Abstract
A PPPY motif within the M protein of vesicular stomatitis virus (VSV) functions as a late-budding domain (L-domain); however, L-domain activity has yet to be associated with a downstream PSAP motif. VSV recombinants with mutations in the PPPY and/or PSAP motif were recovered by reverse genetics and examined for growth kinetics, plaque size, and budding efficiency by electron microscopy. Results indicate that unlike the PPPY motif, the PSAP motif alone does not possess L-domain activity. Finally, the insertion of the human immunodeficiency virus type 1 p6 L-domain and flanking sequences into the PSAP region of M protein rescued budding of a PPPY mutant of VSV to wild-type levels.
Late-budding domains (L-domains) have been identified in the matrix or matrix-like proteins of several different RNA virus families (for a review, see reference 7). L-domains are thought to facilitate virus budding by interacting with and usurping specific host proteins or machinery during late stages of virus replication. The PPPY (PY) motif within the vesicular stomatitis virus (VSV) M protein has been shown to function as an L-domain (4, 11, 14). For example, mutations that disrupt the PY motif result in the defective budding of both the M protein alone and the infectious virus (4, 11, 14). The PY motif of VSV has been shown to mediate interactions in vitro with WW-containing cellular ubiquitin ligases, such as Nedd4 (9, 11). Such virus-host interactions have been postulated to occur in vivo to promote the efficient budding of virus from infected cells.
It has been noted recently that many of the RNA viruses that possess functional L-domains may also possess redundant or secondary L-domain motifs. For example, a bipartite L-domain has recently been identified in the p6 Gag protein of human immunodeficiency virus type 1 (HIV-1) (17). In addition, the VP40 protein of Ebola virus possesses two functional, overlapping L-domains (16). A potential secondary L-domain having the core consensus sequence of PSAP (PS) is also present in the M protein of VSV (Indiana serotype) just downstream of the PY motif. The PS motif is similar in sequence to functional L-domains identified in HIV-1 and Ebola virus (1, 6, 10, 12, 16-19, 22). Since mutations that disrupt the PY motif of VSV M never completely abolished budding (14), a possible role for PS as a secondary L-domain was hypothesized.
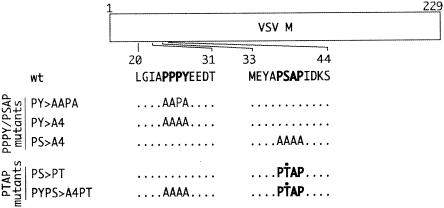
To determine whether the PS motif within M protein possesses L-domain activity and contributes to the budding of VSV, we introduced changes into the PS sequence within the full-length cDNA clone of VSV (Fig. 1). In addition to the PY>AAPA mutant described previously (14), we constructed a second mutant in which the PY motif was mutated to four alanines (PY>A4) (Fig. 1). A similar four-alanine substitution for the PSAP motif yielded the PS>A4 mutant (Fig. 1). Two additional constructs in which the PS motif was converted to a PTAP (PT) motif were generated (Fig. 1). The S-to-T substitution was engineered into both the wild-type (wt) VSV M protein background to yield the PS>PT mutant and the PY>A4 background to yield the PYPS>A4PT mutant (Fig. 1). The serine-to-threonine substitution was constructed, since functional L-domains within HIV-1 p6 Gag and Ebola virus VP40 have the core sequence PT rather than PS (7). All mutations introduced into the genomic cDNA were confirmed by automated DNA sequencing. All cDNA constructs (Fig. 1) were employed in the VSV reverse-genetics system (15, 24), and all constructs yielded infectious viruses. The resultant VSV recombinants were plaque purified two times, and the introduced mutations were confirmed by reverse transcription-PCR followed by automated DNA sequencing.
FIG. 1.
Diagram of VSV M protein gene highlighting both the PY (amino acids 20 to 31) and PS (amino acids 33 to 40) regions. The dotted line alone indicates that the wt amino acid sequence is retained. The amino acid changes for the various mutants are indicated by the single-letter code.
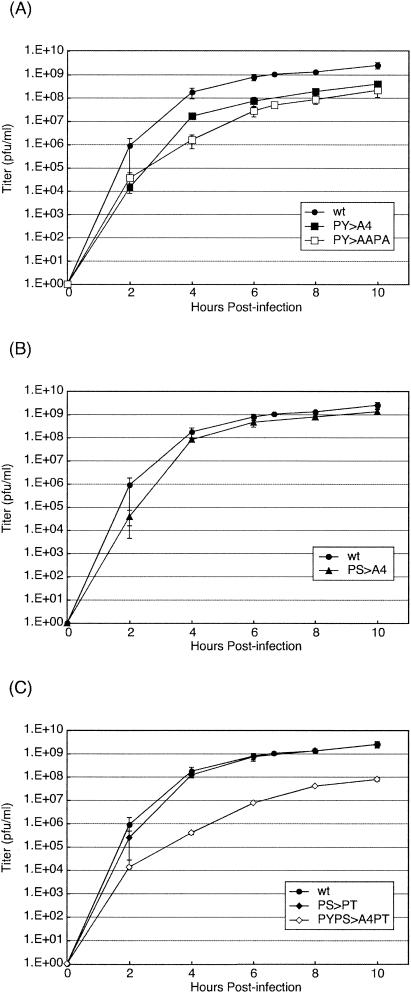
To determine the effect of PS mutations on virus replication, recombinants were assayed for growth kinetics in cell culture (Fig. 2). BHK-21 cells were infected with the indicated viruses at a multiplicity of infection (MOI) of 10. Culture medium was harvested at 2, 4, 6, 8, and 10 h postinfection (p.i.), and viral titers were determined by duplicate plaque assays with BHK-21 cells (Fig. 2). wt VSV served as a positive control, and the PY>AAPA recombinant served as an L-domain-defective control (Fig. 2A), as reported previously (14). As expected, titers of the PY>AAPA mutant were reduced by 1 to 2 logs compared to those obtained for wt VSV (Fig. 2A). Similarly, titers of the PY>A4 recombinant were virtually identical to those of the PY>AAPA recombinant, confirming that the substitution of alanines for the PY L-domain resulted in reduced virus yield (Fig. 2A). Unlike with the PY mutants, titers of the PS>A4 recombinant were on average only twofold lower than those of wt VSV (Fig. 2B). These data suggest that the disruption of the PS motif does not impair the budding of VSV as severely as does disruption of the PY motif.
FIG. 2.
Growth kinetics of wt and recombinant viruses. BHK-21 cells were infected at an MOI of 10, and samples were harvested for titration at the indicated times p.i. (A to C) Graphs of virus titers relative to time p.i. for the wt, PY>A4, and PY>AAPA viruses (A); the wt and PS>A4 viruses (B); and the wt, PS>PT, and PYPS>A4PT viruses (C). Viral titers represent averages of titers from at least two independent experiments.
We next sought to determine whether a serine-to-threonine (PS>PT) change would alter the growth kinetics of VSV (Fig. 2C). Viral titers of the PS>PT recombinant were virtually indistinguishable from those of wt VSV and were slightly higher (twofold) than those of the PS>A4 recombinant (compare Fig. 2B and C). Although the PS>PT recombinant virus replicated to titers virtually identical to those of wt VSV, the presence of PT was not sufficient to rescue budding of a PY mutant virus to wt levels (Fig. 2C). Indeed, titers of the PYPS>A4PT recombinant remained an average of 1 to 2 logs lower than those of wt VSV (Fig. 2C).
We next examined the profiles of virion proteins encoded by wt virus and the various VSV recombinants. Virions were harvested from infected BHK-21 cells and lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Virion proteins were fractionated by SDS-PAGE and visualized by staining with Coomassie brilliant blue (Fig. 3). The quantity of virion proteins correlated well with titers obtained in the growth curve assays. For example, the level of M protein present in wt VSV virions was >10-fold higher than in the PY>AAPA and PY>A4 mutants (Fig. 3A, lanes 1 to 3). In addition, the level of M protein present in PS>A4 virions was approximately twofold lower than that in wt VSV (Fig. 3A, compare lanes 1 and 4), whereas the amount of M protein present in the PS>PT mutant was virtually identical to that in wt virions (Fig. 3A, compare lanes 1 and 5). Finally, the amount of virion proteins present in the PYPS>A4PT mutant was identical to that in both the PY>AAPA and PY>A4 mutants (Fig. 3A, lane 6). The amounts of M protein present in cells infected with wt VSV and all recombinants were equivalent, indicating that mutations in the PY and PS motifs did not disrupt viral protein synthesis.
FIG. 3.
Protein profiles for wt and recombinant viruses. (A) Virions were harvested and lysed from supernatants of infected BHK-21 cells at 8 h p.i. Virion proteins were analyzed by SDS-PAGE and Coomassie blue staining. The protein profiles for wt VSV (lane 1), the PY>A4 mutant (lane 2), the PY>AAPA mutant (lane 3), the PS>A4 mutant (lane 4), the PS>PT mutant (lane 5), and the PYPS>A4PT mutant (lane 6) are shown. L protein is not shown on this gel. (B) Radiolabeled lysates from infected cells were immunoprecipitated with anti-M monoclonal antibody, and immunoprecipitates were analyzed by SDS-PAGE.
Electron microscopy was used previously to demonstrate conclusively that PY mutants of VSV were defective in the ability to separate from the plasma membrane of infected cells (14). The ability of the PS>A4 mutant to separate from infected cells was assessed by electron microscopy (Fig. 4A). Briefly, BHK-21 cells were infected with either wt VSV or the PS>A4 mutant at an MOI of 3.0. At 6 h p.i., cells were washed extensively and resuspended in 1.0% low-melting-point agarose in 1× phosphate-buffered saline. After chilling on ice, the cells in small agarose blocks were fixed overnight in 2.5% glutaraldehyde-2.0% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), washed in cacodylate buffer, and incubated in 2.0% OsO4 for 2 h. The samples were then stained for 1 h with 1.0% uranyl acetate in 150 mM maleate buffer (pH 6.0), washed, dehydrated with ethanol, and finally embedded in resin (LX-112; Ladd Research Industries). Ultrathin sections (80 nm thick) were stained with uranyl acetate and lead citrate according to standard protocols, and samples were examined with a Philips CM-100 transmission electron microscope. As expected, wt VSV was found to separate efficiently from cellular membranes with the typical bullet morphology (Fig. 4A). Similarly, the PS>A4 mutant was also observed to separate efficiently as a bullet-shaped virion. A representative micrograph for both wt VSV and the PS>A4 mutant is shown in Fig. 4A. Examination of numerous micrographs revealed no significant quantitative or qualitative evidence to suggest that the PS>A4 mutant was defective in budding.
FIG. 4.
Electron microscopy and plaque size determination of wt and recombinant viruses. (A) Electron micrographs of wt VSV and the PS>A4 mutant virus showing bullet-shaped virions budding from infected BHK-21 cells. Bar, 200 nm. (B) Bar graph showing average measurements of plaque size for wt VSV and the PY>AAPA mutant, the PY>A4 mutant, the PS>A4 mutant, the PS>PT mutant, and the PYPS>A4PT mutant viruses. The data represent the average plaque size determined from at least two independent experiments.
The PY mutants of VSV were previously reported to exhibit a small-plaque phenotype which correlated with their budding defect (14). To determine whether PS mutants displayed a similar small-plaque phenotype, the average plaque size of each recombinant virus was measured (Fig. 4B). Briefly, BHK-21 cells in 100-mm-diameter dishes were infected to yield approximately 50 plaques per dish. At 24 h p.i., cells were stained with crystal violet, and the areas of 10 plaques on each plate were measured with NIH Image 1.52 software. As expected, the average plaque size of the PY mutants was approximately 50% smaller than that observed for wt VSV (Fig. 4B). In contrast, the PS>A4 and PS>PT mutants yielded plaques that were virtually identical in size to those of wt VSV (Fig. 4B). Finally, the small-plaque phenotype remained associated with the PYPS>A4PT mutant. Thus, the small-plaque phenotype correlated precisely with those recombinants that exhibited a defect in budding due to mutations in the PY motif.
Taken together, these data strongly suggest that the PS motif within VSV M does not possess L-domain activity similar to that of the PY motif. Attempts to recover a double mutant having four alanine substitutions in both the PY and PS motifs are currently under way. Mutations that disrupt both the PY and PS regions of M may be too disruptive to normal M protein structure and/or function. In an attempt to address this possibility, we engineered and rescued a recombinant virus (M6) containing the known functional L-domain and flanking sequences of the p6 Gag protein of HIV-1 in place of the PS motif of VSV M protein (Fig. 5A). Moreover, the p6 L-domain and flanking sequences were inserted into the PY>A4 background (Fig. 5A). Interestingly, the presence of the HIV-1 p6 L-domain was sufficient to rescue the budding defect of the PY>A4 mutant and thus allow the M6 recombinant virus to achieve titers identical to those of wt VSV (Fig. 5B). These data indicate that alterations of the PS core sequence and surrounding amino acids are not lethal and that the insertion of a heterologous, functional L-domain at this site can effectively restore the L-domain activity absent in the PY>A4 mutant. Furthermore, these data indicate that amino acids flanking the PT core motif must contribute to the restoration of budding to the M6 recombinant virus. Thus, the recovery of the M6 recombinant lends further support to our conclusion that the PS region of wt VSV M protein does not possess significant L-domain activity.
FIG. 5.
Diagram and growth kinetics of recombinant M6 virus. (A) Diagram of the M protein of VSV illustrating amino acids present between positions 20 and 31 and 33 and 44 in wt VSV and M6 recombinant virus. All sequences were confirmed by automated DNA sequencing. (B) Growth kinetics of wt VSV and M6 recombinant virus. The graph shows virus titers relative to time p.i. for wt and M6 viruses. Viral titers represent averages of results from more than two independent experiments.
The possibility that the PS region is important for either the structure or functions of M protein other than budding remains. It is also possible that the PS region may play a role in virus replication in cells derived from animal hosts (e.g., equine or bovine cells) or during VSV replication in insect vectors (e.g., mosquito or sandfly) at low temperatures. The replication of wt and PS mutant viruses in bovine, equine, mosquito, and sandfly cell lines maintained at various temperatures is currently being examined (T. Irie and R. N. Harty, unpublished data).
The PT- and PS-type L-domains identified in the p6 Gag and VP40 proteins of HIV-1 and Ebola virus, respectively, have been shown to interact with host protein TSG101 (2, 3, 8, 16, 18, 20, 21, 23). This PT/PS-TSG101 interaction appears to be important for the efficient budding of both HIV-1 and Ebola virus. Indeed, the overexpression of the N-terminal region of TSG101 or the depletion of TSG101 by small interfering RNA resulted in a decrease in the budding efficiency of p6 Gag and VP40 (5, 16, 18, 21). In contrast, the expression of small interfering RNA specific for TSG101 did not significantly reduce the budding of wt VSV M protein but did reduce the budding of a chimeric M protein containing the PT-type L-domain of VP40 (12a). The fact that the PYPS>A4PT mutant remained defective in budding is consistent with our notion that VSV buds in a TSG101-independent manner. It will now be of interest to determine whether the M6 recombinant virus, for example, has been redirected into the HIV-1 budding pathway and as a result become sensitive to the depletion of host TSG101.
The precise mechanisms by which viral L-domains interact with host proteins to promote budding during late stages of replication remain unclear. The VSV model system and VSV recombinants represent ideal tools to help elucidate these mechanisms and to understand the biological relevance of these virus-host interactions. Further understanding of virus budding will hopefully lead to the development of novel therapeutics to block this late stage of replication.
Acknowledgments
We thank Shiho Irie for technical assistance, Jason Paragas for critical reading of the manuscript, and all members of the Harty lab for fruitful discussions.
This work was supported in part by NIH grant AI46499 to R.N.H. J.M.L. is supported by NIH training grant AI-07324.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 3.Carter, C. A. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203-205. [DOI] [PubMed] [Google Scholar]
- 4.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 9.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Irie, T., J. M. Licata, J. P. McGettigan, M. J. Schnell, and R. N. Harty. 2004. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TSG101 and VPS4A. J. Virol. 78:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. (Erratum, 92:9009.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Serrano, J., and P. D. Bieniasz. 2003. A bipartite late-budding domain in human immunodeficiency virus type 1. J. Virol. 77:12373-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 19.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile: assimilation of cellular machinery by HIV-1. Immunity 15:687-690. [DOI] [PubMed] [Google Scholar]
- 21.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of ebola virus matrix protein VP40. Virology 283:1-6. [DOI] [PubMed] [Google Scholar]
- 23.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whelan, S. P., L. A. Ball, J. N. Barr, and G. T. Wertz. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. USA 92:8388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]