Abstract
Rotavirus plus-strand RNAs not only direct protein synthesis but also serve as templates for the synthesis of the segmented double-stranded RNA (dsRNA) genome. In this study, we identified short-interfering RNAs (siRNAs) for viral genes 5, 8, and 9 that suppressed the expression of NSP1, a nonessential protein; NSP2, a component of viral replication factories (viroplasms); and VP7, an outer capsid protein, respectively. The loss of NSP2 expression inhibited viroplasm formation, genome replication, virion assembly, and synthesis of the other viral proteins. In contrast, the loss of VP7 expression had no effect on genome replication; instead, it inhibited only outer-capsid morphogenesis. Similarly, neither genome replication nor any other event of the viral life cycle was affected by the loss of NSP1. The data indicate that plus-strand RNAs templating dsRNA synthesis within viroplasms are not susceptible to siRNA-induced RNase degradation. In contrast, plus-strand RNAs templating protein synthesis in the cytosol are susceptible to degradation and thus are not the likely source of plus-strand RNAs for dsRNA synthesis in viroplasms. Indeed, immunofluorescence analysis of bromouridine (BrU)-labeled RNA made in infected cells provided evidence that plus-strand RNAs are synthesized within viroplasms. Furthermore, transfection of BrU-labeled viral plus-strand RNA into infected cells suggested that plus-strand RNAs introduced into the cytosol do not localize to viroplasms. From these results, we propose that plus-strand RNAs synthesized within viroplasms are the primary source of templates for genome replication and that trafficking pathways do not exist within the cytosol that transport plus-strand RNAs to viroplasms. The lack of such pathways confounds the development of reverse genetics systems for rotavirus.
Rotaviruses, members of the Reoviridae, are an important cause of acute gastroenteritis in infants and young children (15). The virion is an icosahedron composed of three concentric layers of protein with a genome of 11 segments of double-stranded RNA (dsRNA) (30). The outer layer of the infectious triple-layered particle (TLP) is made up of the glycoprotein, VP7, and the spike protein, VP4. The intermediate layer is formed by VP6 trimers, and the inner layer is formed by the core lattice protein, VP2, arranged with T=1 icosahedral symmetry. Positioned at the vertices of the VP2 lattice are individual copies of the RNA-dependent RNA polymerase (RdRp) VP1, and the mRNA-capping enzyme VP3. Together, VP1, VP2, VP3, and the dsRNA genome make up the core of the virion (19).
Rotavirus entry is accompanied by the loss of the VP4 and VP7 outer layer, thereby converting TLPs to double-layered particles (DLPs). The RdRp of the DLP functions as a transcriptase to synthesize the 11 viral plus-strand RNAs (18). The plus-strand RNAs are extruded from DLPs through channels at the vertices that extend through both the VP2 and VP6 protein layers. The plus-strand RNAs contain 5′ caps but lack 3′ poly(A) tails and are translated to give rise to six structural proteins (VPs) and six nonstructural proteins (NSPs). The plus-strand RNAs also function as templates for the synthesis of the dsRNA genome segments. RNA replication occurs concurrently with the packaging of the genome segments into newly formed cores (25) and is coordinated such that the 11 segments are produced at equimolar levels (24).
Rotavirus infection leads to the formation of perinuclear, non-membrane-bound cytoplasmic inclusions (viroplasms) (28). Evidence that NSP2 and NSP5 have critical roles in viroplasm formation was provided by Fabbretti et al. (8), who demonstrated that these proteins, coexpressed in uninfected cells, give rise to viroplasm-like structures. Viroplasms are the putative sites of RNA replication (minus-strand synthesis) and core and DLP assembly. This hypothesis is based on immunofluorescence (IF) analysis showing that the protein components of cores and DLPs accumulate in viroplasms and on electron microscopic analysis showing that newly made cores and DLPs assemble near viroplasms (1, 7, 28). These inclusions are also electron dense, as though rich in RNA. As potential sources of plus-strand RNAs that can stimulate increased levels of translation and replication, the assembly of progeny DLPs in the infected cell plays a critical role in virus amplification. Since viroplasms represent the likely sites of DLP assembly, amplification can be predicted to be dependent on the formation of viroplasms.
The mechanism of protein trafficking that leads to the accumulation of viral proteins in viroplasms has not been delineated. Similarly, the source of the plus-strand RNAs that accumulate in viroplasms and function as templates for dsRNA synthesis has not been established. However, a favored idea has been that plus-strand RNAs are made by DLPs located outside of viroplasms. Subsequently, the plus-strand RNAs are either incorporated into polysomes, thereby driving protein synthesis, or are transported by viral RNA-binding proteins to viroplasms, where they undergo replication.
The ubiquitous cellular defense mechanism, RNA interference (RNAi) (9, 14), has been developed into an important tool for the study of protein function in virus replication. In RNAi technology, small 21- to 22-nucleotide RNA duplexes termed “short interfering RNAs” (siRNAs) are used to target the degradation of mRNAs through sequence recognition (6, 34). This process is mediated by the incorporation of the siRNA into a nuclease complex (RNA-induced silencing complex [RISC]) in which the duplex is unwound (23), facilitating the binding of the siRNA to a complementary mRNA. The bound mRNA is degraded, resulting in a subsequent loss of the encoded protein product.
We show here that rotavirus gene-specific siRNAs induced formation of silencing complexes in infected cells that caused near-complete reductions in the expression of the nonessential protein NSP1, the viroplasm protein NSP2, and the outer capsid protein VP7. The loss of NSP2 expression inhibited multiple events, including genome replication. In contrast, the loss of either NSP1 or VP7 had no effect on genome replication, an unexpected result given that the degradation of the gene 5 (g5) and g9 plus-strand RNAs necessary for NSP1 and VP7 translation was expected to lead to a corresponding loss of plus-strand RNAs necessary for genome replication. Instead, our results indicate that plus-strand RNAs, which are located outside viroplasms and undergo translation are susceptible to siRNA-induced degradation, whereas those plus-strand RNAs which are located inside viroplasms and undergo replication are not. Thus, plus-strand RNAs residing outside viroplasms probably cannot serve as the source of plus-strand RNAs that template dsRNA synthesis within the viroplasm. Indeed, IF detection of newly made RNAs in infected cells indicate that the plus-strand RNA templates for dsRNA synthesis are made within viroplasms. If no mechanism exists for the intracellular transport of plus-strand RNAs from the cytosol to the viroplasm, then the approach of transfecting recombinant plus-strand RNAs into rotavirus-infected cells as a means of achieving reverse genetics may fail because of the plus-strand RNAs' inability to reach the site of RNA replication.
MATERIALS AND METHODS
Cell culture and viruses.
Fetal rhesus monkey kidney (MA104) cells were maintained in minimal essential medium with Earle's salts (E-MEM) containing 5% fetal bovine serum (HyClone) and 1% GASP (Quality Biological). Rotavirus strains SA11-5N (27) and DxRRV (22) were propagated, and titers were determined in MA104 cells. Rotaviruses were activated by incubation with 10 μg of trypsin per ml at 37°C for 30 min.
siRNA transfection.
Duplex siRNAs containing sequences positioned within the open reading frame of g9 of DxRRV, g5 and g8 of SA11-5N, and g5 of Wa were obtained from Dharmacon Research (Table 1). All siRNAs were synthesized with 3′-dTdT overhangs. Typically in siRNA experiments, MA104 cells were grown to near confluency in 12-well plates. After two rinses with E-MEM, 1 ml of the same was placed on each monolayer. A 200-μl volume of Opti-MEM I (Invitrogen) containing 2% Lipofectamine 2000 (Invitrogen) and 0.3 μM duplex siRNA was then added to the medium overlaying each of the monolayers. After incubation for 4 h, 1 ml of E-MEM containing 20% fetal bovine serum was added to each of the monolayers. Incubation was then continued for 26 to 28 h.
TABLE 1.
siRNA names and mRNA target sequences
| Strain | Gene/protein | siRNA name | mRNA target (nucleotide numbers)a |
|---|---|---|---|
| SA11-5N | g5/NSP1 | g5A | AAUAGCUGAUUUAACCCAUUG (171-191) |
| g5B | AACCACUAUGCAAAGAAAUAC (640-660) | ||
| g5C | AAUAUUUGAGUUGGGACAUCA (924-944) | ||
| g5D | AAAUUACAGUGUAUCAUAGAC (310-330) | ||
| g5E | AAGUAAUGUUAACGUUGGACA (1101-1121) | ||
| SA11-5N | g8/NSP2 | g8A | AAAGCUAUGCUGACAGCUAAA (128-148) |
| g8B | AAAGUGACGCAAGCAAACGUC (320-340) | ||
| g8C | AACACUGAUUGCUAUUGGACA (451-461) | ||
| g8D | AAAGAAUUAGUCGCUGAACUU (653-673) | ||
| DxRRV | g9/VP7 | g9A | AAUUACAGGAUCAAUGGACGC (222-242) |
| g9B | AAUGAUGGUGACUGGAAAGAC (327-347) | ||
| g9C | AAACGUUAGGGAUAGGUUGUC (650-670) | ||
| g9D | AACAGCAGAUCCAACAACUAA (861-881) | ||
| WA | g5/NSP1 | IR | AACUGCUUCUUGGCUGGAAUG (782-802) |
Target sequences are located within the coding region of the indicated genes. Nucleotide numbers correspond to the 5′ and 3′ ends of the mRNA target sequence.
Virus infection.
siRNA-transfected cells were rinsed twice with E-MEM and infected with DxRRV or SA11-5N at a multiplicity of infection (MOI) of 1. At 1 h postinfection (p.i.) the inoculum was removed and replaced with E-MEM. To radiolabel proteins, the inoculum was replaced instead with 80% methionine, cysteine-free MEM and 20% E-MEM, containing 25 μCi of [35S]-Express Protein Labeling Mix (Perkin-Elmer Life Sciences) per ml. To radiolabel RNAs, the inoculum was replaced with 80% phosphate-free Dulbecco MEM and 20% E-MEM. At 3 h p.i., the medium was supplemented with 25 μCi of [32P]orthophosphate (8,000 to 9,000 Ci/mmol) and 5 μg of actinomycin D per ml.
Analysis of proteins produced in vivo.
Rotavirus-infected MA104 cells were scraped into the medium, pelleted, and resuspended in water containing 1 μg per ml each of leupeptin and aprotinin. After three cycles of freeze-thawing, the lysates were analyzed by electrophoresis on 10% NuPAGE Bis-Tris gels (Invitrogen). Proteins were detected by staining with Coomassie blue and by autoradiography. 35S-labeled proteins were quantified with a Molecular Dynamics PhosphorImager 445 SI.
In Western blot assays, proteins were transferred from gels onto nitrocellulose membranes, and the blots were soaked in phosphate-buffered saline (PBS) containing 5% milk. DxRRV VP7 and VP6, SA11-5N NSP1, NSP2, and VP2 were detected by using guinea pig antisera prepared against whole RRV (1:2,500), VP6 (1:1,000), NSP2:NSP1 fusion protein (1:500), and VP2 (1:2000), respectively. The NSP2:NSP1 fusion protein was prepared by modification of the NSP2 bacterial expression vector pQE60g8 (31) to link the C-terminal 19 amino acids of RRV NSP1 to the C terminus of NSP2 via a GPGP hinge. A His tag was placed downstream of the NSP1 sequence. The purified protein was used to prepare guinea pig polyclonal antisera. VP2 was expressed from the recombinant baculovirus rBVg2 (26), purified as described previously (33), and used to prepare guinea pig polyclonal antisera. Actin was detected with goat anti-actin (1:400) antisera (Santa Cruz Biotechnology). Horseradish peroxidase-conjugated goat anti-guinea pig and rabbit anti-goat antisera at dilutions of 1:10,000 were used as secondary antibodies. Blots were developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and exposed to BioMax MR film.
Analysis of virion-derived dsRNA.
Rotavirus-infected MA104 cells were collected, resuspended in water, subjected to three cycles of freeze-thawing, and briefly centrifuged to remove large cellular debris. After treatment with 160 U of DNase I (Ambion) per ml for 30 min at 37°C, dsRNAs were purified by phenol-chloroform extraction, resolved by electrophoresis on 10% polyacrylamide gels, and detected by silver staining.
Electrophoretic analysis of cytoplasmic RNA.
MA104 cells were scraped into the medium, pelleted, washed twice with PBS, and lysed by suspension in 100 mM Tris-HCl (pH 8), 150 mM NaCl, 1.5 mM MgCl2, 30 mM EDTA, 1% Triton X-100 for 5 min. After removal of nuclei by low-speed centrifugation, the lysates were adjusted to 0.5% sodium dodecyl sulfate and 250 mM NaCl. After phenol-chloroform extraction, RNAs were collected by ethanol precipitation and resuspended in water. RNAs were combined with an equal volume of formamide sample buffer (98% formamide containing 10 mM EDTA and 1 mg of bromophenol blue per ml), heated for 5 min at 42°C, and analyzed by electrophoresis on 5% polyacrylamide gels containing 7 M urea. Radiolabeled RNAs were detected by autoradiography and quantified with a phosphorimager.
Synthesis of RNA probes.
The minus-sense probes SA11-g5PR and SA11-g8R were each made by T7 transcription of DNA templates generated by two sequential rounds of PCR. The first round used Pfu Turbo DNA polymerase (Stratagene), the template DNAs pSP65g5 or pSP65g8R, and the g5- or g8-specific primer pairs, 5′-GCTTCCTTGACAATGAACCTC-3′ and GATTAGATGCCACGTAGTTAGG or ATGGCTGAGCTAGCTTGC and CAACTTGAGAAACTTCGTCC, respectively. The second round used the DNA products of the first round as templates and the g5- or g8-specific primer pairs ATTCAGTAATACGACTCACTATAGGGTATATTGATCTTCCTGG and GCTGCATTTAAGTTTGATGAAAATAATC or TACAGCTAATACGACTCACTATAGGTGCCATCGAAGTTCAGC and ACGAGTAGTATCAATAAGGC, respectively. (The T7 promoter is underlined).
To produce the minus-sense probes RRV-g4PR and D-g9PR, g4 and g9 cDNAs were made from DxRRV dsRNAs by using the Titan One-Tube RT-PCR System (Roche) and the primer pairs GTATCGGATCCAACAGACTCACTGTTCG and GTGACTGGATCCGGGGTAATGTGTTTGGAG or TATTGGGATCCTGGTTATGTAACCCAATGG and ATACAGGATCCCCACCAACTTGTATTACAGC, respectively. (The BamHI site is shown in boldface.) After digestion with BamHI, the cDNAs were ligated into the same site of pSP65. Plasmid DNA containing a g4 or g9 insert was used in amplification reactions containing primer pairs TCGACTTAATACGACTCACTATAGGGGTAATGTGTTTGGAG and AACAGACTCACTGTCCG or TCGACTTAATACGACTCACTATAGGACCAACTTGTATTACAGC and TGGTTATGTAACCCAATGG, respectively, to produce templates for T7 transcription.
PCR products were transcribed by using the MegaScript High Yield Transcription kit (Ambion), producing the 300-nucleotide SA11-g5PR, SA11-g8PR, RRV-g4PR, and d-g9PR RNA probes. Reaction mixtures included 1.87 mM UTP and 2.5 μCi of [α-32P]UTP (800 Ci/mmol) per ml and were incubated at 37°C for 18 h. RNA was purified as according to protocols of the supplier (Ambion), and unincorporated nucleotides were removed with G50 Sephadex Quick-Spin columns (Roche).
Northern blot analysis of cytoplasmic RNAs.
Cytoplasmic RNAs were recovered from MA104 cells by using a Qiagen RNeasy Mini kit, and Northern blot assays were performed by using a NorthernMax-Gly kit (Ambion) according to the protocols of the suppliers. After denaturation with Glyoxal, 5 μg per lane of cytoplasmic RNA was resolved by electrophoresis on 1% Agarose-LE gels (Ambion). The mRNAs were passively transferred to nylon membranes for 2 h and then cross-linked to the membranes by using UV light. The membranes were incubated in prehybridization buffer (UltraHyb; Ambion) at 68°C for 30 min and then in hybridization buffer containing 106 cpm of 32P-labeled RNA probe per ml at 68°C for 24 h. After being washed, the membranes were analyzed by autoradiography.
Isolation of virus particles by isopycnic centrifugation.
MA104 cells in 10-cm plates were transfected with siRNAs, infected with DxRRV, and radiolabeled with 35S-labeled amino acids, as described above. At 18 h p.i., the cells were scraped into the medium and subjected to three cycles of freezing-thawing. After the removal of cell debris by low-speed centrifugation, virus was pelleted by centrifugation at 160,000 × g in a Beckman SW40 Ti rotor for 2 h. The pellet was resuspended in TBS buffer (25 mM Tris-HCl [pH 7.4], 137 mM NaCl, 5 mM KCl, 1 mM MgCl2, 0.7 mM CaCl2, 0.7 mM Na2HPO4, 5.5 mM dextrose), and virus was banded by isopycnic centrifugation in CsCl (12). Peak fractions containing TLPs and DLPs were pooled, dialyzed in 2 mM Tris-HCl (pH 7.5)-0.5 mM Na2EDTA-0.5 mM dithiothreitol, and analyzed by electrophoresis on 12% polyacrylamide gels containing sodium dodecyl sulfate.
Detection of viral proteins by IF.
MA104 cells were grown to near confluency on glass coverslips, transfected with siRNAs, infected with rotavirus, and examined by IF as described elsewhere (35). Cells were fixed with 4% paraformaldehyde, permeabilized with 1% Triton X-100, and then incubated with guinea pig polyclonal NSP2 (1:500) (31) or VP6 (1:500) antisera, rabbit polyclonal NSP1 C19 (1:1,000) antisera (13), or mouse monoclonal NSP5 (1:1,000) (29) antibody. Anti-guinea pig Alexa Fluor 488, and anti-mouse and anti-rabbit Alexa Fluor 594-conjugated antibodies were used as secondary antibodies. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole; Pierce). Fluorescence was detected with a Leica TCS NT confocal microscope. Images were processed by using Adobe PhotoShop version 7.0.
Detection of newly made RNA in vivo by IF.
MA104 cells grown on glass coverslips in 12-well dishes were infected with SA11-5N at an MOI of 1. The inoculum was replaced with 1 ml of E-MEM, and at 8 h p.i. the medium was brought to 10 μg of actinomycin D per ml. At 8.5 h, a 170-μl volume of E-MEM containing 10 mM BrUTP and 6% FuGene transfection reagent (Roche) was added to the medium. Cells were fixed at 9 h p.i. and processed for IF analysis with mouse bromodeoxyuridine (BrdU) monoclonal antisera (Sigma) (1:50), guinea-pig NSP2 (1:500) (31) antisera and an Alexa Fluor 594 Signal Amplification Kit (Molecular Probes).
Detection of sites of transcription by IF.
MA104 cells grown on glass coverslips were infected with SA11-5N. At 8.5 h p.i., the medium was adjusted to 10 μg of actinomycin D per ml. At 9 h p.i., the cells were semifixed by incubation in 0.5% paraformaldehyde for 5 min. After a wash with PBS, the cells were permeabilized by incubation in buffer containing 0.025% Triton X-100, 25% glycerol, and 25 U of Anti-RNase (Ambion) per ml. The semifixed permeabilized cells attached to the coverslips were then incubated in transcription reaction mixtures containing 0.5 mM concentrations of ATP and CTP, 1 mM GTP, 3 mM BrUTP, 100 mM Tris-HCl (pH 7.8), 50 mM sodium acetate, 10 mM magnesium acetate, 0.2 mM S-adenosylmethionine, 1 mM dithiothreitol, and 25 U of Anti-RNase per ml for 30 min at 23°C. The cells were then fixed with 4% paraformaldehyde for 15 min and immunostained as described above.
RNase sensitivity of newly made dsRNA.
MA104 cells infected with SA11-5N were maintained in 80% phosphate-free Dulbecco MEM and 20% E-MEM beginning at 6.5 h p.i. The medium was adjusted to 10 μg of actinomycin D per ml at 8 h p.i. and 50 μCi of [32P]orthophosphate per ml at 8.5 h p.i. At 9 h pi., the cells were washed with hypotonic buffer (3 mM Tris-HCl [pH 8.1], 10 mM MgCl2, 10 mM NaCl), resuspended in the same buffer, incubated on ice for 10 min, and disrupted with 15 strokes of a Dounce homogenizer. To obtain the cytoplasmic extract, nuclei were removed from the lysate by low-speed centrifugation. Portions of the extract were treated for 30 min at 37°C with 50 U of RNase V1 per ml.
Synthesis and transfection of BrU-labeled RNA.
Capped RNAs containing BrUMP were synthesized by using an mMessage Machine T7 transcription kit (Ambion), modified such that reaction mixtures contained 7.5 mM concentrations of ATP and CTP, 1.5 mM GTP, 9 mM BrUTP, and 6 mM cap analog. The mixtures were incubated for 6 h at 37°C and then treated with DNase I. The RNA products were extracted with phenol-chloroform, precipitated with ethanol, resuspended in water, and passed twice through G50 spin columns to remove unincorporated nucleotides.
BrU-labeled g9 plus-strand RNA was made by including pSP65g9, linearized with SacII, in transcription reaction mixtures. pSP65g9 was prepared by amplifying SA11 gene 9 dsRNA with the primers GCCTGCAGGTCGACTCTAGATAATACGACTCACTATAGGCTTTAAAAGAGAGAATTTCC (T7 promoter is underlined, and the XbaI site is in boldface) and GGGATCCTCTAGACCGCGGTCACATCATAC (SacII site is underlined, and XbaI site is in boldface). The cDNA was ligated into the XbaI site of pSP65.
Transfection of BrUMP-labeled RNAs was carried out under conditions similar to those used for siRNAs. Briefly, MA104 cells grown on coverslips in 12-well dishes were infected with SA11-5N at an MOI of 1. At 1 h p.i., 1 μg of BrUMP-labeled RNA was transfected into cells by using Lipofectamine 2000. At 9 h p.i., the cells were fixed with 4% paraformaldehyde, permeabilized with Triton X-100, and immunostained with mouse anti-BrdU (1:50) and guinea pig anti-NSP2 (1:500) antisera.
RESULTS
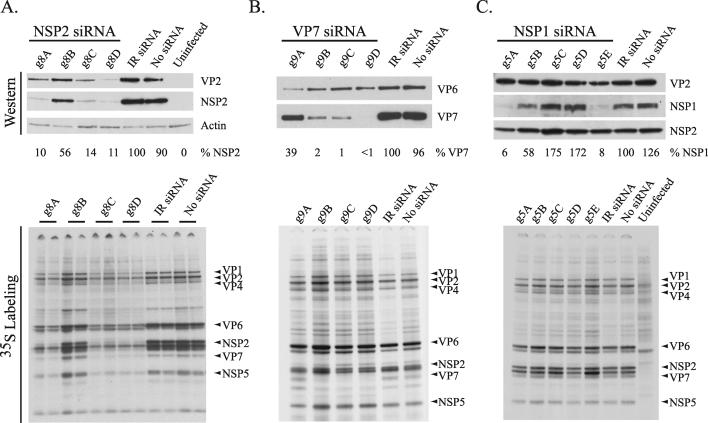
Effect of g8-specific siRNAs on virus replication. (i) Protein synthesis. To test the ability of siRNAs to inhibit the expression of rotavirus proteins, short duplex RNAs containing viral gene-specific sequences were transfected into MA104 cells (Table 1). The following day, the cells were infected with rotavirus. Expression of viral proteins in the cells was monitored by Western blot assay and by radiolabeling in vivo with 35S-labeled amino acids. Western blot analysis of SA11-infected cells transfected with SA11 g8-specific siRNAs showed that three (g8A, g8C, and g8D) were highly effective in inhibiting the expression of NSP2 (Fig. 1A). Based on comparison with cells transfected with an irrelevant (IR) siRNA (Table 1), these three siRNAs reduced NSP2 expression by 80 to 90%. This extent of inhibition parallels the 80 to 90% transfection efficiency that we observed in our experiments (data not shown). Therefore, the failure to reach 100% inhibition of NSP2 expression with any of the g8-specific siRNAs may largely reflect the limitation of transfecting siRNAs into the cells.
FIG. 1.
Viral protein expression in siRNA-transfected cells. MA104 cells were transfected with the indicated siRNA, infected with SA11-5N (A and C) or DxRRV (B), and maintained in 35S-labeled amino acids. Lysates prepared from the cells at 18 h p.i. were analyzed for viral proteins by gel electrophoresis and autoradiography (lower panels) and by Western blot assay (upper panels). The specificity of the antibodies used in Western blot assays is given. Lysates prepared from cells neither transfected with siRNA nor infected with virus (uninfected) or infected with virus but not transfected with siRNA (no siRNA), were also analyzed.
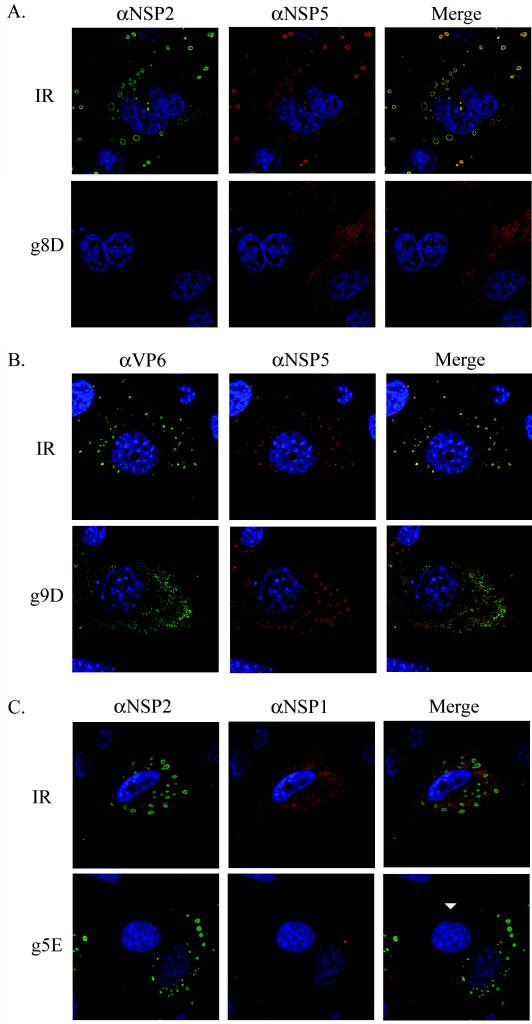
Radiolabeling of MA104 cells transfected with g8-specific siRNAs and infected with rotavirus showed that in cases where NSP2 expression was inhibited, the expression of the other viral proteins was inhibited as well (Fig. 1A). The fact that the IR siRNA failed to suppress viral protein synthesis indicates that the inhibition caused by the g8A, g8C, and g8D siRNAs did not stem from a nonspecific induction of antiviral responses (e.g., interferon and PKR) due to transfection of short duplex RNAs into MA104 cells. IF staining of the infected cells also showed that the IR siRNA did not prevent NSP2 and NSP5 from mediating the formation of viroplasms (Fig. 2A). In contrast, inhibition of NSP2 expression by the g8D siRNA led to a nearly complete inhibition of viroplasm formation. Given the essential contribution of NSP2 to the formation of viroplasms, the absence of these structures in infected cells containing the g8D siRNA is expected.
FIG. 2.
IF analysis of the effect of siRNAs on intracellular accumulation of viral proteins. MA104 cells were transfected with the appropriate siRNA and infected with SA11-5N (A and C) or DxRRV (B). At 12 h p.i., the cells were processed for IF by using the indicated primary antibodies (α) and Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies. Cell nuclei were visualized by DAPI staining. For comparison purposes, panel C includes a cell that was not infected (arrowhead).
(ii) RNA synthesis.
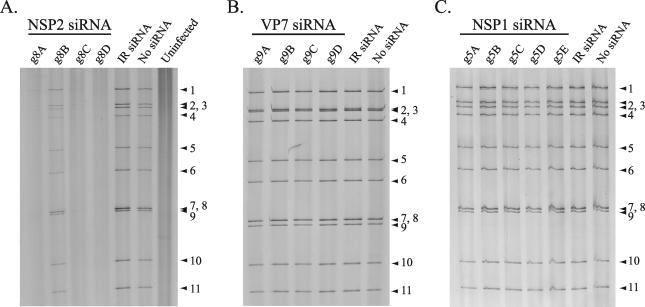
Polyacrylamide gel electrophoresis (PAGE) analysis showed that viral dsRNA synthesis in rotavirus-infected cells was inhibited by the g8A, g8C, and g8D siRNAs (Fig. 3A), the same siRNAs that were efficient in inhibiting protein synthesis (Fig. 1A). The g8B siRNA caused a limited reduction of dsRNA synthesis, a finding consistent with its failure to effectively inhibit protein synthesis (Fig. 1A). Electrophoretic analysis of total RNA synthesized in the infected cells showed that inhibition of protein synthesis by the g8D siRNA was correlated with the lack of accumulation of the g8 plus-strand RNA, as well as all other viral plus-strand RNAs (Fig. 4A). Because the siRNA targeted only the g8 plus-strand RNA for degradation, the decreased accumulation of the other viral mRNAs suggests a need for the g8 plus-strand RNA, or its gene product NSP2, to provide conditions adequate for amplification of viral transcription in the infected cell.
FIG. 3.
Level of viral dsRNAs produced in siRNA-transfected cells. MA104 cells were transfected with the indicated siRNA and infected with SA11-5N (A and C) or DxRRV (B). Cells were harvested at 18 h p.i. and analyzed for the presence of viral dsRNAs by PAGE and silver staining. The positions of the 11 genome segments are labeled.
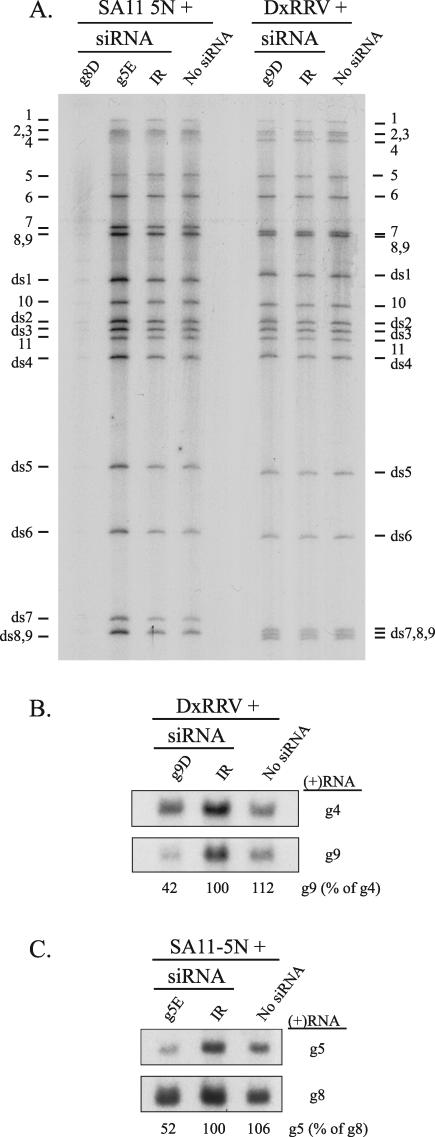
FIG. 4.
Level of viral plus-strand RNAs produced in siRNA-transfected cells. MA104 cells were transfected with the indicated siRNA and infected with SA11-5N or DxRRV. (A) After cells were maintained in [32P]orthophosphate from 3 to 9 h p.i., RNAs were recovered and analyzed by electrophoresis on a urea-polyacrylamide gel and by autoradiography. The positions of viral plus-strand RNAs (1 to 11) and dsRNAs (ds1 to ds9) are shown. (B and C) Unlabeled RNAs were recovered from cells at 6 h p.i., resolved by electrophoresis, and transferred to nylon membranes. Blots were probed with 32P-labeled RNAs specific for SA11 g5 (SA11-g5PR) or g8 (SA11-g8PR) or for DxRRV g4 (RRV-g4PR) or g9 (D-g9PR), and the intensity of the bands was determined with a phosphorimager. The levels of the g9 and g5 plus- strand RNAs were calculated by dividing their band intensities by the corresponding intensities determined for g4 or g8 plus-strand RNAs, respectively. The values were then normalized, with the value for the IR samples taken as 100%.
(iii) Virus assembly.
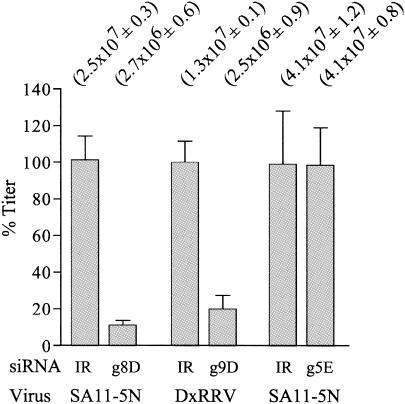
Plaque titrations revealed that the action of the g8D siRNA led to an ∼10-fold decrease in the assembly of infectious virions in SA11-5N-infected cells (Fig. 5). Limitations in transfection efficiency (∼80 to 90%) probably prevent the g8-specific siRNA from causing an even greater decrease in plaque titer.
FIG. 5.
Effect of siRNAs on virus titers. Lysates from MA104 cells infected with rotavirus and transfected with the indicated siRNA were analyzed for virus titer by plaque assay. Titers obtained from three separate assays performed in duplicate were averaged (values in parentheses [with standard errors]). Values were normalized to 100% for lysates from cells transfected with IR siRNAs.
In summary, siRNAs directed against the g8 plus-strand RNA had a negative impact on all phases of the virus replication cycle, including synthesis of all viral proteins, plus-strand RNAs, and dsRNAs, the formation of viroplasms, and the production of infectious virions. The mechanism for this effect may be linked to the role that NSP2 plays in assembling the viroplasms needed for genome replication and morphogenesis, to the role that g8 plus-strand RNAs have as templates for dsRNA synthesis, or to a combination of these.
Effect of g9-specific siRNAs on virus replication. (i) Protein synthesis.
Western blot analysis of cells infected with the reassortant virus DxRRV and transfected with g9-specific siRNAs identified three (g9B, g9C, and g9D) that were highly effective in inhibiting VP7 expression (Fig. 1B). Although the g9B, g9C, and g9D siRNAs suppressed VP7 expression, the analysis also showed that the siRNAs did not cause a reduction in VP6 expression. Similarly, 35S radiolabeling of the transfected cells indicated that the g9-specific siRNAs did not inhibit the expression of viral proteins other than VP7 (Fig. 1B). Hence, unlike the nonspecific reduction of viral protein expression that was observed with the use of g8-specific siRNAs, the effect of the g9-specific siRNAs was apparently limited to the expression of only a single protein. This indicates that efficient expression of the other gene products was not dependent on VP7, the opposite of what was observed for NSP2.
The glycoprotein VP7 accumulates at the rough endoplasmic reticulum (RER) of the infected cell and assembles around DLPs to form TLPs, as the particles bud into the RER (3). This event is believed to be facilitated by NSP4, a viral RER-resident protein, which because of its affinity for VP6 has been suggested to recruit newly formed DLPs from viroplasms to the RER (2, 21). As noted above, IF detection of NSP5 in infected cells showed that the IR siRNA did not interfere with viroplasm formation (Fig. 2B). IF analysis of the IR siRNA-transfected cells also showed that VP6 accumulated in viroplasms, an observation consistent with these structures serving as sites of DLP assembly. Likewise, IF detection of NSP5 in infected cells containing the g9D siRNA showed that suppression of VP7 synthesis did not impede viroplasm formation. However, this siRNA did induce a marked change in the distribution of the VP6 signal detected in the cell, with a large proportion no longer displaying an association with viroplasms. The nature of the VP6 signal localizing outside the viroplasms is not known but most likely represents accumulating DLPs, although the possibility that the signal represents unassembled protein cannot be excluded.
(ii) RNA synthesis.
PAGE analysis showed that the g9-specific siRNAs did not interfere with the accumulation of dsRNAs in infected cells (Fig. 3B). Thus, although the g9B, g9C, and g9D siRNAs were effective in preventing the use of g9 plus-strand RNAs as templates for VP7 expression, these siRNAs did not inhibit the use of g9 plus-strand RNAs as templates for dsRNA synthesis. An electrophoretic analysis of total RNA recovered from infected cells also indicated that the g9D siRNA did not affect the accumulation of the single-stranded forms of viral plus-strand RNAs (Fig. 4A). However, in this analysis, any impact that the g9D siRNA may have had on the level of g9 plus-strand RNAs in the infected cell could have been masked by the near comigration of the g7, g8, and g9 plus-strand RNAs on the gel.
To provide more precise insight into the effect of the g9D siRNA on the intracellular accumulation of the single-stranded form of the g9 plus-strand RNA, total RNA from cells that were mock transfected or transfected with g9D or IR siRNAs were resolved by electrophoresis and transferred to a nylon membrane. Probing of the membrane with radiolabeled RNAs showed that the g9D siRNA caused an ∼60% reduction in the level of g9 plus-strand RNA, relative to a constant amount of g4 plus-strand RNA (Fig. 4B). Together, these results indicated that the mechanism by which the g9-specific siRNAs acted to suppress VP7 expression was via the targeted degradation of g9 plus-strand RNAs. Remarkably, although this targeted degradation suppressed VP7 expression, it had no effect on g9 dsRNA synthesis. Thus, the pool of g9 plus-strand RNA which serves as a source of templates for translation appears to differ in accessibility to RISC complexes from the pool of g9 plus-strand RNA which serves as a source of templates for genome replication.
(iii) Virus assembly.
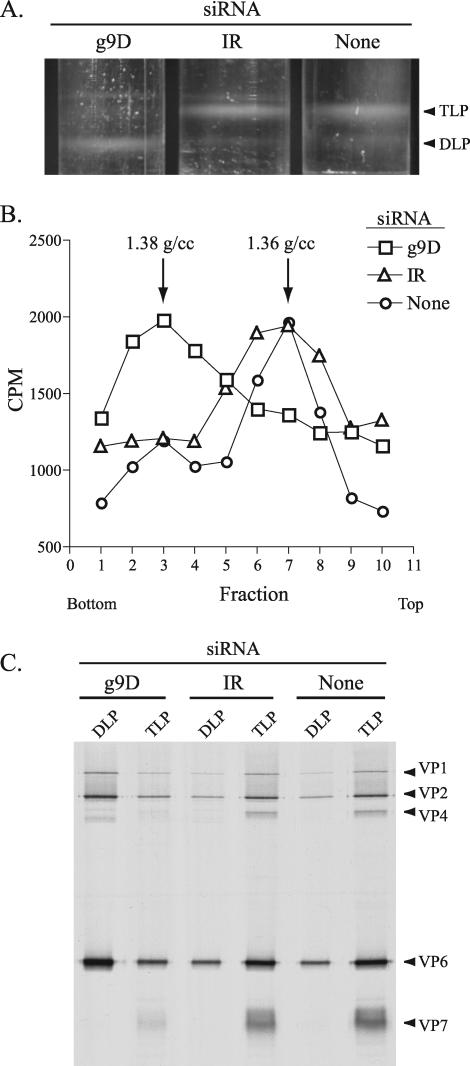
Plaque assays showed that suppression of VP7 synthesis by the g9D siRNA and the resultant inability to form the outer protein shell of the virion led to a six- to sevenfold decrease in infectious titer (Fig. 5). Given that VP7 was the only protein whose expression was suppressed in infected cells containing the g9D siRNA, such cells may be expected to support the assembly of DLPs but not their maturation into TLPs. To evaluate this possibility, lysates were prepared from DxRRV-infected cells that were either mock transfected or transfected with g9D or IR siRNAs and then maintained in 35S-labeled amino acids. Analysis of the lysates by CsCl centrifugation showed that DLPs (ρ = 1.38 g/ml) were the predominant particle type formed in cells containing the g9D siRNA (Fig. 6A). In contrast, TLPs (ρ = 1.36 g/ml) were the predominant particle type formed in cells containing either no siRNA or the IR siRNA. Moreover, the level of DLPs present in cells containing the g9D siRNA was much greater than that of cells containing either the IR siRNA or no siRNA. These results are not only reflected by the bands of virus particles detected in the CsCl gradients but also by analysis of fractions from the gradients for radioactivity and protein content (Fig. 6B and C). Hence, inhibition of VP7 expression by the g9D siRNA led to an accumulation of DLPs.
FIG. 6.
Impact of g9-specific siRNA on virion assembly. MA104 cells were mock transfected or transfected with the g9D or IR siRNA, infected with DxRRV, and then maintained in 35S-labeled amino acids. DLPs and TLPs produced in the cells were resolved by CsCl centrifugation. (A) Bands of virus particles in the gradients were visualized with an inverted light source. (B) Fractions (three drops each) collected from the gradients in regions containing the bands were analyzed for density and for radioactivity in 10-μl aliquots. (C) Fractions containing DLPs (two to four in panel B) and TLPs (six to eight) were pooled and examined for protein content by PAGE and autoradiography.
The RdRp component of DLPs can function as a transcriptase to drive the synthesis of viral plus-strand RNAs. Interestingly, although the levels of DLPs in cells containing the g9D siRNA was several times greater than that observed for cells containing either the IR siRNA or no siRNA, 32P labeling of total RNAs failed to show that greater levels of viral plus-strand RNA were present in cells containing the g9D siRNA (Fig. 4). The majority of DLPs in the siRNA-transfected cell appear to be quiescent with respect to plus-strand RNA synthesis, which suggests that no direct connection exists between levels of DLPs in the cell and levels of transcription.
Effect of g5-specific siRNAs on virus-replication. (i) Protein synthesis.
Studies of rotavirus isolates with sequence rearrangements and nonsense mutations in the g5 genome segment have indicated that NSP1 plays a nonessential role in the replication of the virus in the MA104 cell line (13, 27). The limited available information on NSP1 suggests that the protein functions to inhibit antiviral responses by suppressing the activity of the interferon regulatory factor, IRF3 (11). Western blot analysis of cells infected with SA11-5N and transfected with g5-specific siRNAs identified two (g5A and g5E) that were highly effective in inhibiting NSP1 expression (Fig. 1C). Radiolabeling showed that, although the g5A and E siRNAs significantly reduced NSP1 expression, the siRNAs did not inhibit the synthesis of other viral proteins. IF staining confirmed that the g5E siRNA inhibited NSP1 expression but that this effect had no impact on the formation of viroplasms (Fig. 2C).
(ii) RNA synthesis.
PAGE analysis demonstrated that the accumulation of viral dsRNAs in infected cells was not affected by any of the g5-specific siRNAs (Fig. 3C). Analysis of total RNA from infected cells also indicated that the g5E siRNA did not interfere with the accumulation of any of the single-stranded forms of the viral plus-strand RNAs, with the exception of that of the g5 plus-strand RNA (Fig. 4A). Specifically, quantitation of band intensities with a phosphorimager suggested that in cells transfected with the g5E siRNA, the amount of g5 plus-strand RNA per constant amount of g6 plus-strand RNA was ∼50% of that of cells lacking this siRNA. Northern blot analysis confirmed this observation, showing that the g5E siRNA caused a 50% reduction in the intracellular levels of g5 plus-strand RNA per constant amount of g8 plus-strand RNA (Fig. 4C). Thus, although the g5E siRNA caused the targeted degradation of a significant portion the g5 plus-strand RNA, the loss of the RNA affected NSP1 expression and not genome replication.
(iii) Virus assembly.
Plaque assays performed on infected cell lysates revealed that inhibition of NSP1 synthesis by the g5E siRNA did not affect the assembly of infectious particles (Fig. 5). Although our results with the g5-specific siRNAs do not address whether NSP1 has an essential role in the life cycle of the virus in its natural host, the data clearly support the hypothesis that NSP1 is not essential for replication of the virus in MA104 cells.
Localization of transcription to viroplasms.
The results presented above indicated that the g5E and g9D siRNAs inhibited the expression of NSP1 and VP7, respectively, without affecting genome replication. This indicates that the plus-strand RNAs used as templates for dsRNA synthesis were not accessible to RISC and were therefore located at sites distinct from the plus-strand RNAs directing protein synthesis. Since viroplasms are the probable sites of dsRNA synthesis, the inability of the g5- and g9-specific siRNAs to inhibit genome replication implies that the plus-strand RNAs within these inclusions are inaccessible to RISC. If the ultimate source of the g5 and g9 plus-strand RNAs that function as templates for dsRNA synthesis is outside the viroplasm and it is the same source of plus-strand RNAs that directs protein synthesis, then the g5E and g9D siRNAs should have inhibited genome replication in the cell. Because this was not the case, it can be postulated that the plus-strand RNAs that serve as templates for genome replication originate not from outside viroplasms but from within these structures.
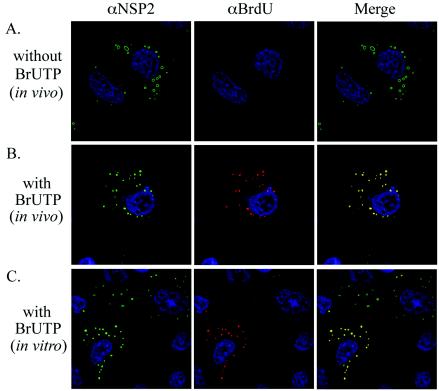
To identify intracellular sites of RNA synthesis, rotavirus-infected cells were treated with actinomycin D to block host replication and transcription and then transfected with BrUTP for 30 min beginning at 8.5 h p.i. After fixation and permeabilization of the cells, the location of newly made RNA was determined by using anti-BrdU antibody and the location of viroplasms was determined by using anti-NSP2 antibody. The analysis showed that BrU-labeled RNA accumulated in viroplasms during the 30-min incubation period with BrUTP, suggesting that these inclusions represented sites of viral RNA synthesis (Fig. 7B). In comparison, weak or no anti-BrdU IF signal (red) was detectable in cells that had been mock transfected and thus lacked BrUTP (Fig. 7A). The absence of signal in the mock-transfected cells confirmed that the anti-BrdU antibody specifically recognized BrU-labeled RNA and did not cross-react with viroplasmic proteins or RNA lacking the nucleotide analog. It is unlikely that the BrU label in the newly synthesized dsRNA in infected cells would be accessible to anti-BrdU antibody because of the coordinated nature of genome packaging and replication. Specifically, the absence of naked dsRNA in infected cells and the observation that replicase activity is associated with core-like replication intermediates that are partly covered with VP6 (10, 25) provides strong evidence for the movement of plus-strand RNAs into cores as minus-strand synthesis occurs. Hence, the anti-BrdU IF signal can be predicted to reflect only the location of newly made plus-strand RNA in the infected cell. The detection of the anti-BrdU signal in viroplasms suggests that these structures represent sites of plus-strand synthesis.
FIG. 7.
Intracellular sites of RNA synthesis in rotavirus-infected cells. MA104 cells were infected with SA11-5N, treated with actinomycin D, and maintained in the absence of BrUTP (A) or in the presence of BrUTP from 8.5 to 9 h p.i. (B). Alternatively, the infected cells were maintained in actinomycin D from 8.5 to 9 h p.i. and then semifixed with paraformaldehyde, permeabilized with Triton X-100, and incubated in transcription buffer containing BrUTP (C). Subsequently, the cells were processed for IF analysis by using NSP2- and BrdU-specific antibodies.
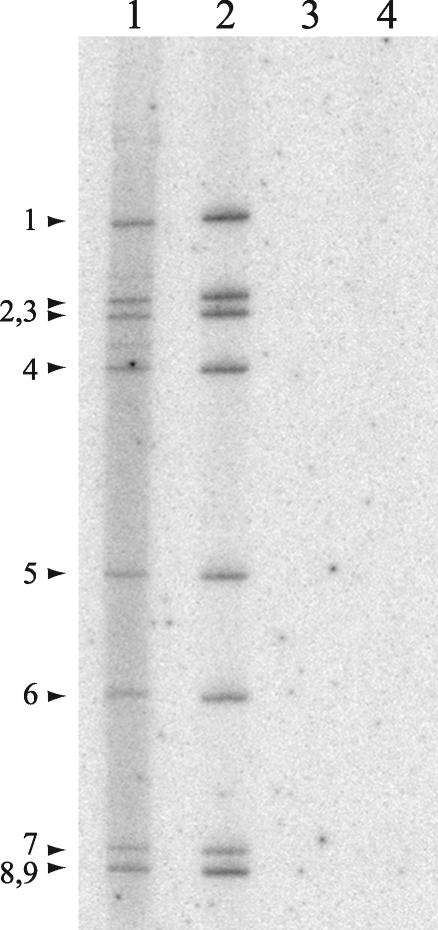
To further support the prediction that dsRNA produced during the BrUTP-labeling period of 8.5 to 9 h p.i. was packaged and thus inaccessible to the anti-BrdU antibody, rotavirus-infected MA104 cells were labeled during the same time period with [32P]orthophosphate. Cytoplasmic extracts recovered from these cells were incubated in the absence or presence of RNase V1, an RNase that specifically degrades duplex RNA (Fig. 8). Analysis of the reaction mixtures by PAGE showed that the newly made 32P-labeled dsRNA was resistant to digestion by the RNase unless deproteinized beforehand by phenol-chloroform extraction. These data are consistent with the concept that the newly made dsRNA was packaged into core-like replication intermediates. Given that RNase V1, a 15.9-kDa protein, did not have access to the newly made dsRNA, the possibility that the much larger anti-BrdU immunoglobulin would have access to the packaged dsRNA is remote.
FIG. 8.
Resistance of newly made viral dsRNA to degradation by dsRNA-specific RNase. MA104 cells were infected with SA11-5N or mock infected, treated with actinomycin D, and maintained in the presence of [32P]orthophosphate from 8.5 to 9 h p.i. Cytoplasmic extracts prepared from infected cells were incubated in the absence (lane 1) or presence (lane 2) of RNase V1 or deproteinized by phenol-chloroform extraction prior to incubation with RNase V1 (lane 3). An extract from mock-infected cells was incubated in the absence of RNase V1 (lane 4).
Although the in vivo BrU-labeling results were consistent with the idea that plus-strand RNAs are made in viroplasms, they did not exclude the possibility that the plus-strand RNAs were initially made elsewhere and then rapidly transported to the viroplasms. To further address the question of whether plus-strand RNAs were made in viroplasms, rotavirus-infected cells were treated with actinomycin D and then, at 9 h p.i., semifixed with paraformaldehyde and permeabilized with Triton X-100. The cells were incubated in transcription buffer containing BrUTP to allow for a brief period of viral RNA synthesis. Subsequently, the cells were completely fixed, and the locations of BrU-labeled RNA and viroplasms were determined by IF assay (Fig. 7C). The analysis showed that BrU-labeled RNA was produced in vitro within viroplasms contained in the semifixed cells. Thus, viroplasms are likely to be an important site of transcription in the infected cells, and plus-strand RNAs made in viroplasms are probably the source of the template RNAs used in genome replication.
Cytoplasmic trafficking of plus-strand RNAs.
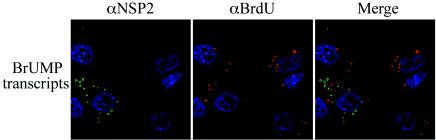
If the source of templates for dsRNA synthesis is plus-strand RNAs synthesized in viroplasms, then the need for pathways to transport plus-strand RNAs to these inclusions seems questionable. To assess whether pathways exist that transport plus-strand RNAs to viroplasms, capped, BrU-labeled g9 plus-strand RNA was made by T7 transcription. The RNA was transfected into rotavirus-infected cells at 1 h p.i., and, at 9 h p.i., the locations of the RNA and viroplasms were determined by IF assay with anti-BrdU and anti-NSP2 antisera. The analysis showed that the BrU-labeled RNA collected into punctate centers within the cytoplasm (Fig. 9) but that these centers of accumulation did not correspond to viroplasms. These data suggest that pathways capable of trafficking viral plus-strand RNAs to viroplasms do not exist in the cytosol of infected cells, a finding consistent with the concept that templates for dsRNA synthesis arise from within and not outside these inclusions. Similar results were obtained regardless of whether the infected cells were transfected at 5 h p.i. instead of at 1 h p.i. or whether the infected cells were transfected with nonviral RNA instead of rotavirus specific RNA (data not shown). The distribution pattern of BrU-labeled RNA transfected into mock-infected cells was the same as that observed upon transfection of infected cells (data not shown).
FIG. 9.
Fate of viral RNA transfected into rotavirus-infected cells. MA104 cells were infected with SA11-5N and, at 1 h p.i., transfected with BrU-labeled g9 plus-strand RNA. Cells were processed at 9 h p.i. for IF analysis with NSP2- and BrdU-specific antibodies.
DISCUSSION
We have identified siRNAs specific for g5, g8, and g9 plus-strand RNAs that efficiently suppressed the production of NSP1, NSP2, and VP7, respectively, in rotavirus-infected cells. The inhibition of NSP1 and VP7 expression was correlated with gene-specific reductions in the levels of the g5 and g9 plus-strand RNAs. Interestingly, the degrees to which the expression of NSP1 and VP7 were reduced were much greater than the degrees to which the levels of the g5 and g9 plus-strand RNAs were reduced. We can conclude from this that the subset of plus-strand RNAs involved in protein expression is more susceptible to siRNA-induced degradation than the population of plus-strand RNAs as a whole and that the nonsusceptible subset of plus-strand RNAs may not represent a source of transcripts for translation, at least at later times of infection (∼9 h p.i.).
Despite their ability to suppress the expression of NSP1 and VP7 and cause reductions in the levels of their template plus-strand RNAs, the g5- and g9-specific siRNAs did not alter the levels of dsRNA synthesis in infected cells. Thus, it is the subset of plus-strand RNAs which directs protein synthesis and not dsRNA synthesis that is susceptible to degradation. Various observations have indicated that viroplasms represent sites in which the viral genome is replicated and packaged within cores and that translation occurs in the cytosol, outside these structures. These observations include recognition that the RNA polymerase VP1 and core lattice protein VP2 accumulate in viroplasms, whereas the viral translation enhancement protein NSP3 and translation initiation factor eIF4G accumulate outside viroplasms (28; unpublished results). Thus, susceptibility of plus-strand RNAs to siRNA-induced degradation can be connected to their intracellular location, with RNAs in viroplasms resistant to degradation and those outside of viroplasms sensitive to degradation.
Although the effect of the g5- and g9-specific siRNAs was to cause an intermediate decrease in the levels of the g5 and g9 plus-strand RNAs, the g8-specific siRNA caused a decrease to near-background levels of all 11 plus-strand RNAs. Significant reductions were also seen in the production of viral dsRNAs, protein, and progeny virions. Hence, all processes in the viral life cycle were inhibited by the siRNA-induced degradation of just the g8 plus-strand RNA. In unpublished experiments, we have observed similar broad suppressive effects on virus replication by siRNAs targeted to plus-strand RNAs encoding VP1, VP2, VP6, and NSP5. Thus, the phenotype produced by the g8-specific siRNA is not unique but apparently common to all siRNAs that induce degradation of plus-strand RNAs encoding proteins necessary for RNA amplification. This includes both the structural proteins needed for assembly of DLPs and the nonstructural proteins needed for formation of viroplasms. In addition to playing an essential role in viroplasm formation, NSP2 also has affinity for single-stranded RNA (17), hydrolyzes NTPs (31), destabilizes RNA-RNA duplexes (32), and may interact with the viral RNA polymerase (16). Because of its multifunctional nature, we cannot exclude the possibility that suppression of virus replication by g8-specific siRNAs stems not only from a loss of viroplasm formation in the infected cell but also from a loss of one or more of the other activities associated with NSP2.
A significant uncertainty in our understanding of rotavirus biology has been the origin of the plus-strand RNAs that serve as templates for dsRNA synthesis. Hypotheses have been put forth for this and other members of the Reoviridae that plus-strand RNAs produced outside viroplasms are transported to these structures via mechanisms mediated by one or more of the viral RNA-binding proteins (4, 20). Suggestions have been made that competition between VP1 and NSP3 dictates whether rotavirus plus-strand RNAs are directed to viroplasms or polysomes, and thus these proteins would govern relative levels of replication and translation. However, given our findings indicating that plus-strand RNAs outside of viroplasms are uniquely sensitive to degradation by RISC, it seems unreasonable to predict that this pool of plus-strand RNAs would serve as the ultimate source of templates for dsRNA synthesis. Otherwise, we would anticipate that the degradation of plus-strand RNAs outside viroplasms would have the downstream effect of reducing the amount of RNA that could be translocated to viroplasms, thereby reducing the level of dsRNA synthesis in the infected cell.
The resistance of plus-strand RNAs templating dsRNA synthesis to siRNA-induced degradation led us to identify sites of RNA synthesis in the infected cell by using the UTP-analog, BrUTP, and a monoclonal antibody recognizing the analog. Because rotavirus dsRNAs do not exist in a naked form within the cell but are sequestered inside of capsid assembly intermediates as they are made (10, 25), the anti-BrdU antibody can interact with newly made naked BrU-labeled plus-strand RNAs but not with newly made packaged BrU-labeled dsRNAs. Hence, BrUTP and its antibody can identify intracellular sites of viral transcription (plus-strand synthesis) but not replication (minus-strand synthesis). Both in vivo pulse-labeling of intact infected cells and in vitro transcription assays performed with semifixed infected cells indicated that viroplasms are sites of plus-strand RNA synthesis. These results, combined with those from experiments with siRNAs, suggest that plus-strand RNAs templating dsRNA synthesis are provided by transcriptionally active DLPs associated with viroplasms. If so, then both plus-strand and minus-strand RNA synthesis occur at the same site in the infected cell, negating the need for viral proteins to transport to the viroplasm the many copies of plus-strand RNAs that would be required to form the genome segments of progeny virions.
From our findings, we propose the following model for events related to plus-strand RNA synthesis. After virion entry and loss of the outer capsid shell, DLPs initiate transcription within the cytoplasm, leading to the synthesis of plus-strand RNAs that are incorporated into polysomes and translated to produce viral proteins. At least some of the DLPs then serve as focal points for the accumulation of newly made proteins into viroplasms and are thereby incorporated into the inclusions. Transcripts produced by DLPs in the viroplasms are captured by one or more of the many viral RNA-binding proteins that accrue in these inclusions (e.g., NSP2, NSP5, VP1, and VP2). The captured plus-strand RNAs serve as sources of templates necessary for genome replication and assembly of progeny cores and DLPs. When the level of transcripts in the viroplasm exceeds the binding capacity of the RNA-binding proteins, the transcripts escape and can become incorporated into polysomes. Thus, viral gene expression is enhanced, and the additional proteins necessary for increasing the sizes and numbers of viroplasms are produced. In this scenario, viroplasm development becomes an autoregulatory process controlled by the capacity of the RNA-binding proteins to capture transcripts made by the DLPs within the inclusion.
The “capture” model requires no trafficking pathways that direct the movement of plus-strand RNAs to viroplasms. Given that BrU-labeled plus-strand RNAs transfected into infected cells failed to localize to viroplasms, the existence of such RNA-trafficking pathways seems doubtful. The lack of such pathways may explain why attempts to develop a reverse genetics system for rotavirus by transfection of recombinant plus-strand RNAs into infected cells have failed. Such experiments have been based on the idea that recombinant plus-strand RNAs, when introduced into the cell, would be translocated to viroplasms, where they would act as templates for dsRNA synthesis and become packaged into progeny virions. The absence of RNA trafficking pathways to viroplasms requires consideration of alternative approaches for engineering the rotavirus genome.
The present study has demonstrated the remarkable usefulness of siRNAs as tools to study the molecular biology of rotaviruses within the infected cell. Combined with an earlier study demonstrating the suppression of VP4 production in infected cells by using a g4-specific siRNA (5), this brings to four the number of rotavirus genes successfully targeted by using siRNAs. Targeting of the other rotavirus genes should lead to additional insight concerning the contribution of the viral proteins to genome replication and virion assembly.
REFERENCES
- 1.Altenburg, B. C., D. Y. Graham, and M. K. Estes. 1980. Ultrastructural study of rotavirus replication in cultured cells. J. Gen. Virol. 46:75-85. [DOI] [PubMed] [Google Scholar]
- 2.Au, K. S., W. K. Chan, J. W. Burns, and M. K. Estes. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chasey, D. 1977. Different particle types in tissue culture and intestinal epithelium infected with rotavirus. J. Gen. Virol. 37:443-451. [Google Scholar]
- 4.Chnaiderman, J., M. Barro, and E. Spencer. 2002. NSP5 phosphorylation regulates the fate of viral mRNA in rotavirus-infected cells. Arch. Virol. 147:1899-1911. [DOI] [PubMed] [Google Scholar]
- 5.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esparza, J., M. Gorziglia, F. Gil, and H. Romer. 1980. Multiplication of human rotavirus in cultured cells: an electron microscopic study. J. Gen. Virol. 47:461-472. [DOI] [PubMed] [Google Scholar]
- 8.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two nonstructural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80(Pt. 2):333-339. [DOI] [PubMed] [Google Scholar]
- 9.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 10.Gallegos, C. O., and J. T. Patton. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172:616-627. [DOI] [PubMed] [Google Scholar]
- 11.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmberger-Jones, M., and J. T. Patton. 1986. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology 155:655-665. [DOI] [PubMed] [Google Scholar]
- 13.Hua, J., and J. T. Patton. 1994. The carboxyl-half of the rotavirus nonstructural protein NS53 (NSP1) is not required for virus replication. Virology 198:567-576. [DOI] [PubMed] [Google Scholar]
- 14.Hutvagner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 15.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 16.Kattoura, M. D., X. Chen, and J. T. Patton. 1994. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology 202:803-813. [DOI] [PubMed] [Google Scholar]
- 17.Kattoura, M. D., L. L. Clapp, and J. T. Patton. 1992. The rotavirus nonstructural protein, NS35, possesses RNA-binding activity in vitro and in vivo. Virology 191:698-708. [DOI] [PubMed] [Google Scholar]
- 18.Lawton, J. A., M. K. Estes, and B. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118-121. [DOI] [PubMed] [Google Scholar]
- 19.Lawton, J. A., C. Q. Zeng, S. K. Mukherjee, J. Cohen, M. K. Estes, and B. V. Prasad. 1997. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J. Virol. 71:7353-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lymperopoulos, K., C. Wirblich, I. Brierley, and P. Roy. 2003. Sequence specificity in the interaction of Bluetongue virus nonstructural protein 2 (NS2) with viral RNA. J. Biol. Chem. 278:31722-31730. [DOI] [PubMed] [Google Scholar]
- 21.Meyer, J. C., C. C. Bergmann, and A. R. Bellamy. 1989. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology 171:98-107. [DOI] [PubMed] [Google Scholar]
- 22.Midthun, K., and A. Z. Kapikian. 1996. Rotavirus vaccines: an overview. Clin. Microbiol. Rev. 9:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 24.Patton, J. T. 1990. Evidence for equimolar synthesis of double-strand RNA and minus-strand RNA in rotavirus-infected cells. Virus Res. 17:199-208. [DOI] [PubMed] [Google Scholar]
- 25.Patton, J. T., and C. O. Gallegos. 1990. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J. Gen. Virol. 71(Pt. 5):1087-1094. [DOI] [PubMed] [Google Scholar]
- 26.Patton, J. T., M. T. Jones, A. N. Kalbach, Y. W. He, and J. Xiaobo. 1997. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 71:9618-9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton, J. T., Z. Taraporewala, D. Chen, V. Chizhikov, M. Jones, A. Elhelu, M. Collins, K. Kearney, M. Wagner, Y. Hoshino, and V. Gouvea. 2001. Effect of intragenic rearrangement and changes in the 3′ consensus sequence on NSP1 expression and rotavirus replication. J. Virol. 75:2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 29.Poncet, D., P. Lindenbaum, R. L'Haridon, and J. Cohen. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 71:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasad, B. V., G. J. Wang, J. P. Clerx, and W. Chiu. 1988. Three-dimensional structure of rotavirus. J. Mol. Biol. 199:269-275. [DOI] [PubMed] [Google Scholar]
- 31.Taraporewala, Z., D. Chen, and J. T. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taraporewala, Z. F., and J. T. Patton. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tortorici, M. A., T. J. Broering, M. L. Nibert, and J. T. Patton. 2003. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J. Biol. Chem. 278:32673-32682. [DOI] [PubMed] [Google Scholar]
- 34.Tuschl, T., P. D. Zamore, R. Lehmann, D. P. Bartel, and P. A. Sharp. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13:3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasquez-Del Carpio, R., F. D. Gonzalez-Nilo, H. Jayaram, E. Spencer, B. V. Venkataram Prasad, J. T. Patton, and Z. F. Taraporewala. 2004. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J. Biol. Chem. 279:10624-10633. [DOI] [PubMed] [Google Scholar]