± 0.47) and additional mutations outside of the variable loops (Fig. 3a). (iii) The difference in both V loop mutations and total mutations (2.50 ± 1.40 versus 6.14 ± 0.40) between the two sets of variants is significant (P = 0.0174 and P = 0.0007, respectively). (iv) The rate of mutation (base substitutions/day) was faster in the 242 and JR-CSF rapid switch variants than predicted by previous studies (35), while the mutation rate for the ADA and BaL late-switch variants was slower and within the expected range (Fig. 3b). (v) Independently isolated coreceptor switch variants derived from the same parental virus showed similar if not identical patterns of mutation in V1/V2 and V3 (Fig. 2). This result suggests unique, context-dependent solutions to coreceptor switch obstacles. (vi) G-to-A base transitions accounted for almost 60% of all mutations (Fig. 4c), and they accounted for 75% of Arg or Lys substitutions (Fig. 4d). This high rate of G-to-A transitions suggests a potential role for Apobec3G/CEM15 (23, 34), the cellular cytosine deaminase whose function is antagonized by viral Vif expression (45). Examination of the nucleotide context of G-to-A mutations showed a preference for GAG and CAG (with the mutated G underlined). This context dependence of mutation is consistent with Apobec3G activity but does not confirm it because of disagreement about the context specificity of the enzyme (23, 34). (vii) There was an obvious bias towards A-rich codons encoding the frequent Arg and Lys mutations (Fig. 4e). There was a highly significant deviation from expected codon use (P < 0.0025, χ2 statistic) calculated by the minimum sequence distance from the parental to the coreceptor switch variant and taking into account the codon options for Arg and Lys. (viii) Finally, every base change identified except two resulted in an amino acid substitution, so the synonymous/nonsynonymous ratio was 0.034. This extraordinarily low value implies high selective pressure for gain-of-function mutations, including those mutations outside of the variable loops.
FIG. 2.
Summary of mutations in V1/V2 and V3 regions of the four HIV-1 isolates studied. All viruses were propagated from molecular clones, and BaL, ADA, and 242 were chimeric clones with unique envelope regions. Each isolate with a different number was recovered from an independent experiment; e.g., 1p and 1 are transitional and full coreceptor switch mutants from the same experiment, but 2, 3, and 4 are mutants from three separate experiments. Sites of N-linked glycosylation are indicated by a small black ball-and-stick symbol; mutations that eliminated glycosylation sites are indicated by the same symbol in grey.
FIG. 3.
Frequency (a) and rate (b) of mutations in V regions and the entire env region. Clonally related sequences are shown by overlapping columns, with V region mutations in black and total mutations in grey. Switch variants are segregated into rapid (242 and JR-CSF) and slow (ADA and BaL) to reflect the timing shown in Fig. 1. Mutations in provisional isolates are indicated by lighter fill. The expected mutation rate (*) is taken from the work of Mansky and Temin (36). The mean number of V region mutations ± standard errors of the mean (excluding the provisional switch mutants) is shown by the horizontal bar and box, and the total mutations ± standard errors of the mean is shown by the filled black circle and vertical bar.
FIG. 4.
Distribution and composition of mutations. Mutations are listed by region (a), amino acid substitution (b), all base changes (c), contribution of G-to-A transitions to frequent substitutions (d), and codon usage for the frequent Arg and Lys substitutions (e). In panel a, V region mutations are indicated by solid fill and C region mutations are indicated by lighter fill. In panel d, the number of amino acid changes is indicated in solid fill and the number generated by G-to-A mutations is indicated in lighter fill. Three E-to-R substitutions were generated by two base changes, one of which was G to A. Panel e shows the observed (solid fill) and expected (lighter fill) codon use for the frequent Arg and Lys substitutions. The expected codon use was calculated based on the minimum number of base changes required to encode Arg or Lys based on the original codon in the parental isolate.
Characterization of switch variant use of CCR5 and CXCR4.
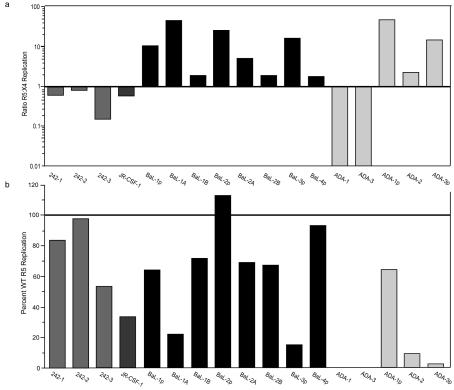
We measured the growth characteristics of each parental virus, transitional switch mutant, and full coreceptor switch mutant on CCR5- or CXCR4-bearing target cells, and we also determined the sensitivity of each isolate to the CCR5-specific entry inhibitor PSC-RANTES (39) or the CXCR4-specific entry inhibitor AMD3100 (16) as an estimate of viral entry efficiency. Most coreceptor switch variants were R5X4 in phenotype, with the exception of two ADA-derived variants that were X4 only (Fig. 2). Most R5X4 viruses showed preferential replication on either CCR5- or CXCR4-expressing target cells (Fig. 5a). Of note, all four rapid switch variants that were derived from HIV-1 242 and JR-CSF showed better replication on CXCR4-expressing target cells, while only ADA-1 and ADA-3 among the slow switch variants preferred CXCR4. However, most R5X4 variants showed poorer replication on CCR5-expressing target cells than their parental R5 isolate (Fig. 5b).
FIG. 5.
Relative replication of each HIV-1 switch variant on U87-CD4-CCR5 cells versus U87-CD4-CXCR4 cells (a). Data are displayed as the ratio of R5:X4 replication, which was calculated by p24 capsid antigen levels on the two target cell lines after 4 to 5 days of culture. Relative replication of each switch variant versus that of the parental R5 isolate on U87-CD4-CCR5 cells is plotted as the percent of parental wild-type (WT) replication (b).
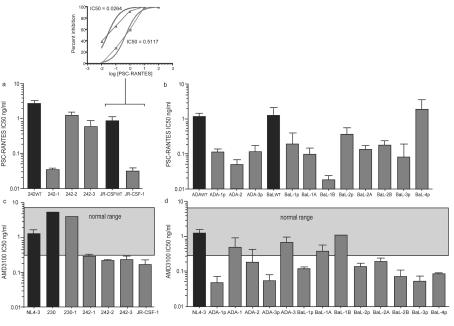
When R5X4 coreceptor switch variants were compared to their parental R5 isolates for sensitivity to the CCR5 entry inhibitor PSC-RANTES (Fig. 6a and b), each variant was significantly more sensitive than the parental isolate with the sole exception of isolate BaL-4p. The IC50s for the switch variants declined from a little as 2-fold (isolate 242-2) to as much as 50-fold (isolate ADA-2). The ability of the coreceptor switch variants to utilize CXCR4 could not be compared to that of the parental R5 isolate but could be compared to historical values (30) and experimental values for X4 (NL4-3, 230) or R5X4 (230-1, a derivative of the X4 isolate 230 with one mutation in V3) isolates (Fig. 6c). Inhibition of these reference isolates by the CXCR4-specific entry inhibitor AMD3100 fell within the expected IC50 range. However, most R5X4 variants had IC50s below the normal range, and transitional variants were extremely sensitive to AMD3100 inhibition. This result is in agreement with their defining phenotype, the ability to grow on U87-CD4-CXCR4 cells but not MT-2 cells. Although most R5X4 switch variants were more sensitive to AMD3100 inhibition, the two ADA variants that had switched to a full X4 phenotype and the two late BaL-1 switch variants (BaL-1A and BaL-1B) had IC50s in the normal range (Fig. 6d). The BaL-1 series of mutants showed a progressive increase in sensitivity to PSC-RANTES inhibition as AMD3100 sensitivity decreased (compare Fig. 6b and d), but this trend was not observed in all coreceptor switch variants.
FIG. 6.
Entry inhibition studies. Entry of parental viruses (wild type [WT]) and coreceptor switch variants was blocked using the potent CCR5 inhibitor PSC-RANTES (39) and the CXCR4 inhibitor AMD3100 (16). Representative data are shown in the small insert, and mean IC50s are shown for PSC-RANTES (a and b) and AMD3100 (c and d). Target cells were U87-CD4-CCR5 in panels a and b and U87-CD4-CXCR4 in panels c and d. Parental isolates in panels a and b are designated by solid fill, and coreceptor switch variants are designated by lighter fill. Since parental isolates were unable to grow on CXCR4 target cells, the normal IC50 range of several X4 isolates is shown (13, 30). NL4-3 and 230 (indicated in filled bars) are well known X4 isolates, and 230-1 is an R5X4 derivative of 230 with a single mutation in V3. Each error bar represents the standard deviation for two to three replicate experiments.
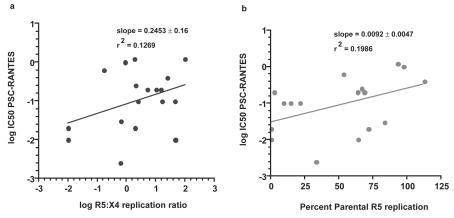
We performed regression analysis of the three parameters of coreceptor switch variants that we measured (sensitivity to PSC-RANTES inhibition, relative replication of R5 versus X4 target cells, and relative replication compared to parental R5 isolates on R5 target cells) to determine if there was a strong correlation between these parameters (Fig. 7a and b). There was only a weak positive correlation between either target cell preference or replication rate on R5 target cells and the PSC-RANTES IC50 value, and neither achieved statistical significance. These results suggest that the assays employed measure independent properties of each virus isolate and that increased sensitivity to coreceptor inhibitors was not caused by loss of replicative fitness.
FIG. 7.
Lack of correlation between sensitivity to PSC-RANTES inhibition (log IC50) and target cell preference (a) or replication relative to the parental R5 isolate (b). The P value for the regression line in panel a is 0.1467 and in panel b is 0.0730, which means that neither slope is significantly different from 0.
DISCUSSION
These results demonstrate that the evolutionary pathway of HIV-1 from CCR5 to CXCR4 coreceptor use involves strong selection of replacement mutations that initially confer high sensitivity to both CCR5 and CXCR4 inhibitors. Subsequent mutations were able to confer more-efficient use of CXCR4, as judged by the ability to infect cells expressing lower levels of CXCR4 and lesser susceptibility to AMD3100 inhibition. Increased sensitivity to RANTES inhibition was a robust indicator of better patient outcome in a recent clinical study (50), suggesting that the increased sensitivity to PSC-RANTES we observed may be related to reduced viral fitness. Another potentially related observation is that viral fitness can be impacted by slower kinetics of entry (41). We hypothesize that the transient inability of coreceptor switch variants to use either CCR5 or CXCR4 as well as their parental viruses would place them at a competitive disadvantage, which should represent a substantial obstacle to completing the process of coreceptor switching. In addition, coreceptor switch variants generally displayed slower replication rates than parental R5 isolates, another component of viral fitness that would place recently generated switch variants at a competitive disadvantage.
Our analysis of coreceptor switching was aided by the capture of a number of transitional switch variants that were able to survive on U87-CD4-CXCR4 cells expressing very high levels of CXCR4 but were not able to survive on MT-2 cells or primary T cells from a CCR5 Δ32/Δ32 donor that expressed less CXCR4. These transitional variants appear to utilize CXCR4 with extremely low efficiency, since they were very sensitive to inhibition with the CXCR4 antagonist AMD3100 (Fig. 6). There has been one recent report that AMD3100-resistant HIV-1 mutants are less fit than their AMD3100-sensitive precursors (2), but we believe that this is the first report that defects in coreceptor use constitute a reproducible feature of the transition from CCR5 to CXCR4 use. There has also been one report of HIV-1 isolates with unusual phenotypes (e.g., R5, syncytium inducing) recovered at the time of coreceptor switching in patients (51), and these may represent the rare in vivo analogues for the transitional variants we have isolated. However, the use of U87-CD4-CXCR4 cells as the selection target may bias the selection process in vitro because of high expression of CXCR4 or cell surface proteoglycans, and the relevance of the mutations we have defined to those that occur in patients remains to be determined.
As expected, evolution to new coreceptor use is driven by mutation. However, the mutational process displays several features that suggest it is not a random walk across sequence space. First, there are a very low number of silent mutations (<4%). Although selection for gain of function is expected to favor replacement mutations, we observed as many as seven replacement mutations (Fig. 3a) with no silent mutations. This absence of silent mutations resembles somatic hypermutation of immunoglobulin V regions during antigen-selected affinity maturation (21) and is much lower than the 0.4 silent/replacement mutation ratio in V3 regions from sequential patient isolates of HIV-1 (33). Codon bias and the high prevalence of G-to-A transition mutations may also contribute to the low frequency of silent mutations (see below). The pattern of mutations in V3 and V1/2 was similar in independent switch variants derived from the same parental isolate, but different parental isolates gave rise to different patterns of mutation (Fig. 2). This observation suggests that the solution to the coreceptor switch problem was context dependent. The high prevalence of Arg and Lys substitutions is expected from previous studies of coreceptor use (20, 36), but it has not been emphasized previously that the vast majority of these substitutions result from G-to-A mutations (Fig. 4c and d). This preference for G-to-A transitions suggests the possible involvement of the cytidine deaminase Apobec3g/CEM15 in the mutation process (23, 34). It was known that G-to-A mutations are the most prevalent during HIV-1 replication (26, 35), but the assumption that this reflects an error-prone reverse transcriptase lacks convincing experimental support (28). It is possible that Apobec3g/CEM15 contributes to the high rate of G-to-A mutations and that the opposing activity of Vif is incomplete. Since three of the four viruses used in these experiments expressed Vif from the common NL4-3 genome, our results cannot be attributed to different Vif alleles. Both U87 cells and MT-2 cells express Apobec3g (C. Pastore, unpublished observations), so it is plausible that this cellular cytidine deaminase influenced the HIV-1 mutants generated in these experiments. There was also a strong codon bias toward A-rich codons encoding Lys or Arg in our data (Fig. 4d and e), as has been noted by others (3). This is expected from the high frequency of G-to-A mutations, but it implies that the HIV-1 genome, which is already the most A rich of all retrovirus genomes (3), may become even more A rich in the process of coreceptor switching. It is possible that HIV-1 is evolving toward an A-rich ceiling and adding more adenosines will prove to be another intrinsic obstacle to switching.
Many of the mutations we observed were in V regions, but there were mutations in C regions and gp41, particularly in the BaL-2 series of variants (Fig. 3a). The low frequency of silent mutations suggests that all of these amino acid substitutions contributed to coreceptor switching, but the importance of each single mutation cannot be assessed until they are introduced by site-directed mutagenesis in future experiments. None of the C-region mutations we observed correspond to the K421D change previously found to reduce fusion efficiency dramatically (41).
The obstacles to coreceptor switching are relevant both for the natural evolution of the disease process and for the development of resistance to antivirals targeting coreceptors. We began these experiments to ascertain if the probability of coreceptor switching differed between R5 isolates, a finding that might explain the development of resistance to CCR5 inhibitors with (37) or without (49) acquisition of CXCR4 use. Our results demonstrate that the mutational distance from CCR5 to CXCR4 use does differ between viral isolates but that the distance is still relatively small (1 to 5 V-region mutations [Fig. 3]). Development of resistance to a CCR5 inhibitor by coreceptor switching for an R5 isolate like ADA or BaL would be predicted to take longer than for 242 or JR-CSF, and the time until switching could be extended even further by increased sensitivity of the transitional intermediates to the inhibitor. Development of resistance to entry inhibitors is thus likely to obey the same general rules as resistance to other classes of antivirals, which reflect the rate of mutation, the number of mutations required for resistance, and the fitness of resistant variants (31).
Acknowledgments
This work was supported by National Institutes of Health grants AI052778 and AI51649 (D.E.M.), The James B. Pendleton Charitable Trust, and National Institutes of Health Grant RR00833 supporting The Scripps Research Institute General Clinical Research Center.
Footnotes
Publication 16057-IMM from The Scripps Research Institute.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Armand-Ugon, M., M. E. Quinones-Mateu, A. Gutierez, J. Barretina, J. Blanco, D. Schols, E. De Clercq, B. Clotet, and J. A. Este. 2003. Reduced fitness of HIV-1 resistant to CXCR4 antagonists. Antivir. Ther. 8:1-8. [PubMed] [Google Scholar]
- 3.Berkhout, B., A. Grigoriev, M. Bakker, and V. V. Lukashov. 2002. Codon and amino acid usage in retroviral genomes is consistent with virus-specific nucleotide pressure. AIDS Res. Hum. Retrovir. 18:133-141. [DOI] [PubMed] [Google Scholar]
- 4.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, M. T., G. R. Simpson, A. J. Cann, M. A. Johnson, and R. A. Weiss. 1993. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol. 67:3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozzette, S., J. McCutchan, S. Spector, B. Wright, and D. Richman. 1993. A cross-sectional comparison of persons with syncytium- and non-syncytium-inducing human immunodeficiency virus. J. Infect. Dis. 168:1374-1379. [DOI] [PubMed] [Google Scholar]
- 7.Cheng-Mayer, C., M. Quiroga, J. W. Tung, D. Dina, and J. A. Levy. 1990. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J. Virol. 64:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1996. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J. Virol. 70:9055-9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., H. Mohri, Y. Cao, and D. D. Ho. 1993. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J. Virol. 67:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq, E., N. Yamamoto, R. Pauwels, J. Balzarini, M. Witvrouw, K. De Vreese, Z. Debyser, B. Rosenwirth, P. Peichl, R. Datema, et al. 1994. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob. Agents Chemother. 38:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dejucq, N., G. Simmons, and P. R. Clapham. 2000. T-cell line adaptation of human immunodeficiency virus type 1 strain SF162: effects on envelope, vpu and macrophage-tropism. J. Gen. Virol. 81:2899-2904. [DOI] [PubMed] [Google Scholar]
- 15.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 16.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 17.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 18.Este, J. A., C. Cabrera, J. Blanco, A. Gutierrez, G. Bridger, G. Henson, B. Clotet, D. Schols, and E. De Clercq. 1999. Shift of clinical human immunodeficiency virus type 1 isolates from X4 to R5 and prevention of emergence of the syncytium-inducing phenotype by blockade of CXCR4. J. Virol. 73:5577-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 20.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannum, L. G., A. M. Haberman, S. M. Anderson, and M. J. Shlomchik. 2000. Germinal center initiation, variable gene region hypermutation, and mutant B cell selection without detectable immune complexes on follicular dendritic cells. J. Exp. Med. 192:931-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harouse, J. M., C. Buckner, A. Gettie, R. Fuller, R. Bohm, J. Blanchard, and C. Cheng-Mayer. 2003. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proc. Natl. Acad. Sci. USA 100:10977-10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 24.Harrowe, G., and C. Cheng-Mayer. 1995. Amino acid substitutions in the V3 loop are responsible for adaptation to growth in transformed T-cell lines of a primary human immunodeficiency virus type 1. Virology 210:490-494. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman, N. G., F. Seillier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, M. A., F. S. Li, A. B. van 't Wout, D. C. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji, J., and L. A. Loeb. 1994. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology 199:323-330. [DOI] [PubMed] [Google Scholar]
- 29.Johnston, E. R., L. S. Zijenah, S. Mutetwa, R. Kantor, C. Kittinunvorakoon, and D. A. Katzenstein. 2003. High frequency of syncytium-inducing and CXCR4-tropic viruses among human immunodeficiency virus type 1 subtype C-infected patients receiving antiretroviral treatment. J. Virol. 77:7682-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labrosse, B., J. L. Labernardiere, E. Dam, V. Trouplin, K. Skrabal, F. Clavel, and F. Mammano. 2003. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J. Virol. 77:1610-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leigh Brown, A. J., S. D. Frost, W. C. Mathews, K. Dawson, N. S. Hellmann, E. S. Daar, D. D. Richman, and S. J. Little. 2003. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J. Infect. Dis. 187:683-686. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukashov, V. V., C. L. Kuiken, and J. Goudsmit. 1995. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J. Virol. 69:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 35.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald, R. A., G. Chang, and N. L. Michael. 2001. Relationship between v3 genotype, biologic phenotype, tropism, and coreceptor use for primary isolates of human immunodeficiency virus type 1. J. Hum. Virol. 4:179-187. [PubMed] [Google Scholar]
- 37.Mosier, D. E., G. R. Picchio, R. J. Gulizia, R. Sabbe, P. Poignard, L. Picard, R. E. Offord, D. A. Thompson, and J. Wilken. 1999. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 73:3544-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogert, R. A., M. K. Lee, W. Ross, A. Buckler-White, M. A. Martin, and M. W. Cho. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastore, C., G. R. Picchio, F. Galimi, R. Fish, O. Hartley, R. E. Offord, and D. E. Mosier. 2003. Two mechanisms for human immunodeficiency virus type 1 inhibition by N-terminal modifications of RANTES. Antimicrob. Agents Chemother. 47:509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 43.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuitemaker, H., N. A. Kootstra, R. E. de Goede, F. de Wolf, F. Miedema, and M. Tersmette. 1991. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 46.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1992. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:9434-9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tersmette, M., J. M. Lange, R. E. de Goede, F. de Wolf, J. K. Eeftink-Schattenkerk, P. T. Schellekens, R. A. Coutinho, J. G. Huisman, J. Goudsmit, and F. Miedema. 1989. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet i:983-985. [DOI] [PubMed] [Google Scholar]
- 48.Trkola, A., T. J. Ketas, K. A. Nagashima, L. Zhao, T. Cilliers, L. Morris, J. P. Moore, P. J. Maddon, and W. C. Olson. 2001. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J. Virol. 75:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola, A., H. Kuster, C. Leemann, C. Ruprecht, B. Joos, A. Telenti, B. Hirschel, R. Weber, S. Bonhoeffer, and H. F. Gunthard. 2003. Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J. Virol. 77:13146-13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A. M. de Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 106:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi, Y., S. Rana, J. D. Turner, N. Gaddis, and R. G. Collman. 1998. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., T. He, Y. Huang, Z. Chen, Y. Guo, S. Wu, K. J. Kunstman, R. C. Brown, J. P. Phair, A. U. Neumann, D. D. Ho, and S. M. Wolinsky. 1998. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J. Virol. 72:9307-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]