Abstract
Recent outbreaks of West Nile Virus (WNV) have been associated with an increase in morbidity and mortality in humans, birds, and many other species. We have initiated studies to define the molecular mechanisms by which a recent pathogenic isolate of WNV evades the host cell innate antiviral response. Biochemical and microarray analyses demonstrated that WNV induced the expression of beta interferon (IFN-β) and several IFN-stimulated genes late in infection of cultured human cells. The late expression of these antiviral genes was due to the delayed activation of the transcription factor IFN regulatory factor 3 (IRF-3). Despite this host response, WNV was still able to replicate efficiently. The effect of the IRF-3 pathway on WNV replication was assessed by examining virus replication and spread in cultures of wild-type or IRF-3-null mouse embryo fibroblasts. The absence of IRF-3 was marked by a significant increase in plaque size and a sustained production of infectious particles. Although the activation of the IRF-3 pathway was not sufficient to block virus replication, our results suggest that IRF-3 target genes function to constrain WNV infection and limit cell-to-cell virus spread.
The emergence of West Nile virus (WNV) in the western hemisphere in 1999 and the dramatic increase in both the incidence and the severity of disease in humans during the subsequent transmission seasons has resulted in the classification of the virus as an emerging pathogen of significant importance. In areas of Asia, the Middle East, and Africa where WNV (lineage II) has been endemic for many years, infections are generally either asymptomatic or associated with a mild childhood febrile illness. Recent WNV epidemics in developed countries in Europe and the United States have been associated with significantly higher rates of morbidity and mortality than previously documented (30). Since its introduction into the United States in 1999, WNV has rapidly spread and has now been detected in nearly every state in the continental United States. The number of documented human cases has continued to rise with every subsequent transmission season. As of 14 January 2004, the Centers for Disease Control and Prevention (CDC) had confirmed 9,006 human cases of WNV during the 2003 transmission season; 220 of these cases were fatal, and at least 30% were associated with either meningitis or encephalitis (information found on the CDC website[http://www.cdc.gov/ncidod/dvbid/westnile/index.htm]). Therapid spread and increasing numbers of cases of WNV over the last five summers suggests that WNV has firmly established itself in the Americas.
WNV is a member of the genus Flavivirus of the family Flaviviridae, which are enveloped single-stranded positive-sense RNA viruses. The WNV genome is ca. 11 kb in length and consists of a single open reading frame bounded by both a 5′ and 3′ nontranslated regions (10). The nontranslated regions contain conserved sequences and predicted secondary structure that function as signals for negative-strand synthesis, genome amplification, translation, and packaging. Translation of the viral genome generates a single polyprotein that is both co- and posttranslationally cleaved by a combination of host and virus-encoded proteases into 10 viral proteins. The first third of the WNV genome encodes the viral structural proteins, i.e., the core (C), membrane (prM/M), and envelope (E) proteins, which are involved in viral assembly and host cell entry. The remaining C-terminal portion of the genome encodes seven nonstructural (NS) proteins (NS1-NS2A/B-NS3-NS4A/B-NS5), which support viral replication (10, 38).
Like other flaviviruses, WNV is an arthropod-borne virus (30). In the case of WNV, major transmission occurs through the bite of an infected mosquito. Recent epidemiological surveillance studies have indicated that WNV can infect a broader range of animals than originally thought; however, wild birds are believed to be the major natural reservoir for WNV due to the high levels of viremia observed in infected birds (30). The level of viremia observed in other animals is below the viral transmission threshold for WNV, making these dead-end infections. The molecular characterization of viral isolates belonging to recent WNV outbreaks in humans indicated that these more pathogenic isolates were subtypes of WNV lineage I (25, 26).
The first line of host defense against an invading viral pathogen is dependent upon the activation of innate antiviral response pathways within the infected cell. Upon sensing the invading viral pathogen, the cell activates multiple distinct signaling pathways by inducing a number of latent transcription factors, one of which is interferon regulatory factor 3 (IRF-3) (3, 34, 44). IRF-3 is unique among the IRF family members in that it is a ubiquitously expressed protein that is present in the cytosol of all cells in an inactive state. Products of viral replication, such as double-stranded RNA (dsRNA) structures within the replicating viral genome or the accumulation of viral proteins trigger the phosphorylation of IRF-3 by a virus-activated protein kinase complex (14, 37, 42, 45, 46). Upon phosphorylation, IRF-3 forms dimers that are retained in the nucleus and interact with the CBP/p300 coactivator to induce the expression of multiple target genes, including beta interferon (IFN-β). Transcription of IFN-β is regulated by a protein complex termed the enhanceosome, which includes NF-κB, ATF2/c-Jun, and IRF-3. However, viral induction of IFN-β expression is absolutely dependent on the activation of IRF-3 and its association with the enhanceosome (33). The binding of secreted IFN-β to the IFN-α/β receptor triggers the activation of the janus kinase and signal transducers and activators of transcription (JAK/STAT) signal transduction pathway, leading to the induction of expression of a wide variety of IFN-stimulated genes (ISGs). The antiproliferative, antiviral, and proapoptotic actions of IFNs are conferred through the action and/or functions of ISGs (34). Thus, ISGs are responsible for the establishment of an antiviral state within the infected cell as well as neighboring tissue.
As the eukaryotic antiviral programs have evolved, so have viruses evolved to evade or escape these programs within the infected cell. Viruses have been shown to regulate IFN signaling at nearly every step of the pathway. The mechanisms used by various viruses to neutralize the antiviral programs are extremely divergent (23). Many viruses have been shown to circumvent the IFN response by preventing the production of IFN-β by such mechanisms as directly sequestering dsRNA activators of the host response and/or regulating IRF-3 function (5, 15, 41, 43). In addition, many viruses attenuate the action of IFN by directly blocking JAK/STAT signaling or by targeting the actions of specific ISGs, such as blocking the function of PKR or 2′-5′ oligoadenylate synthetase (reviewed in reference 34). Viral control of IFN production or ISG action creates a permissive environment for virus replication.
The molecular mechanisms by which WNV overcomes or evades the host cell antiviral response to establish a productive infection have not been extensively characterized. In the present study, the innate immune response to a lineage I isolate from the New York 2000 epidemic was examined (39). We demonstrate that WNV induces an antiviral response in human cells and that this response is triggered, in part, through the viral activation of IRF-3 late during infection. Our results demonstrate that the IRF-3 target gene(s) play a role of in limiting cell-to-cell spread of WNV but that activation of the IRF-3 pathway is not sufficient to block viral replication and cytopathogenesis.
MATERIALS AND METHODS
Cells and viruses.
Vero, HeLa, 293, Huh7, Tel12 human foreskin fibroblast, GRE, U-2 OS, and mouse embryo fibroblast (MEF; kindly provided by Tadatsugu Taniguchi) cell lines were propagated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, antibiotic-antimycotic solution, and nonessential amino acids (complete DMEM). Recovery of infectious particles from plasmid pFLWNV, which encodes the infectious clone WNV-NY, was done as previously described (39). Working stocks of WNV-NY were generated by passing the virus recovered from the infectious clone a single time in 293 cells at a low multiplicity of infection (MOI), and aliquots of this stock were stored at −80°C.
Virus growth curves.
Cultures of 293 cells in six-well plates were infected with WNV-NY at an MOI of 5 for 1 h at 37°C. It should be noted that the amount of virus added to the culture to achieve an MOI of 5 was calculated by using the titer of the viral stock as determined on 293 cells. The inoculum was removed, and 2 ml of complete DMEM was added. Culture supernatants and whole-cell lysates, used in immunoblot assays, were collected at the indicated time points. Cell debris was removed by low-speed centrifugation at 1,500 rpm for 5 min, and supernatants were transferred to new tubes and stored at −80°C until the titers were determined by plaque assay on Vero cells. Growth curves were repeated three times.
Plaque assays.
Monolayers of either 293 or Vero cells in six-well plates were washed two times in serum-free DMEM, followed by the addition of serial dilutions of viral samples. The cells were incubated in a 5% CO2 incubator for 1 h at 37°C with rocking, the inoculum was removed, and a 0.9% agarose-complete DMEM overlay was added. Cell monolayers were incubated for 48 h, and a second overlay of agarose-complete DMEM containing 1% neutral red (ICN Biomedical) was added. The plates were incubated for an additional 48 h prior to counting plaques. The titers of WNV-NY on 293 cells were generally 10-fold lower than titers determined on parallel cultures of Vero cells.
Northern blot analysis.
RNA was extracted from mock- or WNV-infected 293 cells by using TRIzol Reagent as recommended by the manufacturer (Invitrogen Life Technologies, Inc.). Purified RNA was resuspended in water, quantified by spectrometry, and mixed with RNA loading buffer. After being heated at 50°C for 10 min, 10 μg of RNA was separated through a 1% agarose gel containing 2.2 M formaldehyde, 20 mM morpholinepropanesulfonic acid (pH 7.0), 8 mM sodium acetate, and 1 mM EDTA (pH 8.0). To process the gel for transfer of RNA, the gel was soaked in water for 1 h with gentle agitation, followed by incubation in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min. RNA transfer onto Nytran membrane was carried out by using the Schleicher & Schuell Turboblotter downward transfer system as recommended by the manufacturer. DNA probes specific for the WNV NS2A coding region or human ISG6-16, ISG20, ISG15, ISG56, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were generated by using Klenow DNA polymerase and mixed nonomer random primers in a reaction that contained [α-32P]dCTP. Hybridization reactions were carried out with the ULTRAhybe reagent (Ambion) and 106 cpm of radiolabeled probe/ml at 48°C for 16 h. Blots were rinsed twice for 5 min each time with preheated 2× SSC-0.1% sodium dodecyl sulfate (SDS) wash buffer, followed by two 15-min washes with 0.1× SSC-0.1% SDS wash buffer. Blots were imaged by using a Storm 820 phosphorimager (Amersham). In some experiments, probe hybridization was quantified by phosphorimager analysis.
Immunoblot analysis.
Cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris, 150 mM NaCl, 0.02% sodium deoxycholate, 1% Triton X-100, 0.1% SDS) containing protease inhibitors (Sigma) and 1 μM okadaic acid. Proteins (20 μg) were resolved on 10 to 12.5% polyacrylamide gels containing SDS. After electrophoresis, proteins were transferred to NitroPure nitrocellulose transfer membrane (Micron Separations, Inc.), and blots were blocked overnight at 4°C. The following monoclonal or polyclonal antibodies were used to probe the blots: rabbit anti-human IRF-3 serum (kindly provided by Michael David), rabbit anti-phosphoserine 396 IRF-3 (kindly provided by John Hiscott), rabbit anti-P56 (kindly provided by Ganes Sen), rabbit anti-ISG15 (kindly provided by Arthur Haas), rabbit anti-IRF-3 (FL425; Santa Cruz), rabbit anti-IRF-9 (C-20; Santa Cruz), mouse anti-STAT1α (C-111; Santa Cruz), goat anti-human actin (Santa Cruz), mouse anti-Dengue NS1 (this antibody cross-reacts with the WNV NS1 protein and was kindly provided by Peter Mason), and peroxidase-conjugated secondary donkey anti-rabbit, donkey anti-mouse, or donkey anti-goat (Jackson Immunoresearch) antibody. Protein bands were visualized by using the ECL-Plus Immunoblotting detection reagents (Amersham Biosciences), followed by exposure of the blot to film.
Microarray analysis.
Cultures of 293 cells in 10-cm plates were infected with WNV-NY at an MOI of 5 or an MOI of 0.3. At the indicated time points, total RNA was extracted from cells by using TRIzol reagent as recommended by the manufacturer (Invitrogen/Life Technologies, Inc.). Total RNA samples were amplified by using RiboAmp RNA Amplification kit (Arcturus, Mountain View, Calif.) as described by the manufacturer. The quality of amplified RNA was evaluated by capillary electrophoresis by using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif.). The microarray format, protocols for probe labeling, hybridization, slide treatment, and scanning are described online (http://expression.microslu.washington.edu). Microarray expression analyses of RNA recovered from 293 cells infected with an MOI of 0.3 or 5 was conducted. Microarray data from the 5.0 MOI infection are presented here, with the remaining data to be presented elsewhere. Global gene expression was evaluated throughout a time course of infection. Each experiment included six individual RNA samples and was designed to directly compare RNA from mock-infected and WNV-NY-infected cells. RNA samples recovered from mock and WNV-infected cells at each time point were compared by using the dye label reverse technique with four identical microarrays (13,597 unique IMAGE cDNA clones), thus providing mean ratios between the expression levels of each gene in the analyzed sample pair, standard deviations, and P values. All data were entered into a custom-designed database, Expression Array Manager, and then uploaded into Rosetta Resolver System 3.0 (Rosetta Biosoftware, Kirkland, Wash.) and Spotfire software (Spotfire, Somerville, Mass.). Data normalization and the Resolver System error model specifically developed for slide format used in these experiments are described on the website given above. This website is also used to publish all primary data in accordance with the proposed standards (7, 8).
Indirect immunofluorescence analysis.
Cultures of Huh7 (8 × 104) or U-2OS (1.35 × 105) cells were grown on tissue culture chamber slides and infected with either WNV (MOI = 5) or Sendai virus (50 heme agglutination units). At 36 h postinfection, slides were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 30 min at room temperature. Cell monolayers were permeabilized with a solution of PBS-0.2% Triton X-100 for 15 min, followed by a 1-h incubation in PBS containing 10% normal goat serum. After a rinse with PBS, cells were incubated for 1 h in the presence of a 1/500 dilution of rabbit polyclonal anti-human IRF-3 antibody in PBS-0.05% Tween 20-3% bovine serum albumin and washed three times with PBS-0.5% Tween 20. Slides were incubated for 1 h with either a 1/1,000 dilution of goat anti-rabbit immunoglobulin G-fluorescein isothiocyanate antibody conjugate (Jackson Immunoresearch) or a 1/2,000 dilution of goat anti-rabbit immunoglobulin G-Alexa 488 antibody conjugate (Molecular Probes). Cells were washed three times and allowed to dry, and the slides were overlaid with Vectashield solution (Vector Labs), after which coverslips were mounted and sealed prior to visualization with a Zeiss Axiovert fluorescence microscope equipped with a digital camera.
Focus-forming assay.
Subconfluent cultures of wild-type (wt) or IRF-3-null MEFs in a six-well plate were infected with serial dilutions of WNV-NY and overlaid with 1% methylcellulose. At 4 days postinfection, the cells were washed three times with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. After a rinse with PBS-glycine (PBS, 10 mM glycine, 0.5% sodium azide), the cells were incubated for 1 h with a 1/100 dilution horse anti-WNV serum in PBS plus 3% bovine serum albumin, and then cultures were washed three times with PBS and incubated for 1 h with a 1/200 dilution of horseradish peroxidase-conjugated rabbit anti-horse antibody (Jackson Immunoresearch). Cells were washed three times with PBS and exposed to substrate solution (PBS, 0.01% 4-chloro-1-naphthol, 0.003% H2O2) for 5 to 15 min until color developed. Foci were not detected in parallel cultures of mock-infected wt or IRF-3-null MEFs.
Assay for antiviral activity.
Vero cells seeded in six-well plates at a density of 5.5 × 105 cells/well were treated with IFN-α2a (500 U/ml) or clarified supernatants (1 ml) recovered at 96 h postinfection from mock- or WNV-NY-infected wt MEFs for 24 h. Cultures were infected with 0.1 ml of vesicular stomatitis virus encoding green fluorescent protein (VSV-GFP) (kindly provided by Michael Whitt) at an MOI of 0.1. After adsorption for 30 min at 37°C in a 5% CO2 incubator, the inoculum was removed and 2 ml of complete DMEM was added to each well. At 16 h postinfection, culture supernatants were removed and cleared of cell debris by centrifugation. VSV-GFP titers were determined by plaque assay on Vero cells. Briefly, cultures of Vero cells were washed two times with serum-free DMEM, and serial dilutions of VSV-GFP were added to cell monolayers. Cultures were then incubated for 30 min at 37°C with rocking, the inoculum was removed, and an agarose-DMEM overlay was added. The cultures were then incubated for 24 h at 37°C in a 5% CO2 incubator to allow plaques to form.
RESULTS
Virus generated from an infectious clone derived from a lineage I WNV isolate from the 2000 WNV epidemic was used to examine the host response to WNV infection. This clone (WNV-NY) was derived from a low-passage isolate, thus minimizing the possibility of the accumulation of tissue culture adaptive mutations within the viral genome. Virus generated from this clone exhibited similar growth characteristics in mammalian and insect cells to the parental virus and was equally virulent in mice (39). However, like most isolates of WNV the ability of this strain to replicate in human cells has not been examined. Therefore, the replication properties of WNV-NY within cultured human cell lines derived from various tissues were examined.
Replication of WNV in human cell lines.
Several human cell lines were examined for their ability to sustain replication of WNV-NY. Vero cells and MEFs were included as control cell lines that have been previously shown to be susceptible to WNV (9, 39). Cell lines were examined for virus-induced cytopathic effect (CPE), expression of viral proteins, and the ability to generate infectious particles (Table 1). Virtually all cell lines tested were permissive for WNV-NY. However, there was significant variability in the level of virus-induced CPE, as well as the kinetics and level of accumulation of viral proteins (data not shown). Thus, WNV-NY is capable of infecting and replicating in a wide range of human cell lines generated from a variety of tissues/organs.
TABLE 1.
Replication of WNV-NY in human cell lines
| Cell line | Tissue/morphology | CPE | WNV protein expressiona | Release of viral particlesb |
|---|---|---|---|---|
| Vero | Monkey kidney/epithelial | Yes | Yes | Yes |
| HeLa | Human cervix/epithelial | No | Yes | ND |
| 293 | Human kidney/epithelial | Yes | Yes | Yes |
| Huh7 | Human liver/hepatocyte | Yes | Yes | Yes |
| Tel12 | Human foreskin/fibroblast | Yes | NDc | ND |
| HFF | Human primary foreskin/ fibroblast | Yes | Yes | ND |
| GRE | Human brain/glial | Yes | Yes | Yes |
| U-2 OS | Human bone/epithelial | Yes | Yes | Yes |
| MEF | Mouse embryo/fibroblast | Yes | Yes | Yes |
Viral protein production was assessed by either immunofluorescence assay or Western blot analysis.
The production of infectious particles was confirmed by plaque assay on Vero cells.
ND, not determined.
Characterization of WNV replication in human embryonic kidney cells.
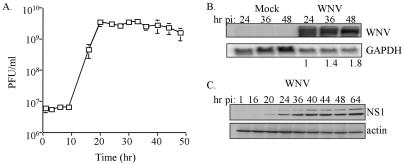
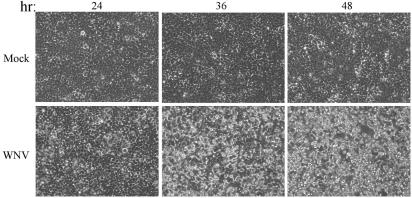
Of the cell lines examined human embryonic kidney cells (293 cells) were among the most susceptible to WNV-NY. A one-step growth curve was used to examine viral growth characteristics in 293 cells. Cells were infected at an MOI of 5 to ensure that all cells within the culture were infected. Under these conditions infectious particles were produced at significant levels within 16 h postinfection and reached peak viral production between 24 and 30 h postinfection, despite the fact that both viral RNA and proteins continued to accumulate throughout the course of the experiment (Fig. 1). The kinetics of the accumulation of viral RNA and protein correlated with the level of observed CPE (Fig. 2). The fact that WNV can readily be isolated from the kidneys of infected animals (1, 24) and that WNV-NY replicates efficiently in 293 cells indicates that this is an appropriate and relevant human cell line for the analysis of WNV-host interactions.
FIG. 1.
Characterization of WNV-NY replication in 293 cells. (A) Growth of WNV-NY in 293 cells. Cultures were inoculated with WNV-NY at an MOI of 5.0. Medium was removed from cultures at the indicated times and cleared of cell debris, and titers were determined by plaque assay on Vero cells. Values represent the number of PFU per milliliter of supernatant and are the results of two separate experiments. (B) Northern blot analysis of WNV-NY-infected 293 cells. The hours postinfection (hr pi) for mock- or WNV-infected cells are indicated above each lane. WNV genomic RNA was detected by using a 32P-labeled probe encoding the WNV NS2A sequence. As a control, the same blot was stripped and reprobed with labeled DNA complementary to GAPDH. Bands were quantified by phosphorimager analysis. The values shown below the samples from WNV-infected cells indicate the abundance of viral RNA detected at 36 and 48 h postinfection relative to the level of viral RNA at 24 h postinfection. The elevated levels of GAPDH expression detected at 36 and 48 h posttreatment in mock-infected cells was due to the overloading of these lanes and does not reflect differences in RNA metabolism. Importantly, the level of GAPDH detected in WNV-NY-infected cultures was similar for all time points examined. (C) Kinetics of expression of the WNV NS1 protein in 293 cells. Extracts, prepared from WNV-NY-infected 293 cells that were harvested at the indicated time points postinfection, were subjected to immunoblot analysis for WNV NS1 and actin expression. The observed doublet is attributed to differential N-linked glycosylation (2).
FIG. 2.
WNV-induced CPE. 293 cells were either mock treated or infected with WNV at an MOI of 5 and then incubated at 37°C for the time in hours (hr) indicated above each panel. Virus-induced CPE presents as a distortion of the cell monolayer and was visualized by using a Zeiss light microscope. The images were captured with a digital camera.
Host ISG expression induced by WNV infection.
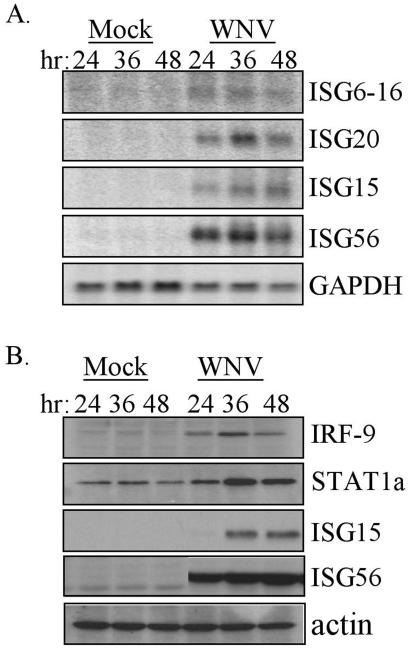
To begin to determine how the host cell responds to infection by WNV, the level of expression of a panel of ISGs was examined. Northern blot analysis of total RNA isolated from 293 cells revealed that WNV infection induced the expression of ISG20, ISG6-16, ISG15, and ISG56 (Fig. 3A). Within the time points examined, the ISG transcripts reached peak abundance at the 36-h time point postinfection. Immunoblot analysis confirmed the WNV-induced increase in transcript levels of both ISG15 and ISG56 also resulted in an increase in the steady-state levels of the encoded proteins (Fig. 3B). In addition, the steady-state protein levels of two other known ISGs, IRF-9 and STAT1α, were also evaluated and showed a marked increase in abundance 36 h postinfection with WNV (Fig. 3B). These results suggest that WNV genome replication and/or the accumulation of viral proteins triggers an innate antiviral response in the host cell that involves induction of ISG expression.
FIG. 3.
Induction of ISG expression during WNV-NY infection. (A) Northern blot analysis of ISG6-16, ISG20, ISG15, ISG56, and GAPDH expression in cells that were mock infected or infected with WNV-NY for the indicated hours (hr). (B) Steady-state protein levels of IRF-9, STAT1α, ISG15, ISG56, and actin in mock- or WNV-infected 293 cells were examined by immunoblot analysis of extracts prepared at the indicated times postinfection.
Analysis of gene expression in WNV-infected 293 cells.
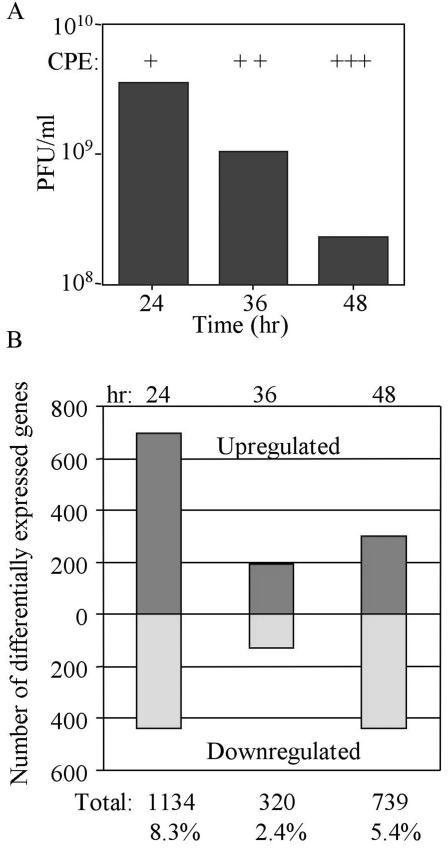
The induction of ISG expression is complex and involves multiple transcription factors (34). For example, the expression of ISG15 and ISG56 is not only induced by IFN, these genes have been identified as direct IRF-3 target genes that are induced in response to viral activation of IRF-3 (21). In contrast, IRF-9, STAT1α, ISG20, and ISG6-16 levels are not direct IRF-3 target genes but are regulated in response to IFN and therefore only indirectly by IRF-3 (11, 21). The observed expression of these genes suggests that WNV infection triggers a complex host response that includes at least the IRF-3 and IFN-α/β signaling pathways. To further dissect this host response, microarray analysis was utilized to evaluate the influence of WNV on global gene expression in cultured cells. 293 cells were infected with WNV-NY at an MOI of 5 to ensure a uniform infection of the cultures. Total RNA was isolated at the times indicated and subjected to microarray analysis. During the course of this experiment, peak infectious virus production occurred at ca. 24 h postinfection and then gradually decreased throughout the remainder of the infection (Fig. 4A). Under these infection conditions WNV-NY induced a biphasic modulation of global gene expression. At 24 h postinfection, 8.3% of the cellular transcripts were differentially regulated by at least twofold in WNV-infected cells compared to mock-infected cells (Fig. 4B). However, at 36 h postinfection only 2.4% of cellular genes were differentially regulated, and at 48 h postinfection the number of differentially regulated host cell genes increased to 5.4% of the total genes analyzed.
FIG. 4.
Microarray analysis of global gene expression in WNV-NY-infected cells. (A) Infectious particle production was monitored by plaque assay on Vero cells by using cell-free supernatants recovered from WNV-NY-infected 293 cells. The level of virus-induced CPE in these cultures is also indicated. The qualitative scale for CPE, as determined visually, is as follows: +, barely detectable; ++, clearly detectable; and +++, extensive CPE throughout the entire culture. (CPE presentation was similar to that shown in Fig. 2). (B) Graphic representation of the total number of host cell genes whose expression was differentially modulated in WNV-infected cells compared to mock-infected cultures. Genes were selected based on two criteria: a >95% probability of being differentially expressed (P < 0.05) and an overall change in expression of ≥2-fold. The total number and frequency of genes that fit these criteria are indicated below each bar.
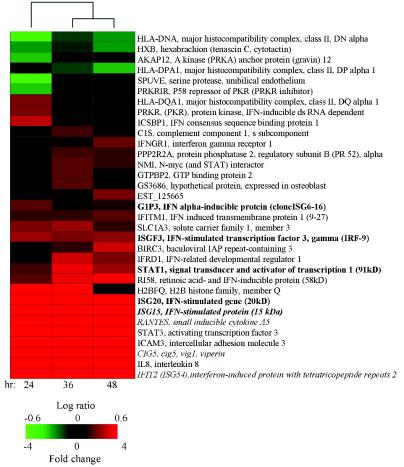
Northern and immunoblot analyses of WNV-NY-infected cells indicated that the antiviral response to this virus was characterized by viral induction of both ISGs and IRF-3 target genes (see Fig. 3). In order to expand the characterization of the host antiviral response to WNV, further microarray analyses were focused on the evaluation of the expression of a spectrum of virus-inducible genes, dsRNA responsive genes, and ISGs that were identified in previous studies (16-19). As shown in Fig. 5, a variety of genes previously shown to be associated with antiviral processes were identified as regulated in response to WNV-NY infection. Although the expression of a minority of the genes was suppressed in response to WNV, the majority of genes, including many ISGs, exhibited increased expression throughout the entire course of the experiment. This pattern of ISG and antiviral gene regulation was in contrast to the biphasic modulation in global gene expression that was observed. The microarray analysis verified that WNV-NY infection triggered the expression of ISG6-16, IRF-9, and STAT1α. In addition, the expression of known IRF-3 target genes—CIG5, RANTES, and ISG54 (21)—was increased in response to WNV-NY infections. These results confirm that the host cell response to WNV-NY infection includes the induction of ISGs and IRF-3 target genes.
FIG. 5.
Regulation of ISGs, virus-inducible, and dsRNA-responsive genes in WNV-NY-infected cells. The effect of WNV replication on the expression of a set of 241 genes was evaluated. The genes in this data set were previously identified as differentially expressed in cultured human cells that were infected with various viruses, treated with dsRNA or IFN (16-19). Among this set, 33 of the genes were differentially expressed in least one of the three time points examined. Two-dimensional hierarchic clustering of genes was preformed by using Spotfire software as described previously (40). Each vertical column represents the relative expression levels of the indicated gene at the indicated time postinfection. The fold changes in mRNA levels in WNV-infected cultures relative to mock-infected cultures are represented by green and red squares, indicating decreased and increased levels of expression, respectively. The color scale shows the magnitude of change. Black bars indicate no change in gene expression level. Genes are indicated by their human genome organization names and brief gene descriptions. Genes previously identified as regulated by WNV are indicated in boldface (see Fig. 3). Italicized genes have been previously identified as regulated by IRF-3 (21).
Induction of the IRF-3 pathway by WNV.
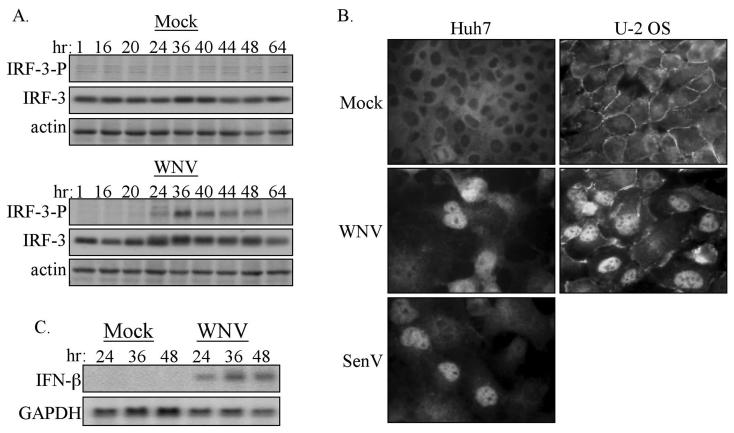
The gene expression data indicated that WNV infection triggers events that lead to the activation of IRF-3, IRF-3 target gene expression and IFN production. Therefore, the activation state of IRF-3 in WNV-NY-infected cells was examined. IRF-3 activation results from phosphorylation at multiple sites within the carboxyl terminus of the protein, which includes but is not limited to serine 396 (27, 35). To examine the phosphorylation state of IRF-3 in 293 cells, whole-cell lysates were recovered from cells that were either mock treated or infected with WNV-NY over a 64-h time course. Immunoblot analysis with a phospho-specific antibody that recognizes the phosphorylated S396 on IRF-3 (35) was used to monitor the phosphorylation state of IRF-3 during WNV-NY infection. As shown in Fig. 6A, the active phosphoserine 396 isoform of IRF-3 was not detected until 24 h postinfection and was then easily detectable throughout all of the later points of the infection time course. To confirm that phosphorylation of IRF-3 results in its nuclear retention within WNV-infected cells, the distribution of IRF-3 within two human cell lines, Huh7 and U-2OS, infected with WNV-NY was evaluated. As a positive control, Huh7 cells were infected in parallel with Sendai virus (SenV), which is known to induce activation and nuclear localization of IRF-3 (36). As expected, IRF-3 was localized to the cytoplasm of mock-infected cells and translocated to the nucleus in response to SenV infection (Fig. 6B). Nuclear localization of IRF-3 was also observed in WNV-NY-infected Huh7 and U-2OS cells. This indicates that the activation of IRF-3 by WNV is not cell type specific and therefore is a general response to WNV infection.
FIG. 6.
Characterization of the activation state of IRF-3 in WNV-NY-infected cells. (A) Phosphorylation state of IRF-3. Whole-cell lysates were recovered from mock- and WNV-NY-infected 293 cells over a 64-h time course, and immunoblot analysis was performed with an antibody specific for the phosphoserine 396 isoform of IRF-3 (IRF-3-P) (upper panels). Blots were stripped and reprobed with anti-sera against total IRF-3 (middle panels) or actin (lower panels). (B) IRF-3 localization in mock-, WNV-, or SenV-infected Huh7 or U-2OS cells was detected by indirect immunofluorescence analysis with IRF-3 polyclonal antiserum and a fluorescein isothiocyanate-conjugated secondary antibody. Images were acquired by using a Ziess Axiovert fluorescence microscope equipped with a digital camera and Axiovision software. (C) Induction of IFN-β expression by WNV-NY. IFN-β and GAPDH expression in mock- or WNV-NY-infected 293 cells was assessed by Northern blot analysis of total RNA harvested at the indicated times postinfection.
Since activation of IRF-3 leads to the expression of IFN-β (29, 33), we examined the influence of WNV infection on the expression of IFN-β in 293 cells. Northern blot analysis of total RNA isolated from infected cells demonstrated that WNV-NY induced the expression of IFN-β in a time course that followed the accumulation of viral products, with only a low level of IFN-β present at 24 h, followed by an increase in expression level at the 36 h time point postinfection (compare Fig. 2 and 6C). The induction of IRF-3 activation and subsequent expression of IFN-β in WNV-infected cultures indicates that the acute phase of the innate antiviral response is intact within 293 cells and that WNV can trigger the activation of this arm of antiviral response, at least at late time points postinfection.
Role of IRF-3 in controlling WNV infection.
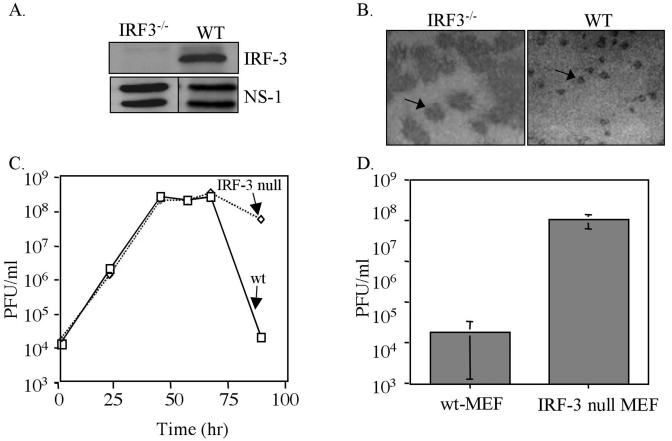
In order to ascertain what role, if any, the IRF-3 pathway plays in controlling WNV infection, MEFs isolated from either wt or IRF-3-null mice (33) were infected with WNV-NY. The NS1 protein of WNV was detected in lysates from both wt virus- and IRF-3-null virus-infected cells, indicating that WNV was able to replicate in either cell line (Fig. 7A). Focus-forming assays were used to assess the ability of WNV-NY to establish a productive infection on a per-cell basis. The number of focus-forming units of WNV-NY stock inoculum was similar on wt and IRF-3-null MEFs. However, the presence of WNV antigens was detected over significantly larger focal areas in monolayers of IRF-3-null MEFs compared to wt MEFs (Fig. 7B). The fact that the number of focus-forming units of WNV-NY were similar on wt and IRF-3-null MEFs suggested that IRF-3 does not play a role in altering the ability of WNV to enter the host cell or to establish the initial productive infection. However, the increased focal area of infected cells within cultures of IRF-3-null MEFs indicates that the IRF-3 pathway plays a role in limiting the cell-to-cell spread of WNV.
FIG. 7.
Replication of WNV-NY in wt and IRF-3-null MEFs. (A) The IRF-3-null genotype was confirmed by immunoblot analysis of lysates prepared from wt and IRF-3-null MEFs with an anti-IRF-3 antibody (FL425; Santa Cruz). The lower panels show WNV NS1 expression in WNV-NY-infected wt and IRF-3-null MEFs. (B) Focus-forming assays were used to visualize foci of replicating WNV-NY in wt and IRF-3-null MEFs at 96 h postinfection. WNV foci are visible as dark-stained areas of the cell monolayer. The arrow points to a single WNV focus in each culture. (C) Infectious particle production by WNV-NY-infected wt and IRF-3-null MEFs. Culture medium was removed from infected MEFs and cleared of cell debris by low-spin centrifugation, and the presence of infectious virus particles was determined as PFU/milliliter by determining the titers of supernatants on Vero cells in duplicate. The results from a representative experiment are shown. Solid line, IRF-3-null MEFs; dashed line, wt MEFs. (D) Viral titer of supernatants from WNV-NY-infected wt or IRF-3-null MEFs at 96 h postinfection. The graph represents the average titer from three independent experiments with two individual clones of IRF-3-null MEFs. The error bars represent ± the standard deviation.
The effect of the IRF-3 pathway on the production of infectious WNV particles was also examined. wt and IRF-3-null MEFs were infected with WNV-NY, and culture supernatants were recovered at the indicated times postinfection. The production of infectious virus particles was monitored by determining the titers of the supernatants on Vero cells. The titers of virus particles generated in wt and IRF-3-null MEFs were similar through 72 h postinfection (Fig. 7C). However, at 96 h postinfection there was a significant decrease in the number of infectious WNV particles present in supernatants of wt MEFs, while in contrast only a slight drop in infectious particles was observed in media recovered from IRF-3-null MEFs at this time point. The large focus-forming phenotype (data not shown) and the sustained production of infectious WNV-NY in the IRF-3-null MEFs at 96 h postinfection was confirmed by using a second clonal cell line derived from IRF-3-null mice (Fig. 7D), indicating the observed results were not specific to an individual clone.
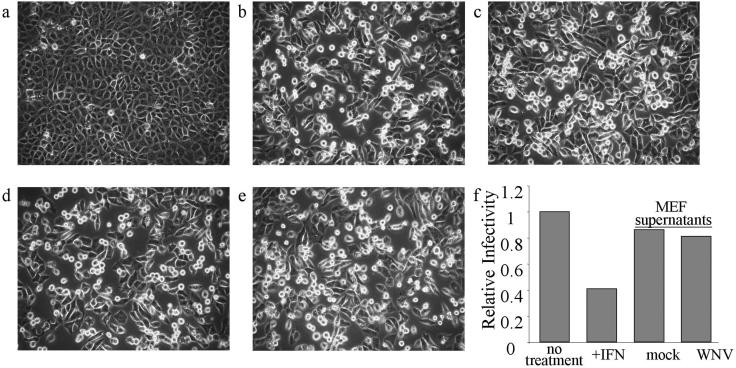
Because the bioactivity of the mouse isoform of IFN upon primate cells is negligible, it is unlikely that the reduction in viral titers observed for particles produced from wt MEFs was due to the actions mouse IFN. However, other MEF-produced soluble factors could possibly influence the infectivity of WNV on Vero cells. To explore this possibility, cleared culture supernatants from WNV-infected wt MEFs were tested for their ability to protect Vero cells from virus challenge. WNV particles were removed from culture media recovered from mock- or WNV-NY-infected wt MEFs at 96 h postinfection by ultracentrifugation, and the clarified supernatants were then exposed to UV irradiation to inactivate any remaining virus. The clarified supernatants from mock- or WNV-infected wt MEF cells were added to Vero cells, followed by infection with VSV-GFP 24 h later. The effect of pretreating Vero cells with these supernatants was determined by monitoring the level of VSV-GFP-induced CPE and the level of viral production. Exposure of Vero cells to supernatants recovered from the WNV-infected MEFs at 96 h postinfection did not reduce VSV-GFP-induced CPE (Fig. 8a to e). In addition, pretreatment of Vero cells with supernatants recovered from WNV-infected MEF cultures had no effect on VSV-GFP infectivity compared to cultures pretreated with supernatants recovered from mock-infected MEF cultures (Fig. 8f). This indicates that the reduction in WNV titers in the culture supernatant of wt MEFs at 96 h postinfection was not due to the actions of soluble factors that influence Vero cell permissiveness to virus infectivity.
FIG. 8.
Soluble factors in MEF culture supernatants do not interfere with VSV-GFP challenge of Vero cells. (a to e) VSV-GFP-induced CPE. Images of mock (a) and VSV-GFP-infected cells (b to e) were acquired by using a light microscope. Cultures were treated as follows: a and b, untreated cultures of Vero cells; c, Vero cells pretreated with IFN-α 2a (500 U/ml); d and e, cultures pretreated with clarified supernatants (1 ml) recovered at 96 h postinfection from WNV-NY-infected or mock-infected wt MEFs, respectively. Vero cells were pretreated for 24 h and then infected with VSV-GFP at an MOI of 0.1 in panels b to e. At 16 h postinfection the supernatants were removed, and VSV-GFP titers were assessed by plaque formation assays. (f) The relative infectivity of viral particles produced under each condition was derived by dividing the number of PFU/milliliter in supernatants from each sample by the number of PFU/milliliter in supernatants from untreated control cultures. For comparison, the relative infectivity of the nontreated control culture was set at 1.0.
DISCUSSION
In the present study the cellular response to WNV infection was examined by using a clonal virus stock, WNV-NY, that was derived from an infectious clone generated from a viral stock isolated from the New York 2000 epidemic. A previous characterization of WNV-NY demonstrated that this cloned virus exhibited similar replication to the original viral isolate and was pathogenic in both cell culture and in a mouse challenge model (39). We found that WNV-NY was also pathogenic in cultured human cells, although the level of virus-induced CPE varied greatly between the various cell lines. This raises the possibility that the CPE of WNV may contribute to disease outcome in vivo.
WNV modulation of global gene expression.
The impact of WNV-NY upon global host cell gene expression over the course of an infection was examined by microarray analysis. Our results show that at a high MOI, WNV-NY induced a biphasic modulation of gene expression. The observed second phase of modulation of host cell gene expression was not due to a new round of infection initiated by virus spread, since all cells were infected during the initial viral inoculation. Instead, this reflects a second wave of cellular responsiveness to the infection. This biphasic response to WNV was only observed when cells were infected at a high MOI (data not shown), suggesting that the second wave of regulation of host cell gene expression may constitute a response to the accumulation of viral replication products and/or an increase in CPEs. Evaluation of the pattern of expression of specific genes from several functional categories in WNV-NY-infected cells suggested that WNV replication modulated the expression of genes across a broad range of functional categories. WNV replication had a prominent impact on genes whose products are involved in stress response, signal transduction, and transcriptional regulation programs. At the 24-h time point postinfection we noted the largest number of genes whose expression was reduced in infected cells, with the chief component of these belonging to signal transduction genes (data not shown). This trend was reversed as the infection progressed, and at 48 h postinfection there was a marked predominance of genes within these three categories whose expression increased in virus-infected cells. This pattern of gene regulation could reflect an acute response to WNV infection that was followed by a period of stabilization and gene expression compensation.
Northern blot and immunoblot analyses revealed that WNV induced the expression of several known ISGs, demonstrating that virus replication was signaling the host cell innate antiviral response pathways. To extend our analysis of the WNV effect on the innate antiviral response, microarray data were examined for the modulation of genes previously shown to respond to dsRNA, IFN, and/or viral stimuli. These analyses confirmed that WNV infection stimulated the expression of several known ISGs, thus extending our initial analysis of ISG mRNA and protein expression in the infected cells. The WNV induction of the majority of ISGs did not follow the same biphasic modulation as the global gene expression profile but instead were variably induced throughout the entire course of the experiment. The expression of at least a subset of these ISGs reached higher levels at 36 h postinfection or later (see Fig. 2 and 6C), suggesting that this arm of the host response to WNV infection could be a characteristic event of late infection. Taken together, our data suggest that ISG expression was effected by different and distinct stimuli than that of the majority of WNV-regulated host genes and that WNV replication sets off a complex host response that includes a variety of gene expression pathways within the host cell.
Activation of the interferon response by WNV.
Our analysis of the innate antiviral response to WNV has established that viral infection induced the expression of several known IRF-3 target genes. We confirmed that WNV activated the transcription effector function of IRF-3 at time points past 24 h postinfection and that this correlated with the expression of IFN-β, a known IRF-3 target gene (21, 33). We also observed an induction in ISG56 protein expression in cultures of 293 cells treated with IFN (data not shown), indicating that the JAK/STAT pathway is intact in these cells and capable of responding to IFN. This raises the possibility that a combination of IRF-3 and IFN-mediated mechanisms are responsible for the ISG expression observed in WNV-NY-infected cultures. Indeed, the increased expression of several IFN-responsive genes, including ISG6-16, ISG20, STAT1α, and IRF-9, has been shown to be stimulated through IFN-dependent mechanisms (11). The fact that an increase in expression of these genes was observed in WNV-infected cells suggests that WNV induced the secretion of IFN-α/β and subsequently signaled the activation of the JAK/STAT pathway through the IFN-α/β receptor. Importantly, there are hundreds of genes that have been shown to respond to IFN-α/β (11), but only a subset of these genes were induced during WNV infection. This raises the possibility that WNV might attenuate the spectrum of ISG expression in an attempt to limit the establishment of an antiviral state within the host cell. It is noteworthy that recent work has identified the NS4B protein of dengue virus, a related flavivirus, as an antagonist of STAT1 function in infected cells (28). In this context dengue virus strains encoding NS4B with the potential to regulate STAT1 would be expected to attenuate the host response to IFN that otherwise limits dengue virus replication (12, 13). It is possible that WNV may similarly direct mechanisms of viral control that limit IFN action and ISG expression.
Induction of IRF-3 activity.
Extracellular dsRNA has been shown to activate IRF-3 through Toll-like receptor 3 (TLR3) (4), but there is a growing body of evidence that intracellular stimuli can activate IRF-3 through a TLR3-independent process (22). The fact that 293 cells, a cell line that also lacks TLR3 expression (14, 32), still activate IRF-3 in response to WNV infection (see Fig. 6) suggests that WNV signals the action of IRF-3 through an as-yet-undefined, TLR-3-independent mechanism. In this respect, WNV replication products may interact with a “viral sensor” factor to initiate the intracellular IRF-3 activation pathway. The kinetics of activation of IRF-3 for many viral infections, including human cytomegalovirus, herpes simplex virus type 1, and Newcastle virus, has been observed to occur between 2 and 6 h postinfection (31, 46). Virus binding to cell surface receptor(s) or the accumulation of viral RNA constitute potential triggers for this rapid activation of IRF-3 (6, 22). In contrast, measles virus infection has been shown to exhibit a comparably delayed activation of IRF-3, which does not occur until 16 to 20 h postinfection, and is triggered in part by the production of the viral nucleocapsid protein (42). Like measles virus, activation of IRF-3 in response to WNV did not occur until late in the infection. A relatively small faction of total IRF-3 was phosphorylated on serine 396 within 20 h postinfection; however, high levels of phosphorylated IRF-3 were not detected until ca. 36 h postinfection. The lack of activation of IRF-3 until later times in infection suggests that an accumulation of WNV specific proteins and/or viral RNA is responsible for stimulating the IRF-3 pathway, although the actual viral component(s) responsible for triggering IRF-3 remain to be determined. Such a delayed activation of IRF-3 in WNV-infected cells could be attributed to a threshold effect in which the activator(s) of IRF-3 must first be produced at sufficient levels to effect IRF-3 activation. Alternatively, WNV may sequester the viral stimulus or actively prevent IRF-3 phosphorylation to preclude activation of IRF-3 until late in infection. The identification of the viral component(s) responsible for the regulation of IRF-3 will aid in distinguishing between these two possibilities. These results suggest the possibility that WNV is unable to block the induction of the host cell antiviral response but may regulate the time of induction by actively avoiding triggering IRF-3 during early infection.
The IRF-3 response limits the spread of WNV but is not sufficient to block viral replication.
The importance of IRF-3 in defending against viral pathogens is attested to by the fact that divergent pathogens have acquired the ability to disrupt the IRF-3 response. Two members of the Flaviviridae family, bovine diarrhea virus (BVDV) and hepatitis C virus, have recently been shown to block the IRF-3 pathway. Hepatitis C virus prevents the virus-induced phosphorylation of IRF-3 (15), whereas BVDV specifically blocks the transcriptional activity of activated IRF-3 (2). In both cases, these viruses effectively prevent the expression of IRF-3 target genes and thereby limit the host cell's response to the infection. In contrast, WNV activates IRF-3 late in infection, but the actions of the IRF-3 pathway are insufficient to prevent WNV replication. However, our results from experiments with IRF-3-null MEFs indicate that the IRF-3 pathway does play a role in limiting the cell-to-cell spread of WNV. Interestingly, a small plaque phenotype was observed for rotavirus NS1 mutants that are no longer able to interact with nor regulate IRF-3 (20). In another study, the plaque size of Semliki Forest virus was found to increase when cell cultures were coinfected with BVDV, an IRF-3 antagonist (2). Taken together, these data suggest that one function of the IRF-3 pathway is to limit the transmission of viral particles between cells during acute infection. The simplest explanation for the IRF-3-dependent suppression of WNV spread is that the accumulation of IRF-3 target genes and/or the induction of ISG expression due to secreted IFN leads to the establishment of an antiviral state within both the host and the neighboring cells. Our preliminary data suggest that the actions of either direct and/or indirect IRF-3-responsive genes do not block the production of virus particles but instead influence the generation and/or accumulation of apparently defective viral particles (B. L. Fredericksen and M. Gale, unpublished observations), which could contribute the inability of WNV to establish a productive infection in neighboring cells. Further characterization of these defective particles, as well as comparisons of the innate antiviral response to pathogenic lineage I and nonpathogenic lineage II isolates, is needed to clearly define the role of the IRF-3 pathway in WNV-induced pathogenesis.
Acknowledgments
We thank P Mason, T. Taniguchi, W. Bresnahan, R. Krug, M. David, J. Hiscott, M. Whitt, G. Sen, and A. Haas for reagents.
This study was supported the UT Southwestern Endowed Scholars for Biomedical Research program. B.L.F. was supported by NIH training grant T32AI07520. M.G. is the Nancy C and Jeffery A. Marcus Scholar in Medical Research in Honor of Bill S. Vowell.
REFERENCES
- 1.Armali, Z., R. Ramadan, A. Chlebowski, and Z. S. Azzam. 2003. West Nile meningo-encephalitis infection in a kidney transplant recipient. Transplant. Proc. 35:2935-2936. [DOI] [PubMed] [Google Scholar]
- 2.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 4.Barton, G. M., and R. Medzhitov. 2003. Linking Toll-like receptors to IFN-alpha/beta expression. Nat. Immunol. 4:432-433. [DOI] [PubMed] [Google Scholar]
- 5.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 8.Brazma, A., U. Sarkans, A. Robinson, J. Vilo, M. Vingron, J. Hoheisel, and K. Fellenberg. 2002. Microarray data representation, annotation, and storage. Adv. Biochem. Eng Biotechnol. 77:113-139. [DOI] [PubMed] [Google Scholar]
- 9.Brinton, M. A. 1983. Analysis of extracellular West Nile virus particles produced by cell cultures from genetically resistant and susceptible mice indicates enhanced amplification of defective interfering particles by resistant cultures. J. Virol. 46:860-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinton, M. A. 2002. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371-402. [DOI] [PubMed] [Google Scholar]
- 11.Der, S. D., A. Zhou, B. R. G. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 13.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 15.Foy, E., K. li, C. Wang, R. Sumpter, M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 16.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiss, G. K., R. E. Bumgarner, M. An, M. B. Agy, A. van't Wout, E. Hammersmark, V. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 18.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 77:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiss, G. K., G. Jin, J. Guo, R. E. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 20.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoebe, K., E. M. Janssen, S. O. Kim, L. Alexopoulou, R. A. Flavell, J. Han, and B. Beutler. 2003. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat. Immunol. 4:1223-1229. [DOI] [PubMed] [Google Scholar]
- 23.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-677. [DOI] [PubMed] [Google Scholar]
- 24.Kramer, L. D., and K. A. Bernard. 2001. West Nile virus infection in birds and mammals. Ann. N. Y. Acad. Sci. 951:84-93. [DOI] [PubMed] [Google Scholar]
- 25.Lanciotti, R. S., G. D. Ebel, V. Deubel, A. J. Kerst, S. Murri, R. Meyer, M. Bowen, N. McKinney, W. E. Morrill, M. B. Crabtree, L. D. Kramer, and J. T. Roehrig. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298:96-105. [DOI] [PubMed] [Google Scholar]
- 26.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. Mackenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 27.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2003. Identification of Ser 386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 11:9698-9702. [DOI] [PubMed] [Google Scholar]
- 28.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 30.Petersen, L. R., and J. T. Roehrig. 2001. West Nile virus: a reemerging global pathogen. Emerg. Infect. Dis. 7:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar, S. N., H. L. Smith, T. M. Rowe, and G. C. Sen. 2003. Double-stranded RNA signaling by Toll-like receptor 3 requires specific tyrosine residues in its cytoplasmic domain. J. Biol. Chem. 278:4393-4396. [DOI] [PubMed] [Google Scholar]
- 33.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 34.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 35.Servant, M. J., N. Grandvaux, B. R. TenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 36.Servant, M. J., B. tenOever, C. LePage, L. Conti, S. Gessani, I. Juljunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 37.Sharma, S., R. Benjamin, B. tenOever, N. Grandvaux, G.-P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through a novel IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 38.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 39.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, M. W., Z. N. Yue, M. J. Korth, H. A. Do, L. Boix, N. Fausto, J. Bruix, R. L. Carithers, Jr., and M. G. Katze. 2003. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology 38:1458-1467. [DOI] [PubMed] [Google Scholar]
- 41.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.TenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA-binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 45.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]
- 46.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]