Abstract
Background
Gastric cancer exhibits familial clustering, and gastric cancer familial relatives (GCF) tend to present with corpus-predominant gastritis and precancerous lesions as SPEM or IM after H. pylori infection. The study determined whether the children of gastric cancer patients (GCA) had genomic single nucleotide polymorphisms (SNPs) predisposed to the gastric precancerous lesions as spasmolytic polypeptide-expressing metaplasia (SPEM) or intestinal metaplasia (IM).
Results
There were 389 family relatives of 193 non-cardiac GCA and 173 duodenal ulcer patients (DU), received blood sampling for DNA collection. The differences of the risk alleles of SNPs in the ITGA5, ITGB1, IL-10, COX-2, RUNX3, and TFF2 genes were compared between 195 children of GCA and 143 DU. The children of GCA had higher allele frequencies of ITGA5-1160 T-carrier (P = 0.006, OR[95% CI] = 2.2[1.2-4]), ITGB1-1949 A-carrier (P = 0.047; OR[95% CI] = 2.8[1.4-5.3]), ITGB1 + 31804 C-carrier (P = 0.013; OR[95% CI] = 4.7[1.7-13.0]), IL-10-592 AA (P = 0.014; OR[95% CI] = 2.3[1.4-4.0]) and COX-2-1195 G-carrier (P = 0.019; OR[95% CI] = 1.7[0.9-3.2]) than DU. The combined genotype with ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA was more prevalent in the children of GCA than in DU (P < 1×10−4), and predisposed with a 5.3-fold risk of getting SPEM in the H. pylori-infected children of GCA (P = 0.016). Such risk of getting SPEM increased to 112 folds, if combined with RUNX3 + 492/TFF2-308 as A-carrier/CC in this limited study scale (P = 1×10−4).
Conclusions
The SNPs of ITGA5-1160/ITGB1-1949/ ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA, or more specific to combine RUNX3 + 492/TFF2-308 as A-carrier/CC shall be host factor predisposing to gastric cancer during H. pylori infection, and serve as marker to identify high-risk subjects for H. pylori eradication.
Electronic supplementary material
The online version of this article (doi:10.1186/s12929-015-0121-7) contains supplementary material, which is available to authorized users.
Keywords: Gastric cancer, Cancer relatives, H. pylori, Corpus predominant gastritis index (CGI), Spasmolytic polypeptide-expressing metaplasia (SPEM), Single nucleotide polymorphisms, COX-2, IL-10, Integrin a5b1, COX-2, Trefoil factor 2
Background
Helicobacter pylori is defined as a World Health Organization type I carcinogen for gastric cancer [1,2]. H. pylori-infected patients may develop gastric cancer after chronic gastritis with gastric atrophy, precancerous lesions such as spasmolytic polypeptide expressing metaplasia (SPEM) or intestinal metaplasia (IM), and dysplasia [3-8]. Gastric cancer typically exhibits familial clustering, and gastric cancer familial relatives (GCF) tend to present with corpus-predominant gastritis and precancerous lesions such as SPEM or IM after H. pylori infection [9-13]. Therefore, GCF can be recognized as being at risk of gastric cancer, and such individuals should receive H. pylori screening for early bacterial eradication in order to prevent gastric cancer.
The reason why GCF can be at a higher risk of developing gastric cancer after H. pylori infection remains uncertain. A more adverse interaction between host and H. pylori virulence may exist in GCF to mediate gastric carcinogenesis. The cagA-vacA-babA2 triple-positive H. pylori isolates have been reported as carrying a high risk of gastric cancer, and almost 100% of the H. pylori strains are cagA-vacA-babA2 triple-positive in Taiwan [14-18]. Therefore, GCF in Taiwan are optimal candidates to investigate whether there is any host genomic predisposition in such a high risk group for gastric cancer to determine the specific process of gastric carcinogenesis. In contrast, the duodenal ulcer patients (DU) developed by H. pylori infection is well recognized to have a protective host phenotype having a low risk of gastric adenocarcinoma [19]. By examining differences in the selected candidate genes’ single nucleotide polymorphisms (SNPs) between GCF and DU, the study could validate the SNPs potentially correlated to the higher gastric cancer risk in GCF.
This study selected several potential candidate genes for an analysis of SNPs, including those involved in the specific interactions of bacterial virulence and host receptors related with gastric inflammation, precancerous lesions, or even gastric carcinogenesis. We first included the candidate genes ITGA5 and ITGB1, which encode integrin α5β1 gastric epithelial cell surface receptors to adhere with CagL of H. pylori to construct injection apparatus to translocate CagA of H. pylori via the type IV secretion system (T4SS) [20-22]. As H. pylori isolates can directly prime α5β1 integrin of the host, and as the activity of T4SS determines the risk of cancer [20], we investigated whether any host genomic predisposition in ITGA5 and ITGB1 existed in high-risk GCF. We also included the interleukin-10 gene (IL-10) and cyclooxygenase-2 gene (COX-2), both of which participate in chronic inflammation and have been reported to affect gastric cancer outcomes after H. pylori infection [16,21-25]. There were two additional SNPs being included, with the runt-related transcription factor 3 gene (RUNX3) related to SPEM formation and intestinal-type gastric cancer [26,27], and the trefoil factor 2 (TFF2) gene expressed in SPEM during H. pylori-related gastric carcinogenesis [5,28].
The results showed that several candidate genes (including ITGA5, ITGB1, IL-10, COX-2, RUNX3 and TFF2) differed in the specific genotypes of SNPs between GCF and DU, and predisposed to pre-cancerous lesion, SPEM, within GCF. These specific genotypes of SNPs may offer candidate markers to screen out those at high risk of gastric cancer for early H. pylori eradication in a large-scale study.
Methods
Patients and study designs
This study prospectively and consequently enrolled 389 family relatives of 193 non-cardiac gastric adenocarcinoma (GCA) index patients and 173 benign DU, all of whom joined a screening project for H. pylori infection (NHRI-EX101-9908 BI). A number of the subjects have been referred from the recently published study by our group [12], and the emerging increase of the additional subjects were collected from the funding of the same multi-component grant to fulfill the statistical power required to answer the question whether or not any host genomic predisposition in several candidate genes can exist in the high-risk gastric cancer relatives different to low-risk gastric cancer patients, and to correlate with the presence of precancerous lesions with an increased risk of gastric cancer, especially after H. pylori infection. All of the enrolled study subjects provided written informed consent before entering the study. Both the consent form and study design were reviewed by the Research Ethics Committee of National Cheng Kung University Hospital (certification code: HR-98-023). A flow chart of patient enrollment and the familial relationships to the GCA index patients are shown in Figure 1. Among the 389 GCF, there were 61 spouses, 43 siblings, 205 children, 49 grandchildren and 31 other relatives. Each of the enrolled subjects provided blood samples to collect DNA.
Figure 1.
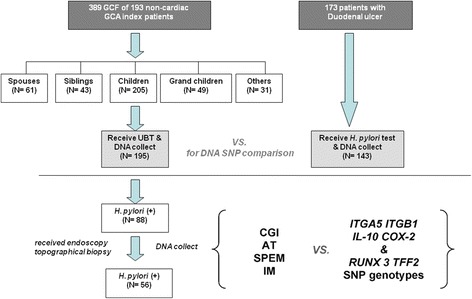
The study flow chart of case recruitment to screen H. pylori infection. In total, 389 family relatives (GCF) from 195 non-cardiac gastric cancer (GCA) index patients, including different familial relations, were enrolled. Blood sampling for DNA extraction and H. pylori screening with the urea breath test (UBT) were performed in all 389 GCF. There were also 143 of 173 duodenal ulcer patients to receive blood sampling and H. pylori screening. The differences in single nucleotide polymorphisms (SNPs) were compared between the DNA of 195 GCA children and 143 duodenal ulcer controls. Of the 88 H. pylori-infected children of GCA, 56 received endoscopy and topographical biopsies to assess whether the corpus predominant gastritis index (CGI), atrophy (AT), spasmolytic polypeptide expressing metaplasia (SPEM) and intestinal metaplasia (IM) correlated to the SNPs of ITGA5, ITGB1, IL-10, COX-2, RUNX3, and TFF2 identified from the comparison between the children of GCA and the duodenal ulcer controls.
The study at first selected the DNA of 195 children of GCA and 143 DU controls to validate whether there were any differences between these two groups with regards to the predisposing risk alleles of SNPs in the ITGA5, ITGB1, IL-10, COX-2, RUNX3, and TFF2 genes. All of the 195 children of GCA were also screened for H. pylori infection by the 13C-urea breath test. For DU, the presence of H. pylori infection was confirmed by positive histology results.
Of the 88 H. pylori-infected children of GCA with a positive 13C-urea breath test, 56 were invited to receive gastric endoscopy to provide topographic gastric biopsy specimens for histological assessments including the corpus predominant gastritis index (CGI), atrophy, SPEM, and IM as described in our previous studies [12,17,29,30]. We further analyzed whether any SNPs associated with specific histology features including CGI, atrophy, SPEM and IM in the children of GCA.
Blood sampling and SNP identification
The genomic DNA of the participants was extracted from peripheral blood mononuclear cells by the manufacturer’s instructions (Viogene, Taipei, Taiwan). The genes of interest were divided into three groups: 1. integrin α5β1 encoding genes ITGA5 and ITGB1; 2. IL10 and COX-2 genes that participate in the inflammation process; and 3. RUNX3, and TFF2 genes relating to late carcinogenesis processes and metaplasia. Because no reports about the relations of diseases for SNPs in ITGA5, ITGB1, and TFF2 genes in previous literatures and their allele frequencies in our population, we performed direct sequencing of the promoters of ITGA5, ITGB1, and TFF2 genes in 20 randomly-selected GCA offspring and found the candidate SNPs for our study. The other candidate SNPs were identified based on those reported in previous papers, tag SNPs, and those located on the promoter, exon or 3’UTR. We assessed the candidate SNPs with two-step polymerase chain reactions (PCR) and direct sequencing or restriction fragment length polymorphism (RFLP), real time PCR or MassARRAY iPLEX Gold-SNP genotyping. The detailed methods of the identification for 24 candidate SNPs were listed in Additional file 1: Table S1, including ITGA5 (−1160 G/T;–240 C/T), ITGB1 (−1949 A/G;–1840 C/T;–1660 -/TTTAA;–1575 A/G;–685-/C; +31804 C/G/T; +32492 A/G), IL-10 (−1082 A/G;–819 C/T;–592 A/C), COX-2 (−1195 A/G; +8473 C/T), RUNX3 (−18508 C/G;–1714 A/C;–1582 C/T;–1166 C/T; +492 A/T; + 73375 A/C) and TFF-2 (−1373 C/T;–503 A/G;–308 A/C; +4649 A/G).
As in our previous study [31,32], we performed two-step PCR with the following cycling conditions: 94°C for 5 minutes then 15 cycles at 94°C for 30 s, 66°C for 30 s, and 72°C for 60 s; then 25 cycles at 94°C for 30 s, 61°C for 30 s, and 72°C for 60 s, and final elongation at 72°C for 10 minutes. The amplified products were sent for direct sequencing (Genomics BioSci & Tech, Taipei, Taiwan, with an ABI3730XL system) or RFLP with specific restriction enzymes (New England Biolabs, Inc., MA, United States). Some SNPs were identified with real-time PCR. The real-time PCR in this study were performed with TaqMan analysis (PE Applied Biosystems, CA, USA), and the primers were designed by File Builder software. PCR were performed in a 10-μl volume containing 10 ng DNA, 2× Master Mix 5 μl, and 0.5 μl probes. The amplification reaction profile included heating at 95°C for 10 minutes followed by 45 cycles at 95°C for 15 s and 60°C for 60 s. The signal was acquired at 60°C during each cycle. The genotypes were analyzed by SDS 21 software.
We also used a MassARRAY iPLEX Gold-SNP genotyping system to perform genotyping in addition to real-time PCR, PCR-RFLP and direct sequencing. Genotyping with the iPLEX assay involved the assay design, DNA isolation, PCR amplification, clean resin, dispensing to a SpectroCHIP bioarray and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis [33]. The assays were performed by Feng Chi Biotech Corp. Taipei, Taiwan, with a Sequenom MassARRAY system and analyzed by the Taipei Institute of Pathology.
Endoscopy and topographic gastric histology grading
The GCA children with H. pylori infection proven by a positive urea breath test were invited to receive endoscopy and topographical biopsies for five specimens from the antrum (x2), corpus (x2) and high corpus near to the cardia (×1) to assess the related gastric histological features [12,34-37]. Each specimen of the topographic site was reviewed in a blinded manner by the pathologist, and the presence of IM (absence: 0; presence: score 1–3) and atrophy (absence: 0; presence: score 1–3) was assessed according to the modified updated Sydney system [38]. In this study, the presence of IM was strictly defined as at least one gastric biopsy sample showing metaplastic cells (goblet cells) involving at least 5% of the upper third of the gastric mucosa [35-37]. Based on the acute and chronic inflammation scores according to the modified updated Sydney system, the presence of CGI was defined as the occurrence of either: 1) the combination of acute and chronic inflammation scores in the high corpus being equal to or larger than the antrum, but not equal to 1; or 2) the combination of scores in the body being larger than the antrum. All of the other conditions were defined as an absence of CGI, including the combination of acute inflammation score and chronic inflammation score of the antrum and high corpus being equal to 1 [12].
TFF2 immunohistochemistry for SPEM
We performed TFF2 immunohistochemistry to assess SPEM in the topographical tissues from the antrum, corpus, and high corpus of the H. pylori-infected children of non-cardiac gastric cancer patients. Immunohistochemical staining was performed using mouse anti-human spasmolytic polypeptide antibody (GE16C, Leica Biosystem Newcastle Ltd., UK) and a commercial kit (NovoLink™ Polymer Detection System, Leica Biosystem Newcastle Ltd., UK). The protocol was modified from the instructions of the manufacturer as used in our previous study [17]. The presence of SPEM was defined as glands formed by TFF2-expressing cells detected in the specimens of either the corpus or high corpus as described previously [12],17]. Because a whole layer (including the upper, middle and lower portion of the gland) of gastric mucosa is needed to assess SPEM. Any cases where SPEM could not be evaluated were treated as missing data [17]. Thus there were fewer case numbers for the comparison of SPEM.
Statistics
The Student’s t test was used when appropriate to determine parametric differences such as age. We verified the non-parametric proportions such as genomic frequencies of specific SNPs in the GCF and DU patients using mixed effect model with SAS PROC MIXED procedure because the genomic frequencies of the GCF were familial dependent while most DU patients had independent data. To correct multiple testing, permutation test by SAS PROC MULTTEST procedure (20000–50000 permutations) was used. We generated the odds ratios (OR) and 95% confidence intervals (95% CI) between the GCF and DU patients by logistic regression with the adjustment of age and H. pylori infection. The linear-by-linear association was applied to assess the frequencies change of the risk SNPs in different familial relation groups. The analyses of the Student’s t test, the logistic regression, and the linear-by-linear association were performed using SPSS version 17.0 for Windows (SPSS Inc., Chicago, USA). The analyses of the mixed effect model and permutation test were performed using SAS software version 9.3 (SAS Institute, Cary NC). All tests were 2-tailed with the significance set at a P value of less than 0.05.
Results
The differences in the selected SNPs between children of GCA and DU
In Figure 1, 195 children of GCA have provided DNA to validate the differences in the SNPs of the ITGA5, ITGB1, IL-10, COX-2, RUNX3, and TFF2 genes, as compared to 143 DU controls. There was no difference in gender between the 195 children of GCA and the 143 DU controls. However, children of GCA had a younger mean age and a lower prevalence of H. pylori infection than DU (Table 1). The children of GCA had a higher allele frequencies of ITGA5-1160 T-carrier (79% vs. 65%; P = 0.006), ITGB-1949 A-carrier (84% vs. 74%; P = 0.047), ITGB1 + 31804 C-carrier (95% vs. 88%; P = 0.013), IL-10-819 TT (52% vs. 38%; P = 0.014), IL-10-592 AA (52% vs. 38%; P = 0.014) and COX-2-1195 G-carrier (83% vs. 71%; P = 0.019), as compared with DU controls. However, there were no differences in the allele frequencies of the SNPs in the RUNX3 and TFF2 genes between the children of GCA and DU controls (Table 1).
Table 1.
The differences of the single nucleotide polymorphisms in the ITGA5 , ITGB1 , IL10 , COX-2 , RUNX3 , and TFF2 genes between 195 children of gastric cancer patients and 143 duodenal ulcer patients
| Parameters N (%) | GCA children | DU patients | P * | P ** | OR (95% CI) |
|---|---|---|---|---|---|
| Age (yr) | 38.2 ± 10.6 | 49.2 ± 13.8 | 1×10-4 | ||
| Gender (F: M) | 99 (51): 96 (49) | 59 (41): 84 (59) | 0.050 | 0.096 | 1.5 (1.0 - 2.3) |
| H. pylori (+: -) | 88 (48): 97 (52) | 110 (79): 29 (21) | <1×10-4 | <1×10-4 | 0.2 (0.1 - 0.4) |
| ITGA5 | |||||
| ITGA5 -1160 | (N = 195) | (N = 139) | |||
| T-carrier: GG | 154 (79): 41 (21) | 90 (65): 49 (35) | 0.017 | 0.006 | 2.2 (1.2 - 4.0) |
| ITGA5 -240 | (N = 193) | (N = 138) | |||
| CC: CT | 190 (98): 3 (2) | 136 (99): 2 (1) | 0.751 | 1.000 | 0.3 (0.0 - 3.1) |
| ITGB1 | |||||
| ITGB1 -1949 | (N = 189) | (N = 136) | |||
| A-carrier: GG | 158 (84): 31 (16) | 101 (74): 35 (26) | 0.047 | 0.047 | 2.8 (1.4 - 5.3) |
| ITGB1 -1840 | (N = 189) | (N = 136) | |||
| CC: T-carrier | 100 (53): 89 (47) | 75 (55): 61 (45) | 0.872 | 0.735 | 0.9 (0.6 - 1.6) |
| ITGB1 -1575 | (N = 189) | (N = 136) | |||
| AA: G-carrier | 10 (5): 179 (95) | 15 (11): 121 (89) | 0.034 | 0.059 | 0.3 (0.1 - 0.8) |
| ITGB1 -685 | (N = 185) | (N = 126) | |||
| -/-: -/C | 26 (14): 159 (86) | 13 (10): 113 (90) | 0.633 | 0.386 | 1.5 (0.7 - 3.4) |
| IGB1 -1660 | (N = 192) | (N = 138) | |||
| TTTAA/TTTAA: others | 165 (86): 27 (14) | 117 (85): 21 (15) | 0.385 | 0.877 | 0.9 (0.5 - 1.7) |
| ITGB1 +31804 | (N = 189) | (N = 136) | |||
| C-carrier: TT | 180 (95): 9 (5) | 119 (88): 17 (13) | 0.003 | 0.013 | 4.7 (1.7 - 13.0) |
| ITGB1 +32492 | (N = 189) | (N = 136) | |||
| G-carrier: AA | 97 (51): 92 (49) | 57 (42): 79 (58) | 0.022 | 0.113 | 1.7 (1.0 - 2.9) |
| IL10 | |||||
| IL10 -1082 | (N = 190) | (N = 138) | |||
| AA: G-carrier | 172 (91): 18 (9) | 121 (88): 17 (12) | 0.216 | 0.839 | 1.7 (0.7 - 3.9) |
| IL10 -819 | (N = 193) | (N = 138) | |||
| TT: C-carrier | 101 (52): 92 (48) | 53 (38): 85 (62) | 0.022 | 0.014 | 2.3 (1.4 - 4.0) |
| IL10 -592 | (N = 193) | (N = 138) | |||
| AA: C-carrier | 101 (52): 92 (48) | 53 (38): 85 (62) | 0.022 | 0.014 | 2.3 (1.4 - 4.0) |
| COX-2 | |||||
| COX-2 -1195 | (N = 189) | (N = 129) | |||
| G-carrier: AA | 156 (83): 33 (17) | 92 (71): 37 (29) | 0.064 | 0.019 | 1.7 (0.9 - 3.2) |
| COX-2 +8473 | (N = 194) | (N = 138) | |||
| TT: C-carrier | 134 (69): 60 (31) | 84 (61): 54 (39) | 0.259 | 0.129 | 1.4 (0.9 - 2.3) |
| RUNX3 | |||||
| RUNX3 -18508 | (N = 186) | (N = 135) | |||
| C-carrier: GG | 130 (70): 56 (30) | 100 (74): 35 (26) | 0.586 | 0.449 | 0.7 (0.4 - 1.2) |
| RUNX3 -1714 | (N = 193) | (N = 137) | |||
| CC: A-carrier | 109 (56): 84 (44) | 68 (50): 69 (50) | 0.302 | 0.261 | 1.3 (0.7 - 2.1) |
| RUNX3 -1582 | (N = 189) | (N = 136) | |||
| T-carrier: CC | 84 (44): 105 (56) | 69 (51): 67 (49) | 0.338 | 0.307 | 0.9 (0.5 - 1.4) |
| RUNX3 -1166 | (N = 193) | (N = 137) | |||
| TT: C-carrier | 109 (56): 84 (44) | 68 (50): 69 (50) | 0.302 | 0.258 | 1.3 (0.7 - 2.1) |
| RUNX3 +492 | (N = 189) | (N = 136) | |||
| TT: A-carrier | 106 (56): 83 (44) | 67 (49): 69 (51) | 0.312 | 0.263 | 1.2 (0.7 - 2.1) |
| RUNX3 +73375 | (N = 189) | (N = 135) | |||
| CC: A-carrier | 45 (24): 144 (76) | 23 (17): 112 (83) | 0.122 | 0.167 | 1.5 (0.8 - 2.9) |
| TFF2 | |||||
| TFF2 -1373 | (N = 189) | (N = 136) | |||
| CC: T-carrier | 139 (74): 50 (26) | 97 (71): 39 (29) | 0.692 | 0.702 | 1.0 (0.6 - 1.8) |
| TFF2 -503 | (N = 189) | (N = 136) | |||
| AA: G-carrier | 139 (74): 50 (26) | 97 (71): 39 (29) | 0.692 | 0.706 | 1.0 (0.6 - 1.8) |
| TFF2 -308 | (N = 189) | (N = 136) | |||
| CC: A-carrier | 85 (45): 104 (55) | 53 (39): 83 (61) | 0.394 | 0.311 | 1.2 (0.7 - 2.1) |
| TFF2 +4649 | (N = 188) | (N = 136) | |||
| GG: A-carrier | 100 (53): 88 (47) | 64 (47): 72 (53) | 0.343 | 0.308 | 1.4 (0.8 - 2.4) |
GCA: gastric cancer. DU: duodenal ulcer. SNPs: single nucleotide polymorphisms. *P value by mixed effect model. **P value by permutation test.
Combined predisposing SNPs to differentiate between GCA children and DU
Based on the P values of permutation tests, we choose SNPs in each gene most significantly different between the case and control groups to derive combined genotypes for further analysis. The study tested the differences of the combined genotypes of the predisposing SNPs in integrin α5β1 encoding genes ITGA5, ITGB1 -1949 and ITGB1 +31804, implicating the influence of integrin α5β1 on host-bacterial interaction processes during gastric carcinogenesis, between the children of GCA and DU controls (Table 2). The children of GCA had a 5-fold higher risk to have all of the three risk SNPs (ITGA5 -1160 T-carrier, ITGB1 -1949 A-carrier, and ITGB1 +31804 C-carrier), as compared with DU (67% vs. 47%; P = 0.05). Because of the linkage of disequilibrium between IL-10 -819 and IL-10 -592, this study selected IL-10 -592 to combine with COX-2 -1195 to indicate a possibly more severe gastric inflammation. This study illustrates that the children of GCA exerted a 3.3-fold higher risk to harbor a combined SNPs in IL-10-592/ COX-2-1195 as AA/G-carrier than DU controls (43% vs. 25%; P = 0.005) (Table 2).
Table 2.
The comparisons of the genomic difference in the combinations of ITGA5 -1160 T-carrier, ITGB1 -1949 A-carrier, ITGB1 +31804 C-carrier, IL10 -592 AA and COX-2 -1195 G-carrier between the children of gastric cancer patients and the duodenal ulcer controls
| Predisposing SNPs | Risk SNPs N (%) | GCA children | DU controls | P * | P ** | OR (95% CI) |
|---|---|---|---|---|---|---|
| ITGA5 -1160 | (N = 189) | (N = 134) | ||||
| T-carrier | 3R:2R:1R:0R | 126(67): 49(26): 12(6): 2(1) | 63(47): 48(36): 18(13): 5(4) | 0.002 | ||
| ITGB1 -1949 | ||||||
| A-carrier | 3R | 126 (67) | 63 (47) | 0.225 | 0.050 | 5 (0.9 - 26.5) |
| ITGB1 +31804 | 2R | 49 (26) | 48 (36) | 0.539 | 0.514 | 2.6 (0.5 - 13.8) |
| C-carrier | 1R | 12 (6) | 18 (23) | 0.575 | 0.756 | 1.7 (0.3 - 10.0) |
| 0R | 2 (21) | 5 (4) | 1 | |||
| COX-2 -1195 | (N = 188) | (N = 125) | ||||
| G-carrier | 2R: 1R: 0R | 80 (43): 93 (49): 15 (8) | 31 (25): 75 (60): 19 (15) | 0.013 | ||
| With or without | ||||||
| IL-10 -592 AA | 2R | 80 (43) | 31 (25) | 0.021 | 0.005 | 3.3 (1.5 - 7.2) |
| 1R | 93 (49) | 75 (60) | 0.457 | 0.344 | 1.6 (0.7 - 3.3) | |
| 0R | 15 (8) | 19 (15) | 1 | |||
| ITGA5-1160 | (N = 185) | (N = 124) | ||||
| T-carrier | 2R: 1R: 0R | 44 (24): 115 (62): 26 (14) | 10 (8): 67 (54): 47 (38) | <1×10-4 | ||
| ITGB1 -1949 | ||||||
| A-carrier | ||||||
| ITGB1 +31804 | 2R | 44 (24) | 10 (8) | <1×10-4 | <1×10-4 | 8.0 (3.4 - 18.4) |
| C-carrier | 1R | 115 (62) | 67 (54) | 2×10-4 | 3×10-4 | 3.1 (1.8 - 5.5) |
| With or without | 0R | 26 (14) | 47 (38) | 1 | ||
| COX-2 -1195 | ||||||
| G-carrier | ||||||
| IL10 -592 AA | ||||||
| (N = 193) | (N = 135) | |||||
| IL-10 -592 AA | 2R: 1R: 0R | 74 (38): 105 (54): 14 (7) | 33 (24): 73 (54): 29 (21) | 4×10-4 | ||
| With or without | ||||||
| ITGA5 -1160 | 2R | 74 (38) | 33 (24) | 1×10-4 | <1×10-4 | 4.6 (2.2 - 9.9) |
| T-carrier | 1R | 105 (54) | 73 (54) | 6×10-4 | 0.003 | 3.0 (1.5 - 6.0) |
| 0R | 14 (7) | 29 (21) | 1 |
GCA: gastric cancer. DU: duodenal ulcer. SNPs: single nucleotide polymorphisms. 3R: the presence of 3 predisposing SNPs with risk alleles. 2R: the presence of 2 predisposing SNPs with risky alleles.
1R: the presence of 1 predisposing SNP with risk allele. 0R: no any predisposing SNPs with risk alleles. *P value by mixed effect model; **P value by permutation test.
We further combined the above two sets to increase the relative risk to differentiate the children of GCA children to DU. Also in Table 2, the children of GCA had a 8.0-fold higher risk (P < 1×10−4) to have a combined genotype with ITGA5-1160/ITGB1-1949/ ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA than DU controls. To simplify the combination, we selected the SNPs most significantly different allele frequencies between GCA children and DU controls, ITGA5 -1160 T-carrier and IL-10 -592 AA, to serve as risk genotypes. The children of GCA had a 4.6-fold risk of having both risk genotypes as compared to DU controls (P < 1×10−4).
The combined SNPs correlate to SPEM in children of GCA
In Figure 1, 56 children of GCA with H. pylori infection received endoscopy to assess the histology features. The study illustrated the combined genotype with ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA correlated to the presence of SPEM (P = 0.016), but not CGI, atrophy or IM (P > 0.05). The children of GCA with the combined genotype with ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA had a 5.25-fold higher risk to have SPEM (P = 0.016) (Table 3). Although the children of GCA had a 4.6-fold higher chance of being ITGA5-1160/IL-10-592 as T-carrier/AA, but such a simplified combined genotype did not correlate to CGI, atrophy, SPEM or IM (P > 0.05).
Table 3.
The difference of the H. pylori-related histological features between the children of gastric cancer patients with and without the risk allele genotypes as ITGA5-1160 T-carrier/ITGB1-1949 A-carrier/ITGB1+31804 C-carrier & COX-2-1195 G-carrier/IL10-592 AA
| Risks N (%) Pathology | ITGA5 -1160/ ITGB1 -1949/ ITGB1 +31804/ COX-2 -1195/ IL10 -592 as T*/A*/C*/G*/AA | Others | P * | P ** | OR (95% CI) |
|---|---|---|---|---|---|
| CGI (N = 56) | |||||
| Present: absent | 14 (58): 10 (42) | 15 (47): 17 (53) | 0.115 | 0.429 | 1.6 (0.5-4.6) |
| Atrophy (N = 56) | |||||
| Present: absent | 17 (71): 7 (29) | 21 (66): 11 (34) | 0.932 | 0.778 | 1.3 (0.4-4.0) |
| SPEM (N = 48) | |||||
| Present: absent | 18 (82): 4 (18) | 12 (46): 14 (54) | 0.010 | 0.016 | 5.3 (1.4-19.8) |
| IM (N = 56) | |||||
| Present: absent | 4 (17): 20 (83) | 6 (19): 26 (81) | 0.831 | 1.000 | 0.9 (0.2-3.5) |
CGI: corpus predominant gastritis index. SPEM: spasmolytic polypeptide-expressing metaplasia. IM: intestinal metaplasia. *Indicated the carrier of the specific allele in SNPs. *P value by mixed effect model; **P value by permutation test.
Since RUNX3 and TFF2 regulate the SPEM formation during gastric carcinogenesis [26-28], we further tested if any specific SNPs in RUNX3 and TFF2 genes correlated to the presence of SPEM in the children of GCA. In Table 4, the children of GCA with SPEM had a higher allele frequencies of RUNX3 + 492 as A-carrier (53% vs. 22%; P = 0.042) and TFF2-308 as CC (60% vs. 28%; P = 0.041) than those without SPEM. Thus we further analyzed the combined genotypes of these 2 SNPs. Nearly 90% of the children of GCA with SPEM were either RUNX3 + 492 A-carrier or TFF2-308 CC and these two genotypes led to a 11.3-fold increased risk of having SPEM in the children of GCA (P = 0.001) (Table 4). However, there was no significant correlation between the ITGA5-1160/ITGB1-1949/ITGB1 + 31804/COX-2-1195/IL-10-592 as T-carrier/A-carrier/C-carrier/G-carriers/AA and the RUNX3 + 492/ TFF2-308 as A-carrier/CC among the children of GCA. By combining these two sets of SNPs, the risk of getting SPEM within children of GCA elevated up to 112 folds (P < 1×10−4) (Table 5).
Table 4.
The differences of the single nucleotide polymorphisms in the RUNX3 , and TFF2 genes between children of gastric cancer patients with (N = 31) or without (N = 18) SPEM
| Parameters N (%) | SPEM (+) | SPEM (-) | P * | P ** | OR (95% CI) |
|---|---|---|---|---|---|
| Age (yr) | 39.9 ± 7.7 | 38.2 ± 11.3 | 0.312 | ||
| Gender (F: M) | 20 (65): 11 (35) | 7 (39): 11 (61) | 0.092 | 0.136 | 2.9 (0.9 - 9.5) |
| RUNX3 | |||||
| RUNX3 -18508 | (N = 29) | (N = 17) | |||
| C-carrier: GG | 21 (72): 8 (28) | 12 (71): 5 (29) | 0.897 | 1.000 | 1.1 (0.3 - 4.1) |
| RUNX3 -1714 | (N = 31) | (N = 18) | |||
| A-carrier: CC | 16 (52): 15 (48) | 4 (22): 14 (78) | 0.044 | 0.067 | 3.7 (1.0 - 13.9) |
| RUNX3 -1582 | (N = 30) | (N = 18) | |||
| T-carrier: CC | 16 (53): 14 (47) | 5 (28): 13 (72) | 0.085 | 0.137 | 3.0 (0.8 - 10.4) |
| RUNX -1166 | (N = 31) | (N = 18) | |||
| C-carrier: TT | 16 (52): 15 (48) | 4 (22): 14 (78) | 0.044 | 0.070 | 3.7 (1.0 - 13.9) |
| RUNX3 +492 | (N = 30) | (N = 18) | |||
| A-carrier: TT | 16 (53): 14 (47) | 4 (22): 14 (78) | 0.035 | 0.042 | 4.0 (1.1 - 15.0) |
| RUNX3 +73375 | (N = 30) | (N = 18) | |||
| C-carrier: AA | 21 (70): 9 (30) | 14 (78): 4 (22) | 0.613 | 0.741 | 0.7 (0.2 - 2.6) |
| TFF2 | |||||
| TFF2 -1373 | (N = 30) | (N = 18) | |||
| C-carrier: TT | 30 (100): 0 (0) | 17 (94): 1 (6) | 0.206 | 0.377 | 2.6 (0.8 - 9.2) |
| TFF2 -503 | (N = 30) | (N = 18) | |||
| AA: G-carrier | 23 (77): 7 (23) | 10 (56): 8 (44) | 0.127 | 0.194 | 2.6 (0.7 - 9.2) |
| TFF2 -308 | (N = 30) | (N = 18) | |||
| CC: A-carrier | 18 (60): 12 (40) | 5 (28): 13 (72) | 0.022 | 0.041 | 3.9 (1.1-13.8) |
| TFF2 +4649 | (N = 30) | (N = 18) | |||
| G-carrier: AA | 26 (87): 4 (13) | 17 (94): 1 (6) | 0.419 | 0.637 | 0.4 (0.1-3.7) |
| RUNX3+492/TFF2-308 | |||||
| A-carrier/CC | |||||
| 2R | 7 (23) | 1 (6) | 0.001 | 0.009 | 23.3 (2.0 - 273.3) |
| 1R | 20 (67) | 7 (39) | 0.001 | 0.004 | 9.5 (2.0 - 44.9) |
| 0R | 3 (10) | 10 (56) | 1 | ||
| 2R+1R:0R | 27 (90): 3 (10) | 8 (44): 10 (56) | 3×10-4 | 0.001 | 11.3 (2.5 - 51.0) |
SPEM: Spasmolytic polypeptide-expressing metaplasia; SNPs: single nucleotide polymorphisms. 2R: the presence of 2 predisposing SNPs with risk alleles. 1R: the presence of 1 predisposing SNP with risk allele. 0R: no any predisposing SNPs with risk alleles. *P value by mixed effect model; **P value by permutation test.
Table 5.
The risk of SPEM increases stepwise for ITGA5 -1160 T-carrier, ITGB1 -1949 A-carrier, ITGB1 +31804 C-carrier, COX-2 -1195 G-carrier, and IL10 -592 AA to combine with RUNX3 +492 as A-carrier & TFF2 -308 as CC in the children of gastric cancer patients
| Predisposing SNPs | Risk SNPs N (%) | SPEM (+) | SPEM (−) | P * | P ** | OR (95% CI) |
|---|---|---|---|---|---|---|
| ITGA5-1160/ITGB1- | 2R: 1R: 0R | 16 (53): 13 (43): 1 (3) | 1 (6): 10 (56): 7 (39) | <1×10−4 | ||
| 1949/ITGB1+31804/COX-2-1195/IL10-592 | 2R: 1R+ 0R | 16 (53): 14 (47) | 1 (6): 17 (94) | 5×104 | 0.001 | 19.4 (2.3-165.2) |
| as T*/A*/C*/G*/AA | ||||||
| 2R+ 1R: 0R | 29 (97): 1 (3) | 11 (61): 7 (39) | 5×10−4 | 0.003 | 18.5 (2.0-167.8) | |
| with or without | ||||||
| 2R | 16 (53) | 1 (6) | 1×10−4 | 1×10−4 | 112.0 (6.1-2057.3) | |
| RUNX3+492/TFF2-308 | 1R | 13 (43) | 10 (56) | 0.045 | 0.0518 | 9.1 (1.0-86.5) |
| as A*/CC | 0R | 1 (3) | 7 (39) | 1 |
SPEM: spasmolytic polypeptide-expressing metaplasia. SNPs: single nucleotide polymorphisms. 2R: the presence of 2 predisposing SNPs with risk alleles. 1R: the presence of 1 predisposing SNPs with risk allele. 0R: no any predisposing SNPs with risk alleles. *Indicated the carrier of the specific allele in SNPs. *P value by mixed effect model; **P value by permutation test.
Combined SNPs predisposed to SPEM among different familial relative groups of GCA
We further assessed whether the different gastric cancer familial relative groups had different proportions of the combined SNPs predisposing to SPEM. In Figure 2A, the offspring, including children and grandchildren, of GCA had a higher rate of ITGA5-1160/ITGB1-1949/ITGB1 + 31804/COX-2-1195/IL-10-592 as T-carrier/A-carrier/C-carrier/G-carriers/AA than other familial groups (P = 0.018). However, in Figure 2B, there was no difference in the combined genotype of RUNX3 + 492/TFF2-308 as A-carrier/CC among the different familial groups of GCA (P = 0.313). Furthermore, children of the GCA index patients carrying the combination sets of ITGA5-1160/ITGB1-1949/ITGB1 + 31804/COX-2-1195/IL-10-592 as T-carrier/A-carrier/C-carrier/G-carriers/AA also had a higher prevalence of the same combination SNPs than children of GCA index patients not carrying such combinations of SNPs (43% vs. 16%; P = 0.003; OR 3.9; 95% CI: 1.6-9.6).
Figure 2.
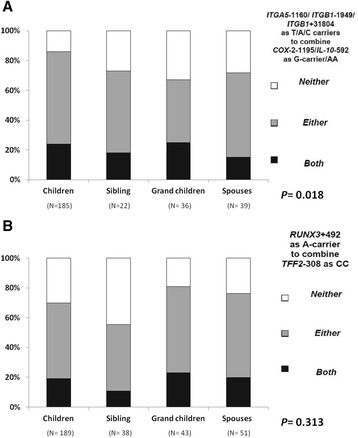
The combined risk genotypes’ distribution in gastric cancer familial relatives. A. The prevalences of the ITGA5-1160/ ITGB1-1949/ITGB1 + 31804 as T-carrier/A-carrier/C-carrier and COX-2-1195/IL10-592 as G-carrier/AA among different GCA familial relative groups were shown. The children of gastric cancer patients had higher prevalence of the presence of either or both risk combination SNPs in ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA than other familial relative groups (P = 0.018). B. There were no difference in the combined genotype of RUNX3 + 492/TFF2-308 as A-carrier/cm3 among different familial groups of gastric cancer patients (P = 0.313).
Discussion
There were genomic predispositions of SNPs in the ITGA5, ITGB1, IL-10 and COX-2 genes in the children of GCA, who are considered to be a risk group of gastric cancer after H. pylori infection. The combined genotype as ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA significantly predisposed to the precancerous lesions such as SPEM after H. pylori infection in children of GCA. Such a predisposing risk of SPEM can be elevated via combination with RUNX3 + 492/TFF2-308 as A-carrier/ CC in children of GCA. The combined genotype such as ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA, or even more specifically combined with RUNX3 + 492/TFF2-308, may thus serve as a host factor to identify the risk group of gastric cancer for an early H. pylori eradication.
The study utilized patients with duodenal ulcers to serve as cohort controls with the least gastric cancer risk. In a review of the related literature, gastric cancer relatives have been recognized as having an increased gastric cancer risk. Gastric cancer relatives exist with a genomic predisposition to gastric cancers, especially after H. pylori infection. The study thus attempted to illustrate whether the possible candidate genomic polymorphisms selected in this study can be different between our high-risk gastric cancer relatives and low-risk duodenal ulcer patients in either Tables 1 and 2. In this study, the children of GCA had a younger mean age than DU patients, whose mean age was compatible to that of DU in a previous study [19]. Thus, our enrolled DU patients were found to be suitable to serve as the low-cancer risk comparison group.
The activity of T4SS mediated by H. pylori positively associated with the risk of gastric cancer [20]. Furthermore, we disclosed that H. pylori isolates with the cagL amino acid sequence polymorphism Y58E59 could exploit higher amounts of gastric integrin α5β1 to mediate stronger T4SS activity [39,40]. Namely, more toxic H. pylori isolates shall have mediated with higher gastric cancer risk via the host integrin α5β1 expressions. The current study showed that the children of GCA, who are at a high risk of gastric cancer, can express with a 5-fold higher risk to be ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers in ITGA5 and ITGB1 than DU (Table 2). This suggests that there should be a host predisposition to regulate the expression of the putative integrin α5β1 in addition to the virulence factor of H. pylori isolates. Thus, it would be worthwhile to validate the correlation of ITGA5 and ITGB1 genes to the H. pylori-related histological precancerous changes, especially among children of GCA.
The current study also identified that the children of GCA patients as having a higher prevalence of IL-10-819 as TT and IL-10-592 as AA than DU, compatible with previous findings to show such genotypes with lower IL-10 excretion and more severe gastritis [21-23,41]. COX-2 over-expression can be evident in pre-malignant gastric lesions after H. pylori infection [11,24,25]. Our data showed that the children of GCA had a higher prevalence of COX-2-1195 G-carrier than DU, but the finding is not compatible with a previous study, which suggests that the COX-2-1195 AA genotype carries a higher risk of GCA [42]. This may also be due to potential ethnic differences, or that the COX-2-1195 G-carrier may just serve to associate with other pathways of gastric carcinogenesis [43].
Because gastric carcinogenesis is a multi-step and multi-factorial process, we anticipate that SNPs on different genes work in concert in the gastric carcinogenesis. We choose TagSNPs in each gene as the most significantly different between the GCF and the DU patients for the analysis of combined genotypes. We found that the children of GCA had an 8-fold risk to have the combined genotypes of either a complicated model (ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA) (Table 2), higher than the risk difference of each single SNP. This suggests that when more risk alleles combine, the higher the risk of gastric cancer. Nevertheless, only the first complicated model can be related to the presence of the precancerous lesions as SPEM in the children of GCA (Table 3). It thus suggested the complicated model with 5 risk alleles shall be a novel host factor to determine the high-risk patients of gastric cancer for early H. pylori eradication.
The combined genotypes ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA can just correlate with SPEM, but not IM. The lack of correlation to IM may be due to the small number of cases with IM or the younger mean age in our series, or to fulfill SPEM is generally earlier than IM [6,44,45].
In Table 4, the combined genotype of RUNX3 + 492/TFF2-308 as A-carrier/cm3 was also correlated to the presence of SPEM in the children of GCA. Accordingly, combining these 2 sets of risk combined genotypes, the risk to get SPEM increased up to 112 folds (Table 5). This indicated these 2 sets of combined genotypes can be synergistic to enhance the SPEM. With regards to no significant difference of allele frequencies of SNPs in RUNX3 and TFF2 genes between the children of GCA with low risk DU, the host-bacterial interaction regulated by integrin α5β1 and the chronic inflammation regulated by IL-10 and COX-2 may be prior to the regulation of RUNX3 and TFF2 in the gastric carcinogenesis process.
The allele frequencies of the combination of ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T-carrier/A-carrier/C-carriers in GCA children was 67%, which was higher than 47% in DU patients (P = 0.0007). When taking more SNPs with COX-2-1195/IL-10-592 and RUNX3/TFF2 into consideration, the allele frequencies of the complicated model (ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers, COX-2-1195/IL-10-592 as G-carrier/AA, and RUNX3/TFF2 as A-carrier/cm3) remained higher in children of GCA than in DU (17% vs. 6%, P = 0.003). This data suggests the greater number of risk SNPs included, the more specific the result to indicate who the high risk group was.
The study has limitations to consider such as that it does not divide the H. pylori-positive and negative offspring of gastric cancer into analysis due to the case scales. Furthermore, another limitation of the study is the small sample size, especially the GCF with available pathology data. This made the significance of the risk combined genotypes to correlate with SPEM limited despite there being a high odds ratio. This was because many GCF were unwilling to receive gastric endoscope examination although we screened 389 GCF and 205 children of non-cardiac GCA patients as outlined in Figure 1. This indicated a lack of insight into the general population, even with GCF having high GCA risks. It also highlights the importance of studying markers to identify high-risk groups of GCA. We do agree if budgeting allows, eradicating H. pylori infection for all offspring of gastric cancer patients remains ideal. If we can understand the genomic predispositions from offspring with a higher risk of having an adverse outcome, such host factors may be helpful to screen out the general dyspepsia patients for early H. pylori eradication to prevent gastric cancer. The relevance of this study is that it answers the reason why offspring of gastric cancer are at risk, and that it also provides clinicians with a potential tool to help screen out the higher risk groups with obvious cost-benefit concerns.
Conclusions
There were SNPs in the ITGA5, ITGB1, IL-10, and COX-2 genes in the children of GCA. The combined genotypes ITGA5-1160/ITGB1-1949/ITGB1 + 31804 as T/A/C carriers and COX-2-1195/IL-10-592 as G-carrier/AA could determine the risk of SPEM after H. pylori infection. Such risk can be elevated up to 112-fold by combining RUNX3 + 492/TFF2-308 as A-carrier/cm3. The host genomic SNPs shall be novel to identify the high-risk patients of gastric cancer for early H. pylori eradication.
Acknowledgements
This work was supported by grants (NSC101-2314-B-006-016-MY3) from National Scientific Council, NHRI-EX99-9908BI from National Health Research Institute, Taiwan. We are also grateful to Shang-Chi Lee for providing the statistical consulting services from the Biostatistics Consulting Center, National Cheng Kung University Hospital.
Abbreviations
- AT
Atrophy
- CGI
Corpus predominant gastritis index
- COX-2
Cycloxygenase-2
- DU
Duodenal ulcer patients
- GCA
Gastric cancer patients
- GCF
Family relatives of gastric cancer patients
- IM
Intestina metaplasia
- IL-10
Interleukin 10
- ITGA
Integrin alpha: ITGB: Integrin beta
- RUXN3
Runt-related transcription factor 3
- SPEM
Spasmolytic polypeptide-expressing metaplasia
- TFF2
Trefoil factor 2
- T4SS
Type four secretion system
- PCR
Polymerase chain reactions
- RFLP
Restriction fragment length polymorphism
Additional file
The identification number, identifying methods and study methods of the target single nucleotide polymorphisms (SNPs) in this study.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BSS coordinated the whole study. YCT conducted the SNP genotypes and prepared the manuscript. WHH screened the H. pylori infection and stained the TFF2. SHL helped to examine the data and perform the statistical analysis. WLC and HCC conducted endoscopic biopsies. HBY reviewed the histological features, and checked the TFF2 stains to define atrophy, SPEM, and IM. CCL helped to design the primers for the SNPs. All authors read and approved the final manuscript.
Contributor Information
Yu-Ching Tsai, Email: dr.aching@msa.hinet.net.
Wei-Hsin Hsiao, Email: p9885221@yahoo.com.tw.
Sheng-Hsiang Lin, Email: shlin922@mail.ncku.edu.tw.
Hsiao-Bai Yang, Email: hbyang@tyh.com.tw.
Hsiu-Chi Cheng, Email: teishuki@mail.ncku.edu.tw.
Wei-Lun Chang, Email: weilun1@mail.ncku.edu.tw.
Cheng-Chan Lu, Email: cclu@mail.ncku.edu.tw.
Bor-Shyang Sheu, Email: sheubs@mail.ncku.edu.tw.
References
- 1.Schistosomes, liver flukes and Helicobacter pylori IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 2.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325(16):1127–31. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 3.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process–First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52(24):6735–40. [PubMed] [Google Scholar]
- 4.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt PH, Lee JR, Joshi V, Playford RJ, Poulsom R, Wright NA, et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest. 1999;79(6):639–46. [PubMed] [Google Scholar]
- 6.Goldenring JR, Nam KT, Wang TC, Mills JC, Wright NA. Spasmolytic polypeptide-expressing metaplasia and intestinal metaplasia: time for reevaluation of metaplasias and the origins of gastric cancer. Gastroenterology. 2010;138(7):2207–10. doi: 10.1053/j.gastro.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis. 2012;13(1):2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291(2):187–94. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 9.Brenner H, Bode G, Boeing H. Helicobacter pylori infection among offspring of patients with stomach cancer. Gastroenterology. 2000;118(1):31–5. doi: 10.1016/S0016-5085(00)70411-2. [DOI] [PubMed] [Google Scholar]
- 10.El-Omar EM, Oien K, Murray LS, El-Nujumi A, Wirz A, Gillen D, et al. Increased prevalence of precancerous changes in relatives of gastric cancer patients: critical role of H. pylori. Gastroenterology. 2000;118(1):22–30. doi: 10.1016/S0016-5085(00)70410-0. [DOI] [PubMed] [Google Scholar]
- 11.Sheu BS, Yang HB, Sheu SM, Huang AH, Wu JJ. Higher gastric cycloxygenase-2 expression and precancerous change in Helicobacter pylori-infected relatives of gastric cancer patients. Clin Cancer Res. 2003;9(14):5245–51. [PubMed] [Google Scholar]
- 12.Tsai YC, Hsiao WH, Yang HB, Cheng HC, Chang WL, Lu CC, et al. The corpus-predominant gastritis index may serve as an early marker of Helicobacter pylori-infected patients at risk of gastric cancer. Aliment Pharmacol Ther. 2013;37(10):969–78. doi: 10.1111/apt.12291. [DOI] [PubMed] [Google Scholar]
- 13.Marcos-Pinto R, Carneiro F, Dinis-Ribeiro M, Wen X, Lopes C, Figueiredo C, et al. First-degree relatives of patients with early-onset gastric carcinoma show even at young ages a high prevalence of advanced OLGA/OLGIM stages and dysplasia. Aliment Pharmacol Ther. 2012;35(12):1451–9. doi: 10.1111/j.1365-2036.2012.05111.x. [DOI] [PubMed] [Google Scholar]
- 14.Hocker M, Hohenberger P. Helicobacter pylori virulence factors–one part of a big picture. Lancet. 2003;362(9391):1231–3. doi: 10.1016/S0140-6736(03)14547-3. [DOI] [PubMed] [Google Scholar]
- 15.Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135(1):91–9. doi: 10.1053/j.gastro.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 16.Sheu SM, Sheu BS, Yang HB, Li C, Chu TC, Wu JJ. Presence of iceA1 but not cagA, cagC, cagE, cagF, cagN, cagT, or orf13 genes of Helicobacter pylori is associated with more severe gastric inflammation in Taiwanese. J Formos Med Assoc. 2002;101(1):18–23. [PubMed] [Google Scholar]
- 17.Sheu BS, Sheu SM, Yang HB, Huang AH, Wu JJ. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut. 2003;52(7):927–32. doi: 10.1136/gut.52.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheu SM, Hung KH, Sheu BS, Yang HB, Wu JJ. Association of nonsynonymous substitutions in the intermediate region of the vacA gene of Helicobacter pylori with gastric diseases in Taiwan. J Clin Microbiol. 2009;47(1):249–51. doi: 10.1128/JCM.01651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansson LE, Nyren O, Hsing AW, Bergstrom R, Josefsson S, Chow WH, et al. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N Engl J Med. 1996;335(4):242–9. doi: 10.1056/NEJM199607253350404. [DOI] [PubMed] [Google Scholar]
- 20.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449(7164):862–6. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 21.Wu MS, Wu CY, Chen CJ, Lin MT, Shun CT, Lin JT. Interleukin-10 genotypes associate with the risk of gastric carcinoma in Taiwanese Chinese. Int J Cancer. 2003;104(5):617–23. doi: 10.1002/ijc.10987. [DOI] [PubMed] [Google Scholar]
- 22.Shanks AM, El-Omar EM. Helicobacter pylori infection, host genetics and gastric cancer. J Dig Dis. 2009;10(3):157–64. doi: 10.1111/j.1751-2980.2009.00380.x. [DOI] [PubMed] [Google Scholar]
- 23.Zambon CF, Basso D, Navaglia F, Belluco C, Falda A, Fogar P, et al. Pro-and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine+ 2005;29(4):141–52. doi: 10.1016/j.cyto.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Lim HY, Joo HJ, Choi JH, Yi JW, Yang MS, Cho DY, et al. Increased expression of cyclooxygenase-2 protein in human gastric carcinoma. Clin Cancer Res. 2000;6(2):519–25. [PubMed] [Google Scholar]
- 25.Sung JJ, Leung WK, Go MY, To KF, Cheng AS, Ng EK, et al. Cyclooxygenase-2 expression in Helicobacter pylori-associated premalignant and malignant gastric lesions. Am J Pathol. 2000;157(3):729–35. doi: 10.1016/S0002-9440(10)64586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim B, Ju H, Kim M, Kang C. Increased genetic susceptibility to intestinal-type gastric cancer is associated with increased activity of the RUNX3 distal promoter. Cancer. 2011;117(22):5161–71. doi: 10.1002/cncr.26161. [DOI] [PubMed] [Google Scholar]
- 27.Ito K, Chuang LS, Ito T, Chang TL, Fukamachi H, Salto-Tellez M, et al. Loss of Runx3 is a key event in inducing precancerous state of the stomach. Gastroenterology. 2011;140(5):1536–46. doi: 10.1053/j.gastro.2011.01.043. [DOI] [PubMed] [Google Scholar]
- 28.Peterson AJ, Menheniott TR, O’Connor L, Walduck AK, Fox JG, Kawakami K, et al. Helicobacter pylori infection promotes methylation and silencing of trefoil factor 2, leading to gastric tumor development in mice and humans. Gastroenterology. 2010;139(6):2005–17. doi: 10.1053/j.gastro.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang HB, Cheng HC, Sheu BS, Hung KH, Liou MF, Wu JJ. Chronic celecoxib users more often show regression of gastric intestinal metaplasia after Helicobacter pylori eradication. Aliment Pharmacol Ther. 2007;25(4):455–61. doi: 10.1111/j.1365-2036.2006.03224.x. [DOI] [PubMed] [Google Scholar]
- 30.Sheu BS, Tsai YC, Wu CT, Chang WL, Cheng HC, Yang HB. Long-term celecoxib can prevent the progression of persistent gastric intestinal metaplasia After H. pylori eradication. Helicobacter. 2013;18(2):117–23. doi: 10.1111/hel.12013. [DOI] [PubMed] [Google Scholar]
- 31.Lu CC, Sheu BS, Chen TW, Yang HB, Hung KH, Kao AW, et al. Host TNF-alpha-1031 and–863 promoter single nucleotide polymorphisms determine the risk of benign ulceration after H. pylori infection. Am J Gastroenterol. 2005;100(6):1274–82. doi: 10.1111/j.1572-0241.2005.40852.x. [DOI] [PubMed] [Google Scholar]
- 32.Hung KH, Hung HW, Yang HB, Lu CC, Wu JJ, Sheu BS. Host single nucleotide polymorphisms of MMP-9–1562/TIMP-1 372 have gender differences in the risk of gastric intestinal metaplasia after Helicobacter pylori infection. Helicobacter. 2009;14(6):580–7. doi: 10.1111/j.1523-5378.2009.00717.x. [DOI] [PubMed] [Google Scholar]
- 33.Fumagalli D, Gavin PG, Taniyama Y, Kim SI, Choi HJ, Paik S, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheu BS, Odenbreit S, Hung KH, Liu CP, Sheu SM, Yang HB, et al. Interaction between host gastric Sialyl-Lewis X and H. pylori SabA enhances H. pylori density in patients lacking gastric Lewis B antigen. Am J Gastroenterol. 2006;101(1):36–44. doi: 10.1111/j.1572-0241.2006.00358.x. [DOI] [PubMed] [Google Scholar]
- 35.Chuang CH, Yang HB, Sheu SM, Hung KH, Wu JJ, Cheng HC, et al. Helicobacter pylori with stronger intensity of CagA phosphorylation lead to an increased risk of gastric intestinal metaplasia and cancer. BMC Microbiol. 2011;11:121. doi: 10.1186/1471-2180-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang HB, Sheu BS, Wang ST, Cheng HC, Chang WL, Chen WY. H. pylori eradication prevents the progression of gastric intestinal metaplasia in reflux esophagitis patients using long-term esomeprazole. Am J Gastroenterol. 2009;104(7):1642–9. doi: 10.1038/ajg.2009.172. [DOI] [PubMed] [Google Scholar]
- 37.Chang WL, Yang HB, Cheng HC, Chuang CH, Lu PJ, Sheu BS. Increased gastric osteopontin expression by Helicobacter pylori Infection can correlate with more severe gastric inflammation and intestinal metaplasia. Helicobacter. 2011;16(3):217–24. doi: 10.1111/j.1523-5378.2011.00832.x. [DOI] [PubMed] [Google Scholar]
- 38.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston. Am J Surg Pathol 1996. 1994;20(10):1161–81. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Yeh YC, Cheng HC, Yang HB, Chang WL, Sheu BS. H. pylori CagL-Y58/E59 prime higher integrin alpha5beta1 in adverse pH condition to enhance hypochlorhydria vicious cycle for gastric carcinogenesis. PLoS One. 2013;8(8):e72735. doi: 10.1371/journal.pone.0072735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh YC, Chang WL, Yang HB, Cheng HC, Wu JJ, Sheu BS. H. pylori cagL amino acid sequence polymorphism Y58E59 induces a corpus shift of gastric integrin alpha5beta1 related with gastric carcinogenesis. Mol Carcinog. 2011;50(10):751–9. doi: 10.1002/mc.20753. [DOI] [PubMed] [Google Scholar]
- 41.Rad R, Dossumbekova A, Neu B, Lang R, Bauer S, Saur D, et al. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during Helicobacter pylori infection. Gut. 2004;53(8):1082–9. doi: 10.1136/gut.2003.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F, Pan K, Zhang X, Zhang Y, Zhang L, Ma J, et al. Genetic variants in cyclooxygenase-2: Expression and risk of gastric cancer and its precursors in a Chinese population. Gastroenterology. 2006;130(7):1975–84. doi: 10.1053/j.gastro.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 43.Hung KH, Wu JJ, Yang HB, Su LJ, Sheu BS. Host Wnt/beta-catenin pathway triggered by Helicobacter pylori correlates with regression of gastric intestinal metaplasia after H. pylori eradication. J Med Microbiol. 2009;58(Pt 5):567–76. doi: 10.1099/jmm.0.007310-0. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi H, Goldenring JR, Kaminishi M, Lee JR. Identification of spasmolytic polypeptide expressing metaplasia (SPEM) in remnant gastric cancer and surveillance postgastrectomy biopsies. Dig Dis Sci. 2002;47(3):573–8. doi: 10.1023/A:1017920220149. [DOI] [PubMed] [Google Scholar]
- 45.Yoshizawa N, Takenaka Y, Yamaguchi H, Tetsuya T, Tanaka H, Tatematsu M, et al. Emergence of spasmolytic polypeptide-expressing metaplasia in Mongolian gerbils infected with Helicobacter pylori. Lab Invest. 2007;87(12):1265–76. doi: 10.1038/labinvest.3700682. [DOI] [PubMed] [Google Scholar]


