Abstract
The human coronavirus 229E (HCoV-229E) replicase gene-encoded nonstructural protein 13 (nsp13) contains an N-terminal zinc-binding domain and a C-terminal superfamily 1 helicase domain. A histidine-tagged form of nsp13, which was expressed in insect cells and purified, is reported to unwind efficiently both partial-duplex RNA and DNA of up to several hundred base pairs. Characterization of the nsp13-associated nucleoside triphosphatase (NTPase) activities revealed that all natural ribonucleotides and nucleotides are substrates of nsp13, with ATP, dATP, and GTP being hydrolyzed most efficiently. Using the NTPase active site, HCoV-229E nsp13 also mediates RNA 5′-triphosphatase activity, which may be involved in the capping of viral RNAs.
Human coronavirus 229E (HCoV-229E), a group 1 coronavirus, is one of the major viral pathogens causing upper respiratory tract illness in humans (7-9, 30). Expression of the 27.3-kb, positive-stranded RNA genome of HCoV-229E involves a series of complex transcriptional, translational, and posttranslational regulatory mechanisms (15, 16, 25, 27, 33, 49, 51, 52). Following receptor-mediated entry (48), two large replicative polyproteins, pp1a (∼450 kDa) and pp1ab (∼750 kDa), are translated. They are encoded by the replicase gene (20,276 bases) that is comprised of open reading frames 1a and 1b (ORF1a and ORF1b) (15). Expression of the ORF1b-encoded region of pp1ab requires ribosomal frameshifting into the −1 frame just upstream of the ORF1a translation termination codon (15, 16). Activation of the viral replication-transcription complex, which also contains a set of enzymes that are rare or even unique among plus-stranded RNA viruses (+RNA viruses) (40), involves extensive autoprocessing of HCoV-229E pp1a and pp1ab by two paralogous papain-like cysteine proteinases (PL1pro and PL2pro) (14, 17, 53) and a 3C-like (main) proteinase (termed 3CLpro or Mpro) (2, 12, 18, 19, 49-52). Together, at least 16 nonstructural proteins (nsp) are released from pp1a/pp1ab by PL1pro, PL2pro, and 3CLpro in a timely regulated manner (13, 51). The replicase complex synthesizes both genome-length RNAs (replication) and a set of subgenomic mRNAs (transcription) (15, 33) that all have a 5′-terminal leader sequence which is acquired from the 5′ end of the genome through a unique discontinuous RNA synthesis mechanism (32, 34-36, 54).
Generally, the coronavirus replicative enzymes are only distantly related to their viral and cellular homologs (10, 11, 24, 40, 52), and in this respect, the viral helicase is no exception. Thus, for example, coronavirus (and other nidovirus) helicase domains are unique among their known +RNA virus homologs in that they are linked in a single protein to an N-terminal binuclear zinc-binding domain consisting of 12 conserved Cys/His residues (11, 19, 37, 44). Also, with respect to their biochemical properties (3, 21, 37, 38, 41, 42), nidovirus helicases differ from other RNA viral helicases, suggesting that the coronavirus enzymes may serve distinct functions in the viral life cycle (37). The HCoV-229E helicase domain is part of nsp13, a 597-residue cleavage product that is released from pp1ab by the activity of 3CLpro (19). Previously, we have shown that the corresponding protein sequence, when expressed as a histidine-tagged fusion protein in insect cells, mediates ATPase, GTPase, and 5′-to-3′ RNA and DNA duplex-unwinding activities (37, 39). In the present study, we used this enzyme, termed HEL, and an inactive control protein, HEL_KA (37), to analyze the biochemical properties of the HCoV-229E nsp13 helicase in more detail.
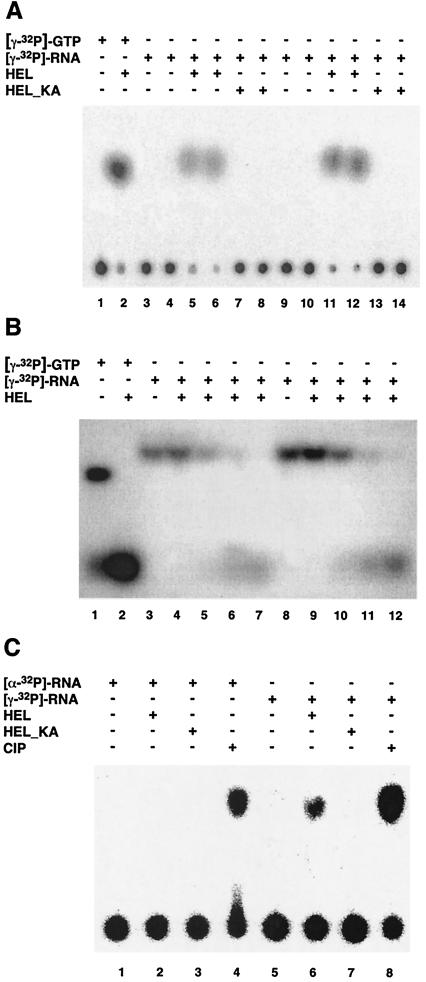
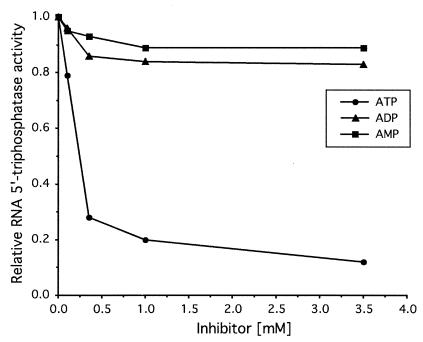
It is well established that the energy required for translocation of helicases along nucleic acids is derived from the binding and hydrolysis of ATP and, in some cases, other (ribo)nucleotides (6, 29, 46). To determine the cofactor requirements of the HCoV-229E nsp13 helicase, we analyzed the nucleoside triphosphatase (NTPase) activities of HCoV-229E nsp13 using a thin-layer chromatography assay (37). Briefly, 0.7 nM HEL was incubated at 22°C with various concentrations of nucleoside triphosphate (NTP) or deoxynucleoside triphosphate (dNTP) (0.5 to 10 μM) in buffer consisting of 20 mM HEPES-KOH (pH 7.4), 10% glycerol, 5 mM magnesium acetate, 2 mM dithiothreitol, 0.1 mg of bovine serum albumin/ml, and the respective α-32P-labeled NTP or dNTP (400 Ci/mmol; Amersham). Following incubation for 2 to 9 min, the reactions were stopped by adding EDTA (pH 8.0) to a final concentration of 100 mM. The reaction products were analyzed by using thin-layer chromatography with polyethyleneimine-cellulose F plates (Merck), using potassium phosphate (pH 4.0) as the liquid phase. Substrate hydrolysis was measured by phosphorimaging using ImageQuant software as described previously (37), and kinetic parameters were determined by using Hofstee plots (20). The data we obtained (Table 1) revealed a high promiscuity of nsp13 with respect to the nucleotide cofactor. All common NTPs and dNTPs were hydrolyzed with Km values in the low micromolar range and with only minor variation in the kinetic constants. ATP, dATP, and GTP were found to be only slightly preferred over other nucleotides. In the presence of poly(U), which was shown previously to stimulate the ATPase activity of HEL (37), both Km and kcat were found to be significantly increased for all NTPs. Also, ATP and GTP were hydrolyzed most efficiently under these conditions, indicating that the allosteric stimulation by the RNA cofactor had no significant effect on the specificity of the HEL NTPase activity. Furthermore, the dNTPase data led us to conclude that the specificity of the dNTP-binding site of HEL is determined primarily by the 5′-phosphate groups rather than the nucleobase or sugar moieties. This finding prompted us to investigate whether HCoV-229E nsp13 (like its severe acute respiratory syndrome coronavirus [SARS-CoV] homolog) (21) can also act as an RNA 5′-triphosphatase. Two 5′-γ-32P-labeled RNA substrates, RNA37 (5′-GACUUAAG-3′) and RNA41 (5′-GACUUAAGUACC-3′), were transcribed in vitro in the presence of [γ-32P]GTP using T7 RNA polymerase. The DNA templates used for the transcription of RNA37 and RNA41 were produced by annealing the following pairs of oligonucleotides (T7 promoter sequences underlined): 5′-AATAATACGACTCACTATAGACTTAAG-3′ was annealed with 5′-CTTAAGTCTATAGTGAGTCGTATTATT-3′, and 5′-AATAATACGACTCACTATAGACTTAAGTACC-3′ was annealed with 5′-GGTACTTAAGTCTATAGTGAGTCGTATTATT-3′. Following incubation of the 5′-γ-32P-labeled RNAs 37 and 41 with HEL, the reaction products were analyzed by thin-layer chromatography. As Fig. 1A shows, HEL released radioactivity from 5′-γ-32P-labeled RNAs, which strongly supported the presumed RNA 5′-triphosphatase activity of HCoV-229E nsp13. The radiolabel comigrated with that of the orthophosphate produced by [γ-32P]GTP hydrolysis, indicating that HEL cleaved the β-γ phosphate bond of the 5′-terminal nucleotide of the 8- and 12-nucleotide (nt) RNA substrates used in this experiment. To formally exclude the possibility that contaminating Escherichia coli RNase activities degraded the RNA substrate and thereby released [γ-32P]GTP that was then hydrolyzed by the GTPase activity of HEL, we analyzed the reactions also by polyacrylamide gel electrophoresis, along with GTP as a control. As shown in Fig. 1B, no [γ-32P]GTP or 5′-[γ-32P]GTP-containing oligoribonucleotides smaller than the substrate RNAs were detectable, confirming that the radiolabel was indeed released from the intact RNA substrate rather than from [γ-32P]GTP. When RNA transcribed in the presence of [α-32P]GTP was used as a substrate, no radioactivity was released (Fig. 1C), which demonstrated that nsp13, unlike alkaline phosphatase, does not have a general phosphohydrolase activity that would also remove the 5′ β- and α-phosphate groups. The NTPase-deficient control protein, HEL_KA, lacked RNA 5′-triphosphatase activity (Fig. 1), and the RNA 5′-triphosphatase activity could be effectively inhibited by ATP (Fig. 2), whereas up to 3.5 mM ADP and AMP had only minor effects on the RNA 5′-triphosphatase activity of HEL. The combined data suggest that (i) the dNTPase and RNA 5′-triphosphatase activities have a common (or overlapping) active site, and (ii) HEL interacts primarily with the γ-phosphate of the 5′-terminal nucleotide of the RNA substrate, which is consistent with the observations reported above for the NTPase activity of HEL.
TABLE 1.
Analysis of HCoV-229E nsp13 NTPase substrate specificitya
| Nucleotide ± poly(U) | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) |
|---|---|---|---|
| ATP − | 2.18 ± 0.07 | 1.85 ± 0.03 | 0.85 |
| UTP − | 4.35 ± 0.01 | 1.31 ± 0.01 | 0.30 |
| GTP − | 2.90 ± 0.05 | 1.67 ± 0.10 | 0.58 |
| CTP − | 7.23 ± 0.24 | 3.08 ± 0.08 | 0.43 |
| ATP + | 100 ± 9 | 215 ± 5 | 2.15 |
| UTP + | 188 ± 8 | 162 ± 2 | 0.86 |
| GTP + | 140 ± 5 | 212 ± 3 | 1.51 |
| CTP + | 290 ± 17 | 213 ± 15 | 0.73 |
| dATP − | 1.62 ± 0.01 | 1.38 ± 0.07 | 0.85 |
| dTTP − | 2.11 ± 0.06 | 0.26 ± 0.01 | 0.12 |
| dGTP − | 1.30 ± 0.04 | 0.59 ± 0.01 | 0.45 |
| dCTP − | 7.34 ± 1.90 | 2.59 ± 0.32 | 0.35 |
The kinetic constants for NTP hydrolysis were determined from Hofstee plots (20). Reactions were performed with 0.7 nM HEL and NTP concentrations ranging from 0.5 to 10 μM (in the absence of polynucleotide cofactor) or from 80 to 300 μM in the presence of 100 nM poly(U), that is, under saturating concentrations of the polynucleotide cofactor.
FIG. 1.
HCoV-229E nsp13 has RNA 5′-triphosphatase activity. In panels A and B, the 5′-γ-32P-labeled RNAs, RNA37 (5′-GACUUAAG-3′) and RNA41 (5′-GACUUAAGUACC-3′), respectively, were incubated with HEL and HEL_KA, and the reaction products were separated by thin-layer chromatography (A) or polyacrylamide gel electrophoresis (B) and visualized by autoradiography. (A) Lane 1, [γ-32P]GTP after incubation for 60 min without protein; lane 2, [γ-32P]GTP after incubation for 60 min with HEL; lanes 3 and 4, RNA37 after incubation for 30 and 60 min, respectively, without protein; lanes 5 and 6, RNA37 after incubation for 30 and 60 min, respectively, with HEL; lanes 7 and 8, RNA37 after incubation for 30 and 60 min, respectively, with HEL_KA; lanes 9 and 10, RNA41 after incubation for 30 and 60 min, respectively, without protein; lanes 11 and 12, RNA41 after incubation for 30 and 60 min, respectively, with HEL; lanes 13 and 14, RNA41 after incubation for 30 and 60 min, respectively, with HEL_KA. (B) Lane 1, [γ-32P]GTP after incubation for 60 min without protein; lane 2, [γ-32P]GTP after incubation for 60 min with HEL; lanes 3 and 8, RNA37 and RNA41, respectively, after incubation for 60 min without protein; lanes 4 to 7, incubation of RNA37 for 5, 10, 30, and 60 min, respectively, with HEL; lanes 9 to 12, incubation of RNA41 for 5, 10, 30, and 60 min, respectively, with HEL. (C) The RNAs used in this experiment had the sequence 5′-GGGAAAAA-3′ and were synthesized by in vitro transcription in the presence of either [α-32P]GTP (lanes 1 to 4) or [γ-32P]GTP (lanes 5 to 8) as described previously (21). The substrate RNAs were incubated without protein (lanes 1 and 5), with HEL (lanes 2 and 6), with HEL_KA (lanes 3 and 7), or with alkaline phosphatase from calf intestine (CIP) (lanes 4 and 8). Reaction products were separated by thin-layer chromatography on polyethyleneimine cellulose F plates and visualized by autoradiography. Reactions were incubated for 60 min (lanes 1, 2, 3, 5, 6, and 7) or 30 min (lanes 4 and 8).
FIG. 2.
Inhibition of the HEL-associated RNA 5′-triphosphatase activity by ATP. The plot illustrates the effect of including various concentrations of ATP, ADP, and AMP on the RNA 5′-triphosphatase activity of HEL. The RNA 5′-triphosphatase activity of HEL in the absence of inhibitor was taken to be 1.0, and all other activities were normalized to this value. The average values of two experiments are plotted.
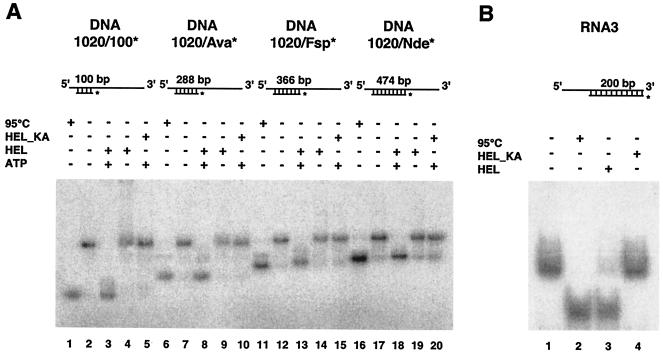
In previous studies, we have shown that nidovirus helicases unwind their substrates in a 5′-to-3′ direction (37, 38). We also demonstrated that the HCoV-229E helicase is able to unwind both RNA and DNA substrates with duplex regions of 33 and 22 bp, respectively (37, 38). To characterize these duplex-unwinding activities in more detail, we now prepared partial-duplex RNA and DNA substrates containing extended double-stranded regions. The structures of the substrates tested in these experiments are represented in Fig. 3. The 5′-tailed partial-duplex RNA3 was produced by annealing single-stranded RNAs of 300 and 200 nt, respectively, which were synthesized by in vitro transcription in the presence of [α-32P]CTP, using previously described methods (37). The template DNAs used for the in vitro transcription reactions were generated by PCR from appropriate HCoV-229E cDNA templates and upstream primers containing the T7 promoter sequence.
FIG. 3.
Effective unwinding by HCoV-229E nsp13 of partial-duplex DNA and RNA substrates containing extended duplex regions. The structures of the substrates are shown schematically, and radiolabeled strands are marked by asterisks. Reaction products were separated on 2% agarose (A) or 6% polyacrylamide (B) gels. (A) Lanes 1, 6, 11, and 16, heat-denatured substrates; lanes 2, 7, 12 and 17, reactions without protein; lanes 3, 8, 13, and 18, reactions containing HEL and ATP; lanes 4, 9, 14, and 19, reactions containing HEL but no ATP; lanes 5, 10, 15, and 20, reactions containing HEL_KA and ATP. (B) Lane 1, reaction without protein; lane 2, heat-denatured substrate; lane 3, reaction containing HEL and ATP; lane 4, reaction containing HEL_KA and ATP.
Partial-duplex DNAs were produced by annealing single-stranded DNAs (ssDNAs) that were obtained in the following way. First, a DNA fragment of desired length was produced by a PCR in which one of the primers was biotinylated at the 5′ end. The purified PCR product was bound to streptavidin-coupled magnetic beads (Dynabeads streptavidin; Dynal) according to the manufacturer's instructions. Following binding, the duplex DNA was denatured using 0.15 M sodium hydroxide, which allowed us to isolate the nonbiotinylated DNA strand from the supernatant while the biotinylated strand remained bound to the streptavidin beads. To produce ssDNAs of different lengths from the same PCR product, the amplicon was digested with different restriction enzymes, such as AvaI, FspI, and NdeI, prior to the incubation with the streptavidin dynabeads. Because the nonbiotinylated restriction fragment(s) did not bind to the streptavidin beads, they could be readily removed with the supernatant. The remaining streptavidin-bound, 5′-biotinylated double-stranded DNA fragment was then denatured as described above, and the nonbiotinylated DNA strand was again isolated from the supernatant. To produce the partial-duplex DNAs 1020/100*, 1020/Ava*, 1020/Fsp*, and 1020/Nde*, which contained bottom strands of increasing length (Fig. 3A), a 1,020-nt ssDNA was annealed with appropriate complementary single-stranded DNAs, namely ssDNA 100* (100 nt), ssDNA Ava* (228 nt), ssDNA Fsp* (366 nt), and ssDNA Nde* (474 nt), respectively. The asterisk indicates the radiolabel that was introduced at the 5′ end by treatment with [γ-32P]ATP and T4 polynucleotide kinase. All these partial-duplex DNAs had a common 5′ single-stranded region of 21 nt, while their double-stranded regions varied from 100 to 474 bp and, accordingly, their 3′ single-stranded regions varied from 899 to 525 nt (Fig. 3A).
The duplex-unwinding reactions shown in Fig. 3 contained 3.5 nM HEL (or HEL_KA) and 10 nM RNA or DNA substrate and were performed in buffer containing 20 mM HEPES-KOH (pH 7.4), 10% glycerol, 5 mM magnesium acetate, 2 mM dithiothreitol, 0.1 mg of bovine serum albumin/ml, and 2 mM ATP. Following incubation for 30 min at 30°C, the samples were mixed with an equal volume of loading buffer (20% glycerol, 0.2% sodium dodecyl sulfate [SDS]) containing bromphenol blue dye. RNA samples were analyzed by electrophoresis in 6% polyacrylamide gels (acrylamide/bisacrylamide ratio of 30/1) buffered with 0.5× Tris-borate-EDTA containing 0.1% SDS. DNA samples were analyzed by electrophoresis in 2% agarose gels buffered with 1× Tris-acetate-EDTA containing 0.1% SDS.
As Fig. 3 shows, HEL was able to unwind very efficiently extended (up to several hundred base pairs) duplex regions in both RNA and DNA substrates. Next, we did an experiment in which a large excess of HEL was mixed at a molar ratio of 35:1 with a partial-duplex DNA substrate (duplex region of 22 bp) (37). Following enzyme-substrate complex formation for 15 min, the unwinding reaction was initiated by the simultaneous addition of ATP and a 3,000-fold excess of trap oligonucleotide over substrate. Under these single-cycle conditions, dissociated helicase cannot rebind to the substrate (1, 22). Analysis of the kinetics of strand separation by gel shift assay revealed no difference in the reaction kinetics between samples containing or lacking the trap oligonucleotide (data not shown), providing preliminary evidence for processivity of the HEL duplex-unwinding activity, although the biochemical details of this property of HEL remain to be determined.
Based on studies on the subcellular localization of coronavirus helicases, which were reported to localize to intracellular membranes throughout the infection cycle (5, 21, 43), we currently do not believe that the DNA helicase activity of nsp13 is of biological significance. The efficient unwinding of long stretches of RNA is consistent with a replicative function for the coronavirus nsp13 helicase which, for example, might be involved in the separation of double-stranded replicative intermediates to provide single-stranded templates for repeated rounds of RNA synthesis. Also, the hepatitis C virus NS3 protein was recently shown to be a processive enzyme (31). Surprisingly however, and in contrast to HCoV-229E nsp13, it was found to be much more active on duplex DNA rather than RNA substrates. This result led to the proposal that in addition to its role in viral RNA replication, NS3 might also act on host cell DNA (31).
In summary, the data demonstrate that HCoV-229E nsp13 (this study) and SARS-CoV nsp13 (21, 41) have a variety of enzymatic functions, including NTPase, dNTPase, RNA 5′-triphosphatase, RNA helicase, and DNA helicase activities. The fact that also in functional details, such as the use of a common active site for the NTPase and RNA 5′-triphosphatase activities and the nucleotide cofactor specificity, no major differences were revealed between the HCoV-229E and SARS-CoV helicases indicates a strong conservation of the key replicative functions among group 1 (HCoV-229E) and group 2 (SARS-CoV) coronaviruses, even though the proteins show only a moderate sequence identity. Since helicase-associated RNA 5′-triphosphatase activities involved in 5′ cap formation have also been identified in several other RNA virus families (4, 28, 45, 47) and coronaviruses are not known to encode a separate RNA 5′-triphosphatase, it seems reasonable to suggest that coronaviruses (and possibly other nidoviruses) employ the helicase to mediate the first step of 5′ cap synthesis. The (viral or cellular) enzymes involved in the guanylylation and (guanine-7) methylation reactions required to synthesize mature coronavirus RNA 5′ cap structures (26) remain to be identified.
As discussed above, both the structure of nidovirus helicases, which involves an N-terminal zinc-binding domain, and their 5′-to-3′ polarity clearly distinguish the nidovirus enzymes from the well-characterized helicases of the Flaviviridae family, which all unwind their substrates in the opposite direction. Nidovirus helicases also differ from the plant +RNA virus helicases involved in cell-to-cell movement, which again have deviant functional properties (23). The profoundly divergent evolution of nidovirus helicases from the homologs of other +RNA virus helicases was probably driven by the unique features of the nidovirus life cycle (25).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (EGK, ZI 618/2-2 and 2-3, SFB 479).
REFERENCES
- 1.Ali, J. A., and T. M. Lohman. 1997. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275:377-380. [DOI] [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Bautista, E. M., K. S. Faaberg, D. Mickelson, and E. D. McGruder. 2002. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology 298:258-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisaillon, M., J. Bergeron, and G. Lemay. 1997. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda1 protein. J. Biol. Chem. 272:18298-18303. [DOI] [PubMed] [Google Scholar]
- 5.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delagoutte, E., and P. H. von Hippel. 2002. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q. Rev. Biophys. 35:431-478. [DOI] [PubMed] [Google Scholar]
- 7.El-Sahly, H. M., R. L. Atmar, W. P. Glezen, and S. B. Greenberg. 2000. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis. 31:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falsey, A. R., E. E. Walsh, and F. G. Hayden. 2002. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J. Infect. Dis. 185:1338-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagneur, A., J. Sizun, S. Vallet, M. C. Legr, B. Picard, and P. J. Talbot. 2002. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J. Hosp. Infect. 51:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E. 2001. Big nidovirus genome. When count and order of domains matter. Adv. Exp. Med. Biol. 494:1-17. [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grötzinger, C., G. Heusipp, J. Ziebuhr, U. Harms, J. Süss, and S. G. Siddell. 1996. Characterization of a 105-kDa polypeptide encoded in gene 1 of the human coronavirus HCV 229E. Virology 222:227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegyi, A., and J. Ziebuhr. 2002. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 83:595-599. [DOI] [PubMed] [Google Scholar]
- 14.Herold, J., A. E. Gorbalenya, V. Thiel, B. Schelle, and S. G. Siddell. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 72:910-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold, J., T. Raabe, B. Schelle-Prinz, and S. G. Siddell. 1993. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology 195:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herold, J., and S. G. Siddell. 1993. An ′elaborated' pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 21:5838-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold, J., S. G. Siddell, and A. E. Gorbalenya. 1999. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 274:14918-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heusipp, G., C. Grötzinger, J. Herold, S. G. Siddell, and J. Ziebuhr. 1997. Identification and subcellular localization of a 41 kDa, polyprotein 1ab processing product in human coronavirus 229E-infected cells. J. Gen. Virol. 78:2789-2794. [DOI] [PubMed] [Google Scholar]
- 19.Heusipp, G., U. Harms, S. G. Siddell, and J. Ziebuhr. 1997. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J. Virol. 71:5631-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofstee, B. H., M. Dixon, and E. C. Webb. 1959. Non-inverted versus inverted plots in enzyme kinetics. Nature 184:1296-1298. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2000. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403:447-451. [DOI] [PubMed] [Google Scholar]
- 23.Kalinina, N. O., D. V. Rakitina, A. G. Solovyev, J. Schiemann, and S. Y. Morozov. 2002. RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296:321-329. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 25.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M., C. D. Patton, and S. A. Stohlman. 1982. Further characterization of mRNA's of mouse hepatitis virus: presence of common 5′-end nucleotides. J. Virol. 41:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 28.Li, Y. I., T. W. Shih, Y. H. Hsu, Y. T. Han, Y. L. Huang, and M. Meng. 2001. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′-triphosphatase activity. J. Virol. 75:12114-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 30.Myint, S. H. 1995. Human coronavirus infections, p. 389-401. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 31.Pang, P. S., E. Jankowsky, P. J. Planet, and A. M. Pyle. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21:1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raabe, T., B. Schelle-Prinz, and S. G. Siddell. 1990. Nucleotide sequence of the gene encoding the spike glycoprotein of human coronavirus HCV 229E. J. Gen. Virol. 71:1065-1073. [DOI] [PubMed] [Google Scholar]
- 34.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 36.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 37.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seybert, A., L. C. van Dinten, E. J. Snijder, and J. Ziebuhr. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 74:9586-9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seybert, A., and J. Ziebuhr. 2001. Guanosine triphosphatase activity of the human coronavirus helicase. Adv. Exp. Med. Biol. 494:255-260. [DOI] [PubMed] [Google Scholar]
- 40.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanner, J. A., R. M. Watt, Y. B. Chai, L. Y. Lu, M. C. Lin, J. S. Peiris, L. L. Poon, H. F. Kung, and J. D. Huang. 2003. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 278:39578-39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 43.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. Krijnse Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dinten, L. C., H. van Tol, A. E. Gorbalenya, and E. J. Snijder. 2000. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 74:5213-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kääriäinen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 46.von Hippel, P. H., and E. Delagoutte. 2001. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell 104:177-190. [DOI] [PubMed] [Google Scholar]
- 47.Wengler, G. 1993. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265-273. [DOI] [PubMed] [Google Scholar]
- 48.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziebuhr, J., J. Herold, and S. G. Siddell. 1995. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 69:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziebuhr, J., G. Heusipp, and S. G. Siddell. 1997. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 71:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziebuhr, J., and S. G. Siddell. 1999. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 73:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 53.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zúñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]