Abstract
Encapsidation of the Moloney murine leukemia virus (MMLV) genome is mediated through a specific interaction between the major viral structural protein, Gag, and an RNA packaging signal, Ψ. Many studies have investigated this process in vivo, although the specific examination of the Gag-RNA interaction in this context is difficult due to the variety of other viral functions involved in virion assembly in vivo. The Saccharomyces cerevisiae three-hybrid assay was used to directly examine the interaction between MMLV Gag and Ψ. In this system, MMLV RNA regions exhibiting high-affinity Gag binding were mapped. All Gag-binding regions were located 3′ to the viral splice donor sequence of the viral RNA transcript. No single short RNA sequence within Ψ supported strong Gag interaction. Instead, an RNA comprised of nearly the entire Ψ region was necessary to demonstrate an appreciable Gag interaction in the yeast three-hybrid system. These finding support the notion that two stem-loops (C and D) are not sufficient to form a core MMLV encapsidation signal.
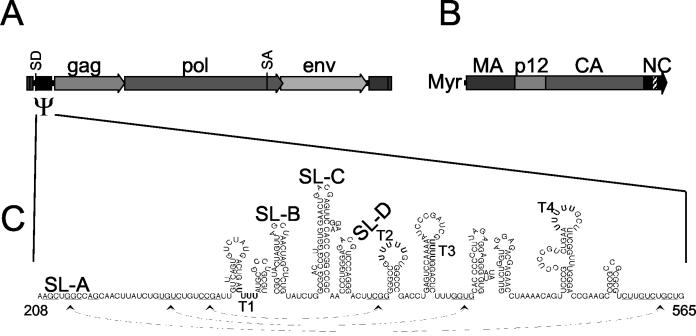
Packaging of retrovirus genomes, which amount to only approximately 1% of the total mRNA in the cytoplasm of an infected cell, requires a mechanism to selectively recognize and recruit viral RNAs during virion assembly (8). This process is mediated by the viral structural protein precursor, Gag, which specifically binds a cis-acting RNA packaging signal (4, 8, 34). For Moloney murine leukemia virus (MMLV), a single zinc finger and surrounding basic residues in the nucleocapsid (NC) region of the Gag protein are thought to possess the major determinants for the specificity and affinity of the interaction (Fig. 1A and B) (11, 44). The MMLV packaging signal, termed Ψ, was originally defined through the characterization of a viral mutant that lacked the ability to package viral RNA and whose genome possessed a large deletion from nucleotides (nt) 210 to 560 (Fig. 1A and C) (28). Analysis of viral mutants indicated that sequences downstream of nucleotide position 400 were dispensable for encapsidation, further defining the essential Ψ region to lie between nt 215 and 400 of the MMLV genome (36). Although these studies defined the minimal Ψ region, other reports have suggested the presence of additional RNA packaging signals throughout the MMLV genome (7, 31, 43).
FIG. 1.
MMLV cis- and trans-acting packaging elements. (A) The MMLV genome. The three MMLV protein-coding sequences are marked, as are the splice donor (SD) and splice acceptor (SA) sites. (B) Diagram of the MMLV Gag protein. Major domains are labeled, as well as the amino-terminal myristyl group (Myr). The position of the zinc finger in NC is indicated by the hatched box. (C) RNA sequence and structures of the extended MMLV packaging signal. SL-A to SL-D and potential Pol III terminator sequences (T1 to T4) are labeled. Sequences that may interact to form tertiary structures are indicated by arrows underneath the sequence.
Numerous techniques, including computer prediction, chemical accessibility, and directed mutagenesis, have been used to determine the secondary structure of the MMLV Ψ region (2, 23, 42) (Fig. 1C). Many stem-loops (SL) and tertiary contacts are predicted in this region, with the major structural features designated SL-A through SL-D. Most functional mapping studies of the MMLV packaging signal have involved mutagenesis of the region and characterization of the effects of these mutations on the viral life cycle (17, 29, 30). These studies have shown a critical role for SL-C and -D, with contributions from upstream elements. Thus, SL-C and -D are considered to contain the core MMLV RNA encapsidation signal.
The region between the MMLV 5′ long terminal repeat and the start of the Gag open reading frame contains sequences required for tRNA primer binding, reverse transcription, and RNA splicing (13). The same region appears to be involved in genomic RNA dimerization (15, 20, 32, 35, 42). The close proximity of all these functional domains to the encapsidation signal may complicate interpretation of in vivo studies of this region, as it is often difficult to ascertain the life cycle stage at which mutations in this area exert their effect. Detailed examination of the MMLV Gag-Ψ interaction would be facilitated by an in vitro system that can directly examine this association out of the context of other viral functions. For other retroviruses, biochemical methods, such as gel shift experiments, have been used to investigate the requirements and kinetics of the protein-RNA association (9, 10). These types of experiments have proven difficult with MMLV, as purification of MMLV Gag precursor protein retaining specific RNA-binding ability has not been achieved.
The Saccharomyces cerevisiae three-hybrid system (37) has been used to investigate the Gag-RNA packaging signal interaction of other retroviruses, such as Rous sarcoma virus (24-26) and human immunodeficiency virus type 1 (HIV-1) (5). In this system, a fusion protein of the LexA DNA-binding domain with the phage MS2 coat protein is used to tether an RNA containing MS2-binding sequences and a sequence of interest to the operator of a reporter gene in yeast. A fusion of the Gal4 activation domain (GAD) and an RNA-binding protein can be recruited to the operator and activate transcription of the reporter if the RNA-binding protein specifically interacts with the tethered RNA (12, 37). Changes to the variable components, the GAD-RNA-binding protein prey or the RNA bait, allow for study of the RNA-binding characteristics of a given protein or the mapping of cis-acting RNA signals. As an example, in the yeast three-hybrid system, a GAD-HIV Gag fusion has been shown to bind specifically to RNA baits containing elements of the HIV encapsidation signal, illustrating which elements are required for this interaction (5).
Initial attempts to utilize the yeast three-hybrid system to study MMLV Gag-ψ binding showed little to no RNA-binding specificity with an MMLV Gag prey (5). Here, this issue is revisited, and the yeast three-hybrid system is successfully used to study the MMLV Gag-Ψ interaction. In this system, MMLV Gag has little affinity for RNA baits containing only SL-C and -D, while RNA baits containing multiple RNA elements or the entire packaging signal display a positive and specific interacting ability. Some mutations to the packaging signal, which have previously been shown to greatly impair packaging in vivo, had no effect on the MMLV Gag-ψ interaction in this system.
MATERIALS AND METHODS
Plasmid construction.
Saccharomyces cerevisiae three-hybrid components were generously provided by Marvin Wickens (12, 37). Standard cloning techniques were employed, and PCR was performed with the ExpandHiFi PCR kit (Roche). RNA baits were PCR amplified from the MMLV proviral DNA in the plasmid pNCA (14) and cloned into the RNA expression vector pIIIA/MS2-2 at SmaI and SphI sites (oligonucleotide sequences available upon request). Site-directed mutagenesis was conducted by two overlapping rounds of PCR with mutation-specific inside oligonucleotides and vector-specific outside oligonucleotides (sequences available upon request). All cloned regions were confirmed by sequencing. The VL30 RNA expression vector (HaMSVψ-MS2) has been previously described (5). The GADMoGag prey expression plasmid contains the complete MMLV Gag gene cloned into the Gal4 activation domain vector, GADNOT, at the BamHI and SalI sites as previously described (27).
Yeast three-hybrid assay.
Saccharomyces cerevisiae strain L40-coat [MATa ura3-52 leu2-3,112 his3-200 trp1-1 ade2 LYS2::(lexA-op)-HIS3 ura3::(LexA-op)-LacZ LexA-MS2coat(TRP1)] is an ura3 derivative of the L40 yeast strain with an integrated LexA-MS2 coat protein expression cassette (37). Plasmids were transformed by the standard lithium acetate method (38). Colonies were qualitatively and quantitatively assayed for β-galactosidase activity by filter lift and o-nitrophenyl-β-d-galactopyranoside (ONPG) liquid assays, respectively, as previously described (40).
Screen for Gag-interacting MMLV RNA regions.
To construct the random MMLV RNA expression library, the plasmid PNCS, containing an MMLV provirus genome, was fragmented by nebulization, where hydrodynamic forces are used to shear DNA into small fragments (39). These fragments were “polished” with polymerases, phosphorylated with T4 polynucleotide kinase (New England Biolabs), and then size fractionated by agarose gel electrophoresis. DNA fragments of approximately 100 to 500 bp were blunt end cloned into the yeast RNA bait expression construct, pIIIA/MS2-2, at the SmaI restriction site. Approximately 20,000 independent clones were generated in this manner, a number large enough to represent the entire 10-kb PNCS plasmid. Yeast colonies cotransformed with this library and GAD-MoGag were looked for by a filter-lift assay. Colonies exhibiting higher than background β-galactosidase activity were isolated and expanded. Total cellular DNA was purified from these yeast cells and electroporated into KC8 bacteria to rescue the RNA expression plasmid for sequence analysis.
Northern blot analysis of RNA bait expression in yeast.
The equivalent of one OD600 (optical density at 600 nm) unit of yeast expressing varied RNA baits, grown in culture to log phase, was pelleted for each RNA preparation. Cell walls were removed by zymolyase (Zymo Research, Orange, Calif.) digestion, and total RNA was extracted using an RNeasy RNA miniprep kit (Qiagen), according to the manufacturer's instructions. Total RNA (1.5 μg) was resolved by denaturing formaldehyde agarose gel electrophoresis and transferred to nylon membranes. For a probe, a PCR fragment of the dual-MS2 coat sequences, generated by amplification of the vector pIIIA/MS2-2 with oligonucleotides MS2-2F (5′ TGG GAG CTG CGA TTG GCA GAA TTC CGG CTA GAA CTA GTG GAT CCC CCG GG) and MS2-2R (5′ CTG CAG ACA TGG GTG ATC CTC ATG TTT TCT AGA GTC GAC CTG CAG GCA TGC), was labeled with the High Prime kit (Roche), according to the manufacturer's instructions. The blot was hybridized with this probe, washed, and exposed to film.
RESULTS
Establishing the yeast three-hybrid assay for the MMLV Gag-RNA interaction.
Previous efforts to study the MMLV Gag-Ψ interaction in the yeast three-hybrid system detected only weak, nonspecific binding between an Gal4AD-MMLV Gag (GAD-MoGag) prey and an RNA bait containing SL-C and SL-D of MMLV Ψ (nt 305 to 381 of the MMLV genome) (5). Since the Gag prey was well expressed and behaved well in yeast two-hybrid assays (3, 18, 27), it seemed likely that the failure of the previous yeast three-hybrid assay was due to the lack of a suitable RNA bait.
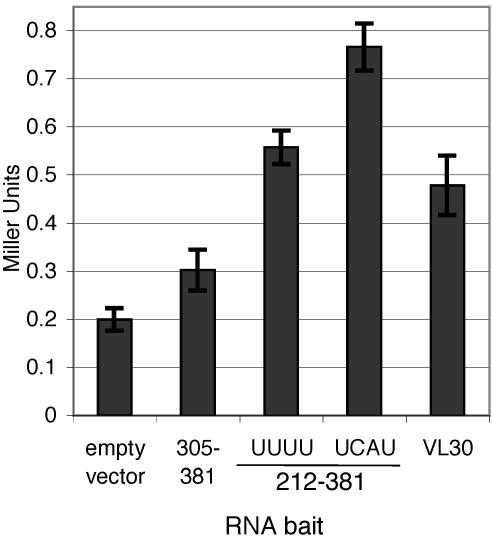
To create a more appropriate RNA bait, various regions of the MMLV genome were cloned as RNA baits and tested for GAD-MoGag-binding activity in yeast. In accordance with the previous study (5), reporter activation with an RNA bait containing SL-C and SL-D was barely above the background level, as determined by an assay against an RNA bait expressed from the empty RNA expression vector (Fig. 2, empty vector and RNA bait 305-381). The nonspecific RNA-binding activity of the Gag prey likely reflects a bona fide characteristic of MMLV Gag and should not disqualify this prey. An RNA bait containing SL-A through -D (MMLV nt 212 to 381) exhibited a significant interaction with the Gag prey, as the reporter activity in this assay was nearly threefold that of the background level (Fig. 2, empty vector and bait 212-381 UUUU). This increase over background levels provided the first evidence of a positive and specific interaction between MMLV Gag and Ψ in the yeast three-hybrid system.
FIG. 2.
Quantitative analysis of yeast three-hybrid interactions. Yeast cells were transformed with the MMLV Gag prey expression vector, along with various RNA bait expression vectors indicated on the x axis. Regions of MMLV present in RNA baits are indicated by nucleotide positions relative to the retroviral genome. Reporter activation was quantified by β-galactosidase assay and given in Miller units (see Materials and Methods). Values are means ± standard deviations (error bars) of at least three independent cultures.
The RNA bait in the yeast three-hybrid system is expressed from an RNA polymerase III (Pol III) promoter. Therefore, it may be difficult to express full-length RNA baits that contain uridine repeats, which function as Pol III transcriptional terminators (19). A grouping of four uridines is generally considered a weak Pol III terminator, while five or more uridines usually result in complete Pol III transcriptional termination. The presence of either a weak or strong Pol III termination signal in an RNA bait would typically reduce or prevent production of a full-length RNA species and thus interfere with examination of interactions with such a bait in the yeast three-hybrid system.
Examination of the MMLV Ψ region showed that four uridine stretches were present, with two regions of four and five uridines each (Fig. 1C). The Ψ bait tested above (bait 212-381) encoded a single group of four uridines at nucleotide positions 262 to 265. To determine whether expression of this RNA bait was impaired by this putative Pol III terminator, this stretch of uridines was disrupted by site-directed mutagenesis (UUUU to UCAU, see Materials and Methods), and the mutant bait was tested in the yeast three-hybrid assay for interaction with MMLV Gag. Disruption of this putative terminator increased Gag-binding ability to nearly fourfold over the background level (Fig. 2, empty vector and bait 212-381 UCAU), a reporter signal 1.4-fold greater than for the unmodified RNA bait (Fig. 2, bait 212-381 UUUU and UCAU). The enhancement of reporter activation caused by disruption of this putative termination signal was viewed as significant, especially considering that the repeat of four uridines may be only a weak terminator. To avoid premature transcriptional termination of RNA baits in subsequent experiments, all baits were constructed with putative Pol III terminators disrupted by site-directed mutagenesis.
To further demonstrate that the reporter activity observed in these yeast three-hybrid assays reflected specific MMLV Gag-RNA-binding activity, an RNA bait containing the rodent VL30 retrotransposable element was tested for interaction with the MMLV Gag prey. The VL30 sequence functions as the packaging signal for the Harvey murine sarcoma virus (HaMSV) and, although it bears little to no sequence or structural similarity to MMLV Ψ, has been shown to allow efficient MMLV packaging of exogenous RNAs (21, 41). Reporter activity when the Gag prey was tested against the VL30 RNA bait was approximately 2.5-fold higher than with the empty RNA expression vector alone (Fig. 2), indicating a specific MMLV Gag-VL30 interaction. This result suggests that the yeast three-hybrid assay faithfully reflected MMLV Gag RNA-binding preferences.
Mapping the Gag-binding region.
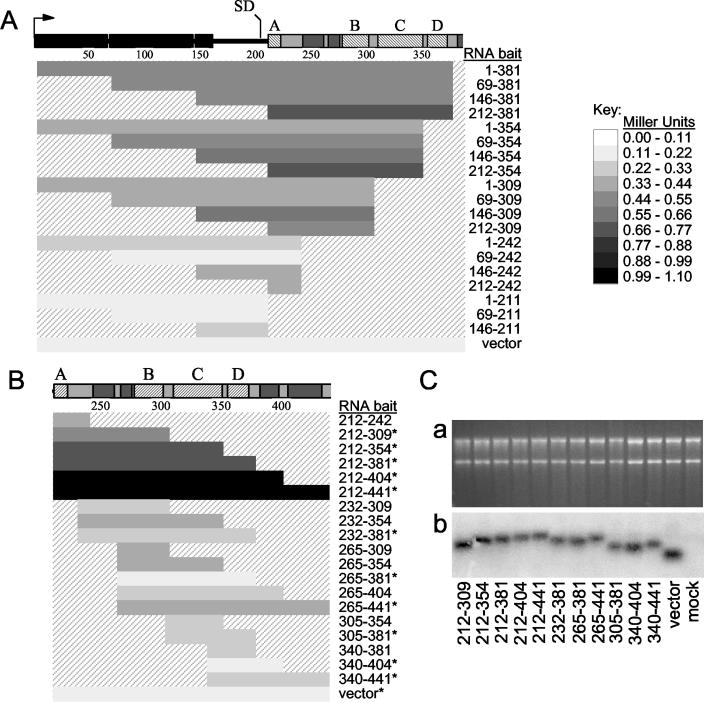
To further define the MMLV RNA regions required for efficient Gag binding, a series of overlapping RNA baits, spanning much of the MMLV 5′ untranslated region from the MMLV transcriptional initiation site to nt 441 (Fig. 3A and B), was constructed and tested for Gag interaction capacity. Putative Pol III terminator sequences were disrupted by minimal nucleotide substitutions. The ability of these RNAs to interact with the Gag prey was scored by reporter activation in the yeast three-hybrid system (Fig. 3A and B).
FIG. 3.
Quantitative mapping of MMLV ψ-binding activity in the yeast three-hybrid system. (A and B) Various RNA baits were tested for Gag interaction in the yeast three-hybrid system. The bars correspond to the MMLV sequences tested in the RNA baits, with nucleotide positions relative to the genome shown to the right of the bars. Reporter activation observed between the RNA bait and Gag prey in yeast, as quantified by β-galactosidase assay, is indicated by the shading of each bar. A scale relating the strength of reporter activation in Miller units to the shading of the bars is indicated to the right of the panel. Values are averages of at least three independent measurements, and standard deviations were within 10%. The RNA expression vector with no cloned MMLV sequences was tested to represent the nonspecific RNA-binding activity of Gag prey. (A) Analysis of RNA regions 5′ of the viral SD with and without Ψ sequences. (B) Mapping of Gag-interacting regions within MMLV Ψ. The RNA baits that were also quantified by Northern blot analysis (as shown in panel C) are indicated by asterisks. (C) Northern blot analysis of RNA bait expression in yeast. (a) Ethidium bromide-stained agarose gel of total yeast RNA analyzed by Northern blotting. The blot was hybridized with a radioactive probe specific for the MS2 coat protein-binding region, present in all baits, and exposed for autoradiography (b). The mock lane contains RNA from yeast cells not transformed with RNA expression vector.
In the yeast three-hybrid system, only RNA baits containing MMLV Ψ sequence demonstrated appreciable GAD-MoGag binding; no Gag-interacting RNA sequences were evident 5′ of the MMLV splice donor (SD) (Fig. 3A). An RNA bait comprised of the entire MMLV genome 5′ of the SD (Fig. 3A, bait 1-211) showed only background reporter activity, indicating that no specific Gag-binding elements were present in this region. In addition, the Gag-binding activity of an RNA bait was never increased, and often decreased, by the inclusion of sequence 5′ of the SD.
To define the sequences within Ψ most important for interaction with MMLV Gag, a series of overlapping RNA baits representing various regions of Ψ was tested against the Gag prey in the yeast three-hybrid system (Fig. 3B). No single RNA structural element appeared capable of strongly interacting with Gag in this system, as RNA baits containing these small MMLV regions within Ψ showed little to no reporter activation above background levels. Larger combinations of multiple RNA elements displayed GAD-MoGag interaction levels significantly above background levels. The highest signal was achieved when the MMLV Gag prey was assayed against the two longest RNA baits (Fig. 3B, baits 212-404 and 212-441). These RNA regions were larger than the previously defined minimal Ψ region (bait 215-400) (1) and included not only SL-A through SL-D, but one or two additional predicted downstream SLs.
To rule out the possibility that RNA bait expression level or stability differences affected the above assays, the steady-state levels of RNA baits were examined by Northern blotting. RNAs were isolated from yeast cells, resolved by agarose gel electrophoresis, and transferred to nitrocellulose membrane, and the Ψ-MS2 RNAs were detected by hybridization with a probe specific for the MS2 coat protein-binding sequences. All RNA baits were expressed at approximately equal levels (Fig. 3C). Importantly, no correlation between RNA abundance and reporter activation was observed. These results indicate that the differences in reporter activity with different RNA baits are not a consequence of steady-state expression differences between these RNAs in yeast.
Mutations known to disrupt RNA packaging have no effect in the yeast three-hybrid assay.
A number of studies have analyzed the MMLV RNA structures required for packaging during viral replication in vivo, but these studies did not directly examine the interaction between Gag and the viral RNA. Therefore, it was of interest to test how mutations previously shown to impair RNA encapsidation in vivo would affect the interaction with MMLV Gag in the yeast three-hybrid system. A report from Fisher and Goff (17) described an analysis of the effects of MMLV Ψ mutations on in vivo RNA encapsidation and viral spread. In this study, SL-C and SL-D mutations were shown to have dramatic effects on viral RNA packaging. Two such mutations were placed in Ψ as RNA baits and tested for their effects on the Gag-Ψ interaction in the yeast three-hybrid system.
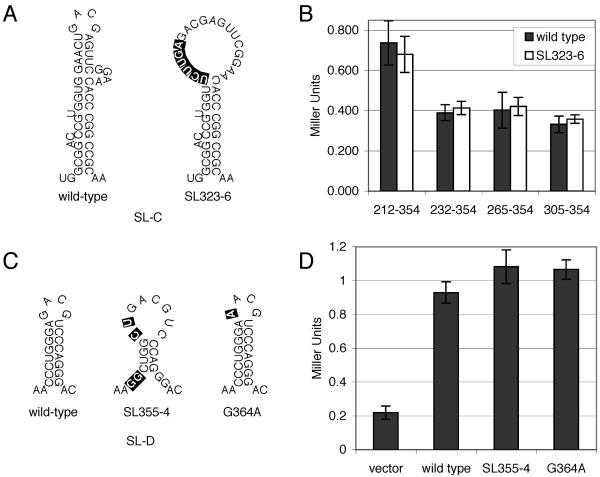
One mutation tested, a disruption of the top of the stem region of SL-C (SL323-6 [Fig. 4A ]), caused an in vivo deficiency in RNA packaging (>10-fold by RNase protection assay of purified virions) and a delay in viral spread (approximately 2 or 3 days by reverse transcriptase assay) (17). To assay the effects of this mutation in the yeast three-hybrid system, this mutation was incorporated into progressively smaller regions of Ψ containing SL-C RNA baits, and these RNAs were assayed for interaction with GAD-MoGag. When compared to wild-type baits, these mutant baits interacted equivalently with the MMLV Gag prey (Fig. 4B). Even a bait that contained only SL-C (nt 305 to 354), although a very weak interactor, bound identically to Gag with or without this mutation, ruling out the possibility that multiple contacts between the protein and RNA bait may have masked the effects of this mutation on the Gag interaction.
FIG. 4.
Analysis of SL-C and SL-D mutant RNA baits. (A) RNA structures of wild-type and SL323-6 mutant (17) SL-C structures. (B) Quantitative analysis of effects of SL323-6 mutation on interaction of MMLV Gag with various MMLV RNA baits. Numbering on the x axis indicates the region of the MMLV genome present in RNA bait encoding either the wild-type or SL323-6 mutant SL-C sequence. (C) RNA structures of wild-type, SL355-4 mutant, and G364A mutant (17) SL-D structures. (D) Quantitative analysis of the effects of SL-D mutations in the MMLV 212-441 RNA bait on interaction with MMLV Gag prey. The RNA baited tested is indicated on the x axis. Reporter activity is quantified in Miller units by β-galactosidase assay. Values are means ± standard deviations (error bars) for three independent cultures.
Two mutations in SL-D (SL355-4 and G364A [Fig. 4C]), each exhibiting dramatic phenotypes in vivo (17), were tested in the context of the strongest Gag-binding RNA bait (nt 212 to 441) in the yeast three-hybrid system. The SL355-4 mutation disrupts the base and top of the SL-D stem. This mutation reduced RNA incorporation into virions by more than 100-fold and severely disrupted replication ability, almost abolishing viral spread as detected by reverse transcriptase assay (17). A second SL-D mutation, G364A, changed the first G in the SL-D tetraloop to an A. This mutation was previously shown to almost completely disrupt viral replication in vivo (17). Surprisingly, neither of these mutations had any detectable effect on the ability of the MMLV RNA bait (bait 212-441) to interact with the Gag prey in the yeast three-hybrid system (Fig. 4D). These results indicate that these Ψ mutations do not affect the interaction with the Gag precursor protein per se and suggest that these sequences, previously thought to constitute the core MMLV encapsidation signal, are dispensable to the MMLV Gag-RNA interaction.
Screen for Gag-interacting RNAs.
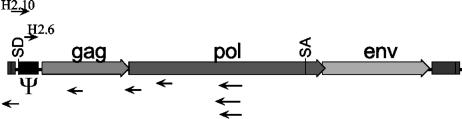
As an alternative approach to define MMLV Ψ and to search for additional cryptic binding sites, the MMLV genome was screened for high-affinity Gag-binding RNA sequences. Randomly generated MMLV genome fragments were cloned into the MS2-RNA expression vector. This RNA expression library was then screened against the Gag prey, with positive clones identified by filter lift assay (see Materials and Methods). Of 1,500 yeast colonies screened, 9 positive clones were isolated (Fig. 5). Seven of these positive clones encoded MMLV RNAs cloned in the reverse orientation, such that they represent antisense RNAs (Fig. 5). The significance of the binding of these RNAs by Gag, if any, is unknown. Since these elements would not be expressed in an infected cell in packageable RNAs, these interactions were considered irrelevant to the normal life cycle of the virus. Two hits contained MMLV sequence cloned in the correct, sense-strand orientation (Fig. 5). These baits contained nonidentical, overlapping portions of the MMLV Ψ region. Hit 2.10 encompassed the RNA region from the viral promoter through the first half of SL-C (MMLV nt 53 to 334) and was measured by β-galactosidase assay to give a signal approximately threefold over background levels. Hit 2.6 contained SL-C and SL-D plus a small amount of flanking RNA (MMLV nt 304 to 376) and interacted weakly over the control. Although these RNAs showed only weak binding, their recovery suggests that, of the short sense RNAs from the viral genome, the 5′ Ψ region contains those sequences with the highest Gag-binding affinity.
FIG. 5.
Diagram indicating location and orientation of RNA sequences recovered from random screen of MMLV genome fragments for Gag binding. Hits containing antisense MMLV sequences are shown below the genome. The positions of two hits, labeled H2.10 and H2.6, identified in the sense orientation are indicated above the genome. SA, splice acceptor site.
DISCUSSION
In this report, the yeast three-hybrid system was used to study the interaction between MMLV Gag and the RNA packaging signal, Ψ. Although the MMLV Gag prey exhibited some nonspecific RNA-binding activity, most likely a characteristic of the native viral protein, a specific interaction between this protein and viral RNA sequences involved in genome packaging could be detected and quantified. By testing defined MMLV sequences in the assay, the minimal Gag-binding sequences of MMLV Ψ were mapped (Fig. 3). No individual short RNA structural elements displayed high MMLV Gag-binding activity (Fig. 3B). The RNA bait exhibiting the strongest MMLV Gag interaction contained the majority of the previously described Ψ region (Fig. 3B, bait 212-441). In addition, no specific MMLV Gag-binding sites were detected 5′ of the MMLV SD (Fig. 3A) in accordance with the notion that packaging elements should be present only in full-length viral RNA and not in the subgenomic env transcript.
Screening the MMLV genome successfully identified fragments of the packaging region as Gag-interacting RNAs, while no other binding sites in the sense orientation of the viral RNA were isolated (Fig. 5). In this screen, isolation of Ψ regions may have been complicated by the presence of Pol III terminators throughout this region, as discussed above. Generation of an RNA expression library from a proviral plasmid with engineered disruptions of these terminators may be required for a more comprehensive screen of Gag-binding sequences. Regardless, this screen suggests that high-affinity Gag-binding sequences in the MMLV genome are predominantly located to the previously defined Ψ region.
Multiple considerations suggest that the RNA structure that serves as the target for Gag binding in the yeast three-hybrid assay may differ significantly from that of the RNA genome during packaging in vivo. In addition, there may well be multiple conformations for the viral RNA, and we cannot know whether the RNA baits have access to all conformations. The RNA being tested is fused to MS2 RNA sequences and thus is not in its normal context. The MS2 sequences could disrupt the folding of the MMLV sequences and might even base pair with them. Although RNA structure predictions made with and without the added MS2 sequences did not indicate significant interactions of MS2 (data not shown), we cannot rule out the possibility that these chimeric RNA baits may fold abnormally.
In addition, the oligomeric state of the MMLV RNA baits in yeast is unknown. These RNA baits are presumably formed as monomers but could dimerize if multiple copies are brought together at high local concentrations by binding to the MS2 protein. The interaction with Gag could also promote dimerization, or otherwise alter the RNA conformation. It should be noted that the importance of dimerization in packaging in vivo is also unclear (15, 16, 22, 33). The viral RNA is thought to form a weak or unstable dimer early in virion assembly, and then a more stable dimer at later times, after the Gag protein is processed to form NC. The sequences involved in dimer formation overlap with the packaging elements, and it has been suggested that the dimerization of viral RNAs can affect the interaction with NC. In spite of these limitations, the yeast three-hybrid system usefully detects protein-RNA interactions that recreate features of the interaction in vivo. Among the various conformations that may be formed, it is likely that the yeast system will report that conformation that provides the tightest binding and the strongest reporter signal.
A number of other retroviral packaging signals have also been studied recently using the yeast three-hybrid system, including those of Rous sarcoma virus (24-26) and HIV-1 (5). For the avian retroviruses, studies in this system have identified a high-affinity interaction between the Gag protein and a large viral RNA element with complex secondary structure (24). This interaction was impaired by a wide range of mutations affecting the RNA, altering not only secondary structures, such as stem and bulge elements, and but also changing sequences in putatively single-stranded regions (24-26). In addition to investigating the RNA elements involved in these interactions, the yeast three-hybrid system has been used to further define the residues of the Gag protein required to bind the viral RNA (24, 25).
The findings presented here suggest that short RNA elements important for packaging in vivo do not function as simple high-affinity binding sites for Gag. Indeed, the RNA elements thought to be most important for packaging in vivo, the SL-C and D sequences, are not sufficient for tight binding of Gag. Rather, a much longer RNA sequence is apparently required for the best interaction. In addition, much of the SL-C and D structures are not even necessary for strong binding. Mutations that significantly disrupt the loops or the base pairing in the stems of these structures, and that have been shown to disrupt packaging, did not affect Gag binding in the yeast assay (Fig. 4).
These results are consistent with recent findings suggesting that NC interacts not with nucleotides at the tips of the SL of Ψ, but rather with sequences at the base of SL-B and SL-C (6, 16). In one of these studies, tests for high-affinity binding of recombinant MMLV NC to RNA oligonucleotides in vitro revealed the requirement for a long sequence extending from SL-B to -D, with only weak interactions observed between MMLV NC and any of several smaller subregions (16). In addition, the GACG loops of SL-C and -D or the tetraloop of SL-B did not did not appreciably contribute to the interaction, much as we observed here. It should be noted that these in vitro binding experiments were performed with NC, while our binding in yeast utilized the Gag precursor protein. Nevertheless, these correlations suggest that binding in vitro is similar in its RNA sequence requirements to binding as assayed in yeast cells.
It remains unclear what the roles the SL-C and SL-D structures may have in RNA packaging, if they are not involved in direct Gag binding. Perhaps these structures are needed for RNA folding or for intracellular transport to the site of assembly, before the interaction with Gag takes place. Another possibility is that they are important after an initial Gag binding to promote conformational changes in the RNA that might be required for retention during virion assembly. Viral RNA may serve to multimerize Gag precursors and so promote virion assembly itself, and although other RNAs can perform this task in its absence, specific viral RNA structures may facilitate assembly. We suspect that the abilities to rapidly score Gag-RNA binding and to screen and select for mutants with altered affinity in the yeast three-hybrid system will be useful in the future in further defining the requirements in both RNA and protein for their interaction.
Acknowledgments
We thank Daniel Shaye and Carina Storrs for helpful discussions and reading early versions of the manuscript.
This work was partially supported by Public Health Service grant CA 30488 from the National Cancer Institute. M.J.E. was supported by an institutional training grant to Columbia University from the NCI. E.B. was an Associate and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1988. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 62:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford, R. L., S. Honda, C. B. Lawrence, and J. W. Belmont. 1991. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology 183:611-619. [DOI] [PubMed] [Google Scholar]
- 3.Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. [DOI] [PubMed] [Google Scholar]
- 4.Aronoff, R., and M. Linial. 1991. Specificity of retroviral RNA packaging. J. Virol. 65:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacharach, E., and S. P. Goff. 1998. Binding of the human immunodeficiency virus type 1 Gag protein to the viral RNA encapsidation signal in the yeast three-hybrid system. J. Virol. 72:6944-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beasley, B. E., and W. S. Hu. 2002. cis-Acting elements important for retroviral RNA packaging specificity. J. Virol. 76:4950-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, M. A., T. D. Palmer, R. E. Gelinas, and A. D. Miller. 1987. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J. Virol. 61:1639-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233-246. [DOI] [PubMed] [Google Scholar]
- 10.Berkowitz, R. D., J. Luban, and S. P. Goff. 1993. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 67:7190-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkowitz, R. D., A. Ohagen, S. Hoglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein, D. S., N. Buter, C. Stumpf, and M. Wickens. 2002. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26:123-141. [DOI] [PubMed] [Google Scholar]
- 13.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 14.Colicelli, J., and S. P. Goff. 1985. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell 42:573-580. [DOI] [PubMed] [Google Scholar]
- 15.De Tapia, M., V. Metzler, M. Mougel, B. Ehresmann, and C. Ehresmann. 1998. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374). Biochemistry 37:6077-6085. [DOI] [PubMed] [Google Scholar]
- 16.D'Souza, V., J. Melamed, D. Habib, K. Pullen, K. Wallace, and M. F. Summers. 2001. Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus ψ-RNA packaging signal: implications for genome recognition. J. Mol. Biol. 314:217-232. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, J., and S. P. Goff. 1998. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology 244:133-145. [DOI] [PubMed] [Google Scholar]
- 18.Franke, E. K., H. E. Yuan, K. L. Bossolt, S. P. Goff, and J. Luban. 1994. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J. Virol. 68:5300-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiduschek, E. P., and G. P. Tocchini-Valentini. 1988. Transcription by RNA polymerase III. Annu. Rev. Biochem. 57:873-914. [DOI] [PubMed] [Google Scholar]
- 20.Girard, P. M., B. Bonnet-Mathoniere, D. Muriaux, and J. Paoletti. 1995. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry 34:9785-9794. [DOI] [PubMed] [Google Scholar]
- 21.Hatzoglou, M., C. P. Hodgson, F. Mularo, and R. W. Hanson. 1990. Efficient packaging of a specific VL30 retroelement by ψ2 cells which produce MoMLV recombinant retroviruses. Hum. Gene Ther. 1:385-397. [DOI] [PubMed] [Google Scholar]
- 22.Kim, C. H., and I. Tinoco, Jr. 2000. A retroviral RNA kissing complex containing only two G · C base pairs. Proc. Natl. Acad. Sci. USA 97:9396-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konings, D. A., M. A. Nash, J. V. Maizel, and R. B. Arlinghaus. 1992. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J. Virol. 66:632-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, E., A. Yeo, B. Kraemer, M. Wickens, and M. L. Linial. 1999. The gag domains required for avian retroviral RNA encapsidation determined by using two independent assays. J. Virol. 73:6282-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, E. G., A. Alidina, C. May, and M. L. Linial. 2003. Importance of basic residues in binding of Rous sarcoma virus nucleocapsid to the RNA packaging signal. J. Virol. 77:2010-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, E. G., and M. L. Linial. 2000. Yeast three-hybrid screening of Rous sarcoma virus mutants with randomly mutagenized minimal packaging signals reveals regions important for gag interactions. J. Virol. 74:9167-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luban, J., K. B. Alin, K. L. Bossolt, T. Humaran, and S. P. Goff. 1992. Genetic assay for multimerization of retroviral Gag polyproteins. J. Virol. 66:5157-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann, R., R. C. Mulligan, and D. Baltimore. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33:153-159. [DOI] [PubMed] [Google Scholar]
- 29.Mougel, M., and E. Barklis. 1997. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 71:8061-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mougel, M., Y. Zhang, and E. Barklis. 1996. cis-Active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J. Virol. 70:5043-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, J. E., and S. P. Goff. 1989. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J. Virol. 63:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paoletti, J., M. Mougel, N. Tounekti, P. M. Girard, C. Ehresmann, and B. Ehresmann. 1993. Spontaneous dimerization of retroviral MoMuLV RNA. Biochimie 75:681-686. [DOI] [PubMed] [Google Scholar]
- 33.Prats, A. C., C. Roy, P. A. Wang, M. Erard, V. Housset, C. Gabus, C. Paoletti, and J. L. Darlix. 1990. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 64:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513-522. [DOI] [PubMed] [Google Scholar]
- 35.Roy, C., N. Tounekti, M. Mougel, J. L. Darlix, C. Paoletti, C. Ehresmann, B. Ehresmann, and J. Paoletti. 1990. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 18:7287-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartzberg, P., J. Colicelli, and S. P. Goff. 1983. Deletion mutants of Moloney murine leukemia virus which lack glycosylated Gag protein are replication competent. J. Virol. 46:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SenGupta, D. J., B. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93:8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi, Y., K. Alin, and S. P. Goff. 1995. Abl-interactor-1, a novel SH3 protein binding to the carboxy-terminal portion of the Abl protein, suppresses v-abl transforming activity. Genes Dev. 9:2583-2597. [DOI] [PubMed] [Google Scholar]
- 39.Surzycki, S. J. 2000. Basic methods in molecular biology. Springer-Verlag, New York, N.Y.
- 40.Tachedjian, G., H. E. Aronson, M. de los Santos, J. Seehra, J. M. McCoy, and S. P. Goff. 2003. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J. Mol. Biol. 326:381-396. [DOI] [PubMed] [Google Scholar]
- 41.Torrent, C., P. Wang, and J. L. Darlix. 1992. A murine leukemia virus derived retroviral vector with a rat VL30 packaging ψ sequence. Bone Marrow Transplant. 9:143-147. [PubMed] [Google Scholar]
- 42.Tounekti, N., M. Mougel, C. Roy, R. Marquet, J. L. Darlix, J. Paoletti, B. Ehresmann, and C. Ehresmann. 1992. Effect of dimerization on the conformation of the encapsidation ψ domain of Moloney murine leukemia virus RNA. J. Mol. Biol. 223:205-220. [DOI] [PubMed] [Google Scholar]
- 43.Yu, S. S., J. M. Kim, and S. Kim. 2000. The 17 nucleotides downstream from the env gene stop codon are important for murine leukemia virus packaging. J. Virol. 74:8775-8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., and E. Barklis. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 69:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]