Abstract
The herpes simplex virus type 1 UL20 protein (UL20p) is an important determinant for cytoplasmic virion morphogenesis and virus-induced cell fusion. To delineate the functional domains of the UL20 protein, we generated a panel of single and multiple (cluster) alanine substitutions as well as UL20p carboxyl-terminal truncations. The UL20 mutant genes could be broadly categorized into four main groups: Group I UL20 mutant genes complemented for both virus production and virus-induced cell fusion; Group II UL20 mutant genes did not complement for either virus-induced cell fusion or infectious virus production; Group III UL20 mutant genes complemented for virus-induced cell fusion to variable extents but exhibited substantially decreased ability to complement UL20-null infectious virus production; Group IV mutant genes complemented for infectious virus production but had variable effects on virus-induced cell fusion; this group included two mutants that efficiently complemented for gBsyn3, but not for gKsyn1, virus-induced cell fusion. In addition, certain recombinant viruses with mutations in either the amino or carboxyl termini of UL20p produced partially syncytial plaques on Vero cells in the absence of any other virally encoded syncytial mutations. These studies indicated that the amino and carboxyl termini of UL20p contained domains that functioned both in infectious virus production and virus-induced cell fusion. Moreover, the data suggested that the UL20p's role in virus-induced cell fusion can be functionally separated from its role in cytoplasmic virion morphogenesis and that certain UL20p domains that function in gB-syn3 virus-induced cell fusion are distinct from those functioning in gKsyn1 virus-induced cell fusion.
Herpes simplex viruses (HSV) specify at least 11 virally encoded glycoproteins, as well as several nonglycosylated membrane-associated proteins, most of which play important roles in multiple membrane fusion events during virus entry and intracellular virion morphogenesis and egress (34, 37-39). Spread of infectious virus occurs either by release of virions to extracellular spaces or through virus-induced cell-to-cell fusion. Mutations that cause extensive virus-induced cell fusion arise predominantly in four genes of the HSV genome: the UL20 gene (6, 28), the UL24 gene (23, 36), the UL27 gene encoding glycoprotein B (gB) (10, 32), and the UL53 gene coding for glycoprotein K (gK) (8, 12, 21, 33, 35). Of these four membrane-associated proteins, only UL20 and gK are essential for the intracellular transport of virions to extracellular spaces in all cell types (6, 16, 18, 22, 24).
Virus morphogenesis and egress of infectious herpes virions is thought to involve sequential de-envelopment and reenvelopment steps in transit to extracellular spaces as follows: (1) primary envelopment by budding of capsids assembled in the nuclei through the inner nuclear leaflet leading to the production of enveloped virions within perinuclear spaces; (ii) de-envelopment by fusion of viral envelopes with the outer nuclear leaflet leading to the accumulation of unenveloped capsids in the cytoplasm; (iii) reenvelopment of cytoplasmic capsids into trans-Golgi network (TGN)-derived vesicles. This final event in cytoplasmic virion envelopment is thought to be mediated by interactions between tegument proteins and cytoplasmic portions of viral glycoproteins embedded within the TGN-derived membranes. Cytoplasmically enveloped viruses are thought to be transported to extracellular spaces within Golgi or TGN-derived vesicles (reviewed in references 25, 31, and 41).
The UL20 gene encodes a 222-amino-acid nonglycosylated transmembrane protein that is conserved by all alphaherpesviruses. The UL20p is a structural component of extracellular enveloped virions, and it is expressed in infected cells assuming a predominantly perinuclear and cytoplasmic distribution (44). A partial deletion of the UL20 gene was reported to form weakly syncytial viral plaques in certain cell types and severe defects in cytoplasmic virion envelopment, implying that UL20 functioned in virus-induced cell fusion and virion morphogenesis. Furthermore, replication of the UL20-null virus could be partially complemented in certain cell types indicating a cellular specificity of UL20 functions (6). This UL20p cellular specificity was phenomenologically associated with disruption of the Golgi apparatus during viral infections, inasmuch as cells that complemented the UL20-null virus retained intact Golgi networks, whereas the Golgi networks of cells failing to complement the UL20-null virus were disrupted and dispersed throughout the cytoplasm (4, 5).
Recently, it was shown that a precise deletion of the UL20 gene caused accumulation of unenveloped capsids into the cytoplasm, indicating that the HSV-1 UL20p functioned in cytoplasmic stages of virion envelopment and that the previously reported partial deletion of the UL20 gene caused a predicted fusion of the UL20p to the adjacent UL20.5 gene. In addition, syncytial mutations in either gB or gK failed to cause fusion in the absence of the UL20 gene, suggesting that the UL20 protein was essential for virus-induced cell fusion (17). Furthermore, it was shown that UL20 is required for cell surface expression of gK, suggesting a functional interdependence between gK and UL20 for virus egress and cell-to-cell fusion (14, 15, 17). In this study we have delineated via site-directed mutagenesis the functional domains of UL20p involved in infectious virus production and virus-induced cell fusion. Importantly, we show that both amino- and carboxyl-terminal portions of UL20p, which are predicted to lie within the cytoplasmic side of cellular membranes, function both in cytoplasmic virion envelopment and virus-induced cell fusion. Overall, the data suggest that UL20p's role in virus-induced cell fusion can be functionally separated from its role in cytoplasmic virion morphogenesis and that certain UL20p domains that function in gB-syn3 virus-induced cell fusion are distinct from those functioning in gKsyn1 virus-induced cell fusion.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Rockville, Md.). The Vero-based UL20 complementing cell line, G5, was a gift of P. Desai, (John Hopkins Medical Center) (13). Cells were maintained as previously described (13, 15, 16). The parental wild-type strain used in this study, HSV-1(KOS), was originally obtained from P. A. Schaffer (Harvard Medical School). Flp-In-CV-1 cells were maintained as directed by the manufacturer (Invitrogen, Inc.). Δ20DIV5, Δ20gBsyn3, and Δ20syn20DIV5 viruses were as described previously (17), and virus stocks were grown on Fd20-1 cells, described below. In this paper, for simplification purposes, the Δ20DIV5 virus is referred to as Δ20 virus and the Δ20syn20DIV5 virus is referred to as Δ20gKsyn1 virus (the syn20 and syn1 mutation are the same, Ala-to-Val at gK amino acid 40).
Construction of transformed Flp-In-CV-1 cell lines.
Generation of stable Flp-In-CV-1 expression cell lines was performed essentially as directed by the manufacturer (Invitrogen). Briefly, confluent Flp-In-CV-1 monolayers in six-well plates were transfected by using Lipofectamine 2000 with 0.3 μg of either pcDNA5/FRT/UL20 or pFRT/gDpro/UL20 and 2.7 μg of pOG44 (Invitrogen), which expresses Flp recombinase. After 2 weeks, hygromycin B (125 μg/ml)-resistant colonies were tested for the ability to complement the growth of Δ20, Δ20gBsyn3, and Δ20gKsyn1 viruses. All colonies derived from each plasmid complemented UL20-null virus. Isolates Fc20-1 and Fd20-1, derived from pcDNA5/FRT/UL20 and pFRT/gDpro/UL20 transfected cells, respectively, were chosen for further use. Fd20-1 cells were used in generating UL20-null virus stocks.
Plasmids.
pCR2.1-UL20, which was used as the parental vector for UL20 mutagenesis, was generated by cloning a 773-bp DNA fragment containing the UL20 gene, obtained by PCR amplification of HSV-1(KOS) viral DNA, into pCR2.1/TOPO (Invitrogen). A 7,185-bp fragment spanning the region comprising UL19, UL20, UL21, and UL22 was PCR amplified from KOS virus DNA by using primers p20F2 5′ (TTTCTTGAATTCACGACGGCGGTGTAGCCCACG) and p20F2 3′ (CTTCTTGAATTCCCCATCACCCACAACGCCAGC), restricted by EcoRI, and ligated into puc19BX (17) to generate p20F2. p20F2 was modified by silencing the BamHI site at genomic nucleotide position 39235 (GenBank accession number NC_001806), conserving the UL19 amino acid sequence, and removing a 767-bp BamHI DNA fragment containing the UL20 gene, resulting in plasmid p20F2BX (see Fig. 2D). UL20 cluster-to-alanine mutants, single point mutants, and truncation mutants were generated by two methods. Mutants CL11, CL23, CL34, CL38, CL121, CL209, 66t, and 149t were generated by using the GeneTailor Site-Directed Mutagenesis kit as directed by the manufacturer (Invitrogen). Mutants CL5, CL16, CL30, CL41, CL46, CL49, CL153, CL173, CL177, 181t, 204t, 211t, 216t, Y49A, S50A, R51A, D123A, R209A, T212A, and R213A were generated by splice-overlap extension PCR (1). Mutant UL20 genes were transferred into the BamHI site of the vector p20F2BX. All p20F2BX mutant constructs were sequenced to verify error free mutagenesis. Plasmid p20R contains a wild-type UL20 gene cloned into p20F2BX. An additional plasmid was constructed from p20F2BX, designated pΔ20NE, to include the same UL20 deletion as pΔ20-EGFP (17) without the EGFP gene cassette. Plasmid pcDNA5/FRT/UL20 was constructed by inserting the UL20 gene, which had been PCR amplified from KOS viral DNA, into pcDNA5/FRT/V5-His-TOPO (Invitrogen). An additional plasmid, pFRT/gDpro/UL20, was constructed by replacing the cytomegalovirus (CMV) promoter in pcDNA5/FRT/UL20 with the HSV-1 KOS gD promoter region.
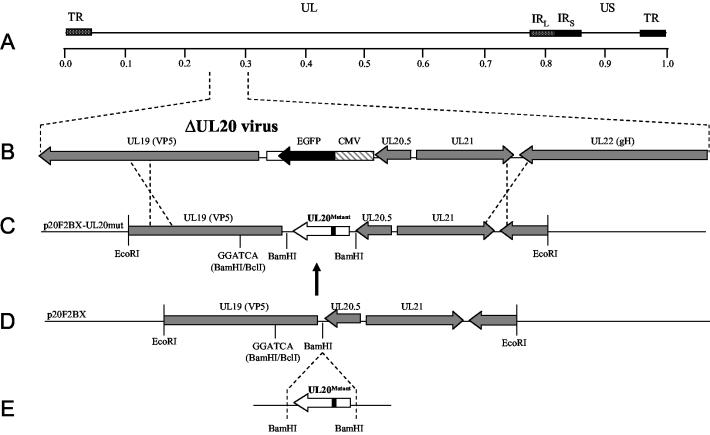
FIG. 2.
Schematic representation of the strategy for the construction of UL20-mutant genes used for complementation and isolation of recombinant viruses. (A) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) Expanded genomic region of the UL20-null virus between map units 0.239 and 0.305 containing the UL19, CMV-EGFP, UL20.5, UL21, and UL22 genes. (C) The p20F2BX-UL20mut plasmid shows the expression and recombination plasmids used for complementation assays and construction of the recombinant viruses carrying each UL20 mutation. (D and E) Mutated UL20 genes were inserted into the BamHI restriction site of p20F2BX.
UL20 complementation assay for infectious virion production.
The complementation assay was modified from a similar assay described previously (7). Confluent Vero monolayers in six-well plates were transfected with 1.5 μg of pcDNA3.1/V5-His-TOPO/lacZ (Invitrogen), expressing β-galactosidase under control of the CMV promoter, and 1.5 μg of wild-type or mutant UL20 plasmid with Lipofectamine 2000 as described by the manufacturer (Invitrogen). Six hours posttransfection, the monolayers were infected with a UL20-null virus at a multiplicity of infection (MOI) of 1. Infections were placed on a rocker for 1 h at 4°C and then transferred to 37°C for 2 h. Residual virus was inactivated by using an acid wash (phosphate-buffered saline containing 0.5 M glycine, pH 3) for 2 min, and monolayers were subsequently washed three times with Dulbecco's modified Eagle's medium to restore the pH to a normal level. Infections were incubated at 37°C for 24 h. After repeated freeze-thaw cycles, the titers of virus stocks were determined in triplicate on G5 cells, which effectively complement the UL20-null defect (17). An aliquot of each virus stock was saved to assay for β-galactosidase expression as described below.
β-Galactosidase expression levels were quantified essentially as described previously (7). One hundred microliters of Z buffer containing 0.27% β-mercaptoethanol and 0.2% Triton X-100 was added to 100 μl of virus stock and vortexed for 30 s. Cell debris was pelleted by centrifugation at 16,100 × g in a microcentrifuge for 30 s. One hundred sixty microliters of the supernatant was transferred to a 96-well plate and allowed to react with 40 μl of 4 mg of ONPG (ο-nitrophenyl-β-d-galactopyranoside) stock/ml at room temperature. The reactions were allowed to proceed for 90 min before absorbance at 405 nm was determined.
Calculation of complementation ratios.
The complementation ratio for each mutant was calculated with the following formula: [(virus titer of mutant/virus titer of negative control)×(experimental β-galactosidase value/β-galactosidase value of negative control)]. With this formula, values obtained from the β-galactosidase assay were used to correct for minor differences in transfection efficiency; the complementation ratio for the negative control (plasmid pΔ20NE) was set at 1.
UL20 complementation assay for virus-induced cell-to-cell fusion.
Confluent Vero monolayers in six-well plates were transfected with 3 μg of wild-type or mutant UL20 plasmid with Lipofectamine 2000 as described by the manufacturer (Invitrogen). Eighteen hours posttransfection, the monolayers were infected at an MOI of 0.1 with either Δ20gKsyn1 or Δ20gBsyn3 viruses. Infections were placed on a rocker at room temperature for 1 h and then transferred to 37°C for 30 min. Cells were overlaid with Dulbecco's modified Eagle's medium containing 1% methylcellulose. Twenty-four hours postinfection (h.p.i.), cell fusion was determined by visualization of syncytia formation by fluorescence microscopy. Cell fusion was blindly scored by two independent observers, and scores were linked to individual mutants after the cell fusion scoring was completed. Cell fusion was scored as completely absent (−), present at very low levels (+), present at moderate levels (++), or present at levels near (+++) or equivalent to (++++) those observed when the wild-type UL20 gene was used. The score of (−) indicates no syncytial formation as evidenced by the absence of cells having more than one nucleus. The score of (+) indicates the presence of syncytia containing on the average five nuclei or fewer. The score of (++) reflects syncytia containing 6 to 10 nuclei per cell. The score of (+++) is assigned to 11 to 20 nuclei per cell. The score of (++++) is for syncytia having >20 nuclei per cell.
Electron microscopy.
Cell monolayers were infected with the indicated virus at an MOI of 5. All cells were prepared for transmission electron microscopy (TEM) examination 24 h p.i. Infected cells were fixed in a mixture of 2% paraformaldehyde and 1.5% glutaraldehyde in 0.1 M NaCaC buffer, pH 7.3. Following osmication (1% OsO4) and dehydration in an ethanol series, the samples were embedded in Epon-Araldyte resin and polymerized at 70°C. Thin sections were made on an MTXL Ultratone (RMC Products), stained with 5% uranyl acetate and CNA lead, and observed with a Zeiss 10 transmission electron microscope as described previously (15).
Generation of recombinant viruses specifying mutant UL20p.
Viruses that contained the UL20 mutations were generated by rescuing the UL20-null viruses (Δ20, Δ20gBsyn3, and Δ20gKsyn1) with plasmids containing the desired UL20 mutant genes bracketed by adjacent viral sequences to facilitate homologous recombination with the viral genome (see Fig. 3). Briefly, confluent Vero cell monolayers were transfected with each plasmid, and transfected cells were infected with the Δ20 virus 6 h after transfection. Virus stocks were prepared at 48 h.p.i., and recombinant viruses were plaque isolated on Vero cells and sequentially plaque purified at least five times. Replacement of the UL20-null resident CMV-enhanced green fluorescent protein (EGFP) gene cassette and the concomitant insertion of the mutated UL20 gene were confirmed by DNA sequencing. Visualization of viral plaques was achieved by either phase contrast microscopy or immunohistochemistry essentially as described previously (17).
FIG. 3.
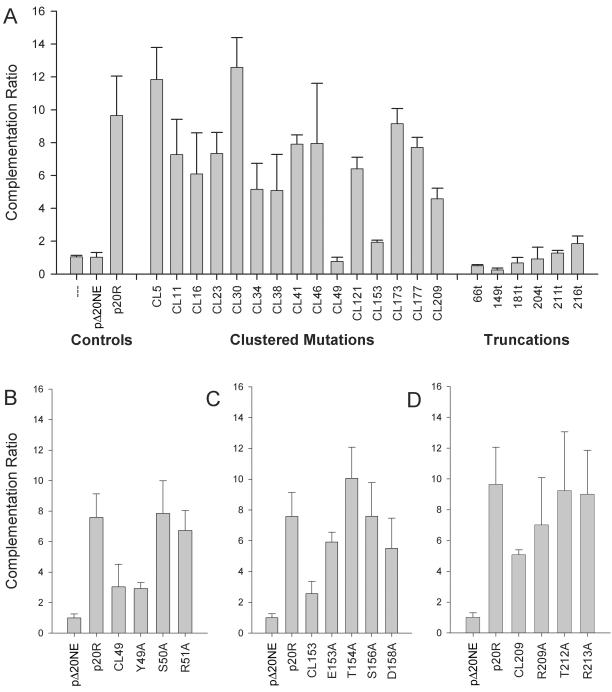
Complementation ratios of mutant UL20p genes. Vero cells were transfected with plasmids encoding wild-type or mutant UL20 genes under the UL20 promoter and then infected with the HSV-1(KOS) UL20-null (Δ20) virus. (A) Bar graph showing complementation ratios for UL20 cluster-to-alanine mutants and carboxyl-terminal truncations. (B) Complementation ratios of the CL49, Y49A, S50A, and R51A mutants. (C) Complementation ratios of the CL153, E153A, T154A, S156A, and D158A mutants. (D) Complementation ratios of the CL209, R209A, T212A, and R213A mutants. The error bars represent the maximum and minimum complementation ratios obtained from three independent experiments, and the bar height represents the average complementation ratio.
RESULTS
Mutagenesis of HSV-1 UL20.
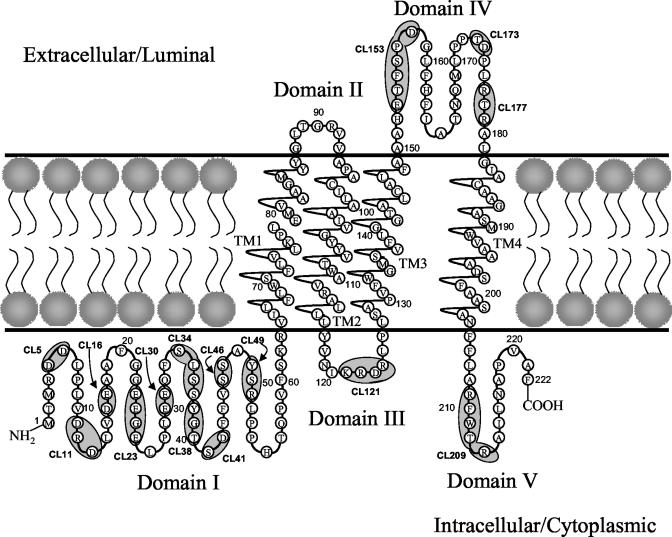
To delineate the functional domains of UL20p required for infectious virus production, virus egress, and virus-induced cell fusion, we constructed a panel of 31 mutations within the UL20 gene by site-directed mutagenesis. These mutations included (i) cluster-to-alanine mutants in which a cluster of proximal amino acids were changed to alanine residues, (ii) single-amino-acid replacement mutants within alanine cluster regions, and (iii) carboxyl-terminal truncations of UL20p. The mutated amino acids for each type of mutation are shown in Table 1. The carboxyl-terminal truncations are identified by the number of the last remaining amino acid (i.e., 66t retains UL20p amino acids 1 to 66). To facilitate placement of each mutation on the predicted luminar or cytoplasmic portions of UL20p, the hydrophobic and membrane-spanning domains of UL20p were calculated by using the TMPred and SOSUI computer algorithms (19, 20) and used to derive a predicted membrane spanning model of UL20p (Fig. 1). This UL20p model features four membrane-spanning regions placing both amino and carboxyl termini of UL20p within the cytoplasm (Domains I and V). In addition, a third small domain is located intracellularly (domain III), while the two other domains are located extracellularly (domains II and IV). The predicted membrane topology of UL20p was used as the guide for selecting alanine scanning mutations within predicted intracellular and extracellular domains of UL20p. Regions containing clusters of charged amino acids as well as potential phosphorylation sites were specifically targeted. The location of each cluster mutation is marked on the UL20p model, starting with the first mutated amino acid of the cluster (Fig. 1; Table 1).
TABLE 1.
Amino acid sequences of mutations
| Mutation or truncation | Domain | Amino acid sequence
|
|
|---|---|---|---|
| Wild type | Mutanta | ||
| Mutations | |||
| CL5 | I | DD | AA |
| CL11 | I | DRD | AAA |
| CL16 | I | DE | AA |
| CL23 | I | EEGE | AAGA |
| CL30 | I | EE | AA |
| CL34 | I | SLSS | ALAA |
| CL38 | I | YGT | AGA |
| CL41 | I | SD | AA |
| CL46 | I | SS | AA |
| CL49 | I | YSR | AAA |
| Y49A | I | ASR | |
| S50A | I | YAR | |
| R51A | I | YSA | |
| CL121 | III | KRDR | AADA |
| CL153 | IV | ETFSPD | AAFAPA |
| E153A | IV | ATFSPD | |
| T154A | IV | EAFSPD | |
| S156A | IV | ETFAPD | |
| D158A | IV | ETFSPA | |
| CL173 | IV | TD | AA |
| CL177 | IV | RTR | AAA |
| CL209 | V | RFWTR | AFWAA |
| R209A | V | AFWTR | |
| T212A | V | RFWAR | |
| R213A | V | RFWTA | |
| Truncations | |||
| 66t | VILFL | VILG* | |
| 149t | LFAAA | LFAG* | |
| 181t | RALGI | RALG* | |
| 204t | SANFF | SANG* | |
| 211t | RFWTR | RFWG* | |
| 216t | AILNA | AILG* | |
*, stop codon.
FIG. 1.
Predicted membrane topology of UL20p and location of the 15 cluster-to-alanine mutations. Membrane topology was predicted by using the TMPred and SOSUI algorithms (19, 20). UL20p domains where cluster-to-alanine mutations are located are indicated by a shaded oval. The naming of cluster mutations is based on the first amino acid mutated in each cluster. TM, transmembrane region; CL, cluster mutant.
Complementation assay for infectious virus production.
It was previously shown that deletion of the HSV-1 UL20 and the PRV UL20 genes resulted in a reduction of up to two logs in infectious virus production relative to their parental wild type strains (6, 17, 18). To delineate functional domains of the UL20 protein involved in infectious virus production, each of the 31 mutated UL20 genes was tested for its ability to complement the HSV-1(KOS) UL20-null virus. The mutated genes were cloned into plasmids under their own promoter control bracketed by upstream and downstream DNA sequences to facilitate complementation experiments as well as isolation of recombinant viruses (Fig. 2D and E) (see Materials and Methods). Complementation experiments involved transfection of Vero cells with plasmids encoding wild-type or mutant UL20 genes, followed by infection with the UL20-null virus (17). A complementation ratio was calculated for each mutant UL20 plasmid, p20R (wild-type UL20, positive control), and pΔ20NE (UL20-null mutation, negative control). The complementation ratio of pΔ20NE was set to 1 for each experiment. The complementation ratios for the UL20 cluster mutants and truncation mutants are shown in Fig. 3A. Mutants CL5, CL11, CL16, CL23, CL30, CL34, CL38, CL41 CL46, CL121, CL173, CL177, and CL209 complemented the UL20-null virus to a variable extent in comparison to the positive plasmid control p20R and the negative plasmid control pΔ20NE (Fig. 3A). Mutants CL49, CL153, 66t, 149t, 181t, 204t, 211t, and 216t failed to efficiently complement the UL20-null defect in infectious virion production producing complementation ratios similar to those of plasmid pΔ20NE (negative control), indicating that the mutated UL20 protein was not able to function properly in virus egress (Fig. 3A). To delineate the specific amino acids responsible for the UL20-associated defects identified by the cluster mutations, we examined the complementation ratios of several point mutants within cluster regions. The CL49 (YSR-to-AAA; Table 1) mutant UL20 gene failed to efficiently complement the Δ20 virus. The Y49A mutant gene, which involves the Y residue within the CL49 target sequence, produced a complementation ratio similar to that of CL49, while the adjacent S50A and R51A mutant genes complemented infectious virus production to nearly wild-type levels (Fig. 3B). These results indicate that the mutation of the Y residue is responsible for the complementation defect exhibited by the UL20 gene carrying the CL49 mutation. In contrast, the UL20 genes carrying the E153A, T154A, S156A, and D158A mutations complemented the UL20-null virus at a greater level than did the gene with the CL153 mutation, indicating that these amino acids were not individually responsible for the lack of complementation by the UL20 gene carrying the CL153 mutation (Fig. 3C). The CL209 UL20 mutant gene as well as mutant genes carrying individual amino acid changes within the CL209 target sequence efficiently complemented Δ20 infectious virus production (Fig. 3D).
Complementation for virus-induced cell-to-cell fusion.
Recently, we showed that syncytial mutations in either gB or gK failed to cause virus-induced cell fusion in the absence of the UL20 gene (17). Therefore, the panel of 31 UL20 mutants was tested for the ability to complement UL20-null viruses containing syncytial mutations in either gB (syn3) or gK (syn1) for virus-induced cell fusion. Confluent Vero monolayers were transfected with plasmids encoding either wild-type or mutant UL20p and subsequently infected with either Δ20gKsyn1 or Δ20gBsyn3 viruses. These viruses contain a CMV-EGFP gene cassette in place of the UL20 gene, which facilitates visualization of syncytia formation by fluorescence microscopy (17). Cell fusion was scored by two observers as detailed in Materials and Methods. Complementation results are summarized in Table 2, and select mutant phenotypes are displayed in Fig. 4.
TABLE 2.
Complementation results for cell fusion
| Mutation or truncation | Domain | Extent of cell fusion by UL20-null complementationa
|
|
|---|---|---|---|
| gK (syn1) | gB (syn3) | ||
| Mutations | |||
| CL5 | I | ++++ | ++++ |
| CL11 | I | ++++ | ++++ |
| CL16 | I | ++++ | ++++ |
| CL23 | I | ++++ | ++++ |
| CL30 | I | +++ | ++++ |
| CL34 | I | +++ | +++ |
| CL38 | I | + | + |
| CL41 | I | ++ | ++++ |
| CL46 | I | ++ | ++++ |
| CL49 | I | ++ | ++ |
| Y49A | I | ++ | ++ |
| S50A | I | ++++ | ++++ |
| R51A | I | ++++ | ++++ |
| CL121 | III | ++ | ++ |
| CL153 | IV | − | − |
| E153A | IV | +++ | ++++ |
| T154A | IV | ++++ | ++++ |
| S156A | IV | ++++ | ++++ |
| D158A | IV | ++++ | ++++ |
| CL173 | IV | +++ | ++++ |
| CL177 | IV | ++++ | ++++ |
| CL209 | V | +++ | ++++ |
| R209A | V | ++++ | ++++ |
| T212A | V | ++++ | ++++ |
| R213A | V | +++ | ++++ |
| Truncations | |||
| 66t | − | − | |
| 149t | − | − | |
| 181t | − | − | |
| 204t | − | − | |
| 211t | − | − | |
| 216t | + | +++ | |
−, completely absent; +, very low levels of cell-fusion; ++, moderate levels; +++, near wild-type levels; ++++, wild-type levels.
FIG. 4.
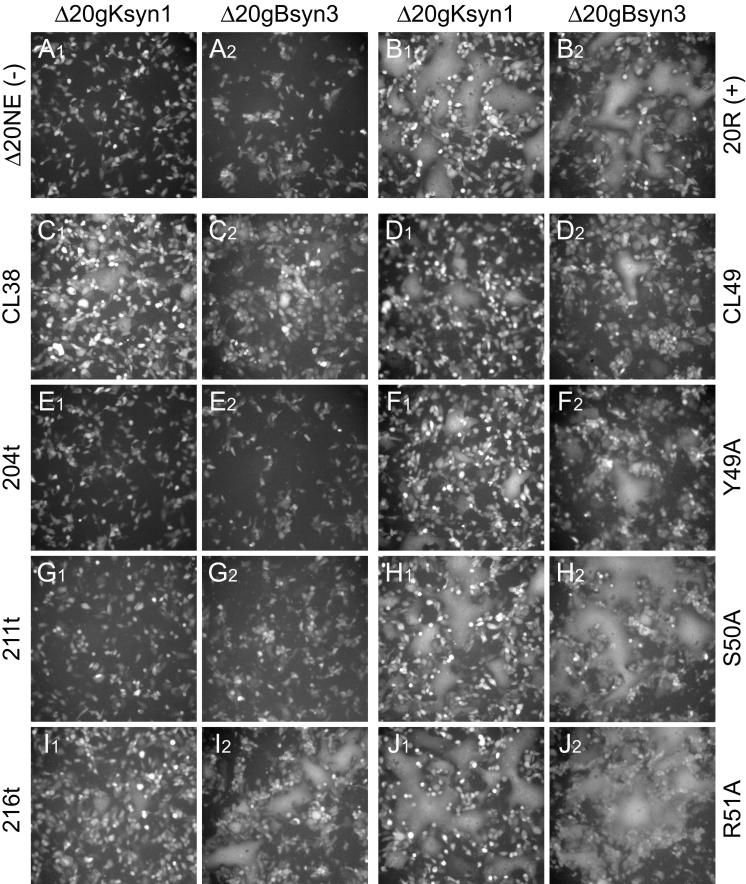
Complementation for virus-induced cell-to-cell fusion of UL20-null viruses containing syncytial mutations in gK(syn1) or gB(syn3). Vero cells were transfected with plasmids encoding wild-type or mutant UL20 genes and then infected with either the Δ20gKsyn1 or the Δ20gBsyn3 viruses. At 24 h.p.i., cell fusion was determined by visualization of syncytia formation by fluorescence microscopy. The extent of syncytial formation (complementation) obtained with the negative control plasmid, pΔ20NE (A), and positive control plasmid, p20R (B), is shown for reference purposes. Representative images for the CL38, 204t, 211t, and 216t mutants (C, E, G, and I, respectively) and for the CL49, Y49A, S50A, and R51A mutants (D, H, F, and J, respectively) are shown.
UL20 genes carrying the CL5, CL11, CL16, CL23, CL30, CL34, CL173, CL177, and CL209 mutations produced syncytia at levels similar to those produced by the positive-control plasmid p20R containing the wild-type UL20 gene. Syncytia formation was completely absent in complementation assays with UL20 genes specifying the CL153 mutation and all UL20p carboxyl-terminal truncations (66t, 149t, 181t, 204t, and 211t) except the 216t truncation, which complemented virus-induced cell fusion (Fig. 4E, G, and I; Table 2). Other UL20 mutations had various phenotypes with regard to the extent of syncytia formation (Table 2). As with the infectious virion production assay, the phenotype produced by the CL49 mutation was similar to that of Y49A, but not S50A or R51A (Fig. 4D, F, H, and J; Table 2). Most mutant UL20 genes were either able or unable to complement both Δ20gBsyn3 and Δ20gKsyn1 viruses, although the level of complementation varied among the different UL20 mutant genes. However, the CL41 and CL46 mutant UL20 genes efficiently complemented gBsyn3, but not gKsyn1, virus-induced cell fusion, indicating that these mutations had a differential effect of gBsyn3 versus gKsyn1-induced cell fusion (Table 2). The CL38 mutation, which is immediately proximal to the CL41 mutation, had an obvious defect in complementation for both gBsyn3 and gKsyn1-induced cell fusion (Fig. 4C; Table 2).
Plaque phenotypes of Δ20gBsyn3 and Δ20gKsyn1 recombinants containing selected UL20 mutations.
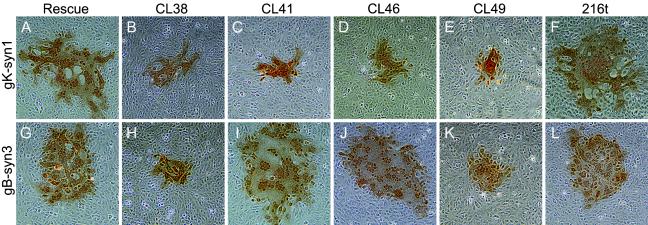
To further investigate the ability of UL20 mutant genes to complement for virus-induced cell fusion caused by the gBsyn3 or gKsyn1 virally encoded mutations, specific recombinant viruses expressing UL20p mutant genes were isolated by rescue of the UL20 gene deletion of the Δ20gBsyn3 and Δ20gKsyn1 viruses. Recombinant viruses were identified by a lack of EGFP expression on Vero cells. Viral plaques produced by each recombinant virus were characterized as small plaques if they contained on the average no more than 75 cells or as large plaques if they contained more than 250 cells per plaque. As expected, we were unable to isolate recombinants expressing the CL153, 66t, 149t, 181t, 204t, or 211t mutations on Vero cells, whereas rescue with the wild-type UL20 gene resulted in viruses that formed large syncytial plaques similar to the viruses carrying the gBsyn3 or gKsyn1 mutations alone (Fig. 5A and G). Recombinant viruses carrying the gKsyn1 mutation and either the CL41 or CL46 UL20 mutation produced small syncytial plaques on Vero cells (Fig. 5C and D), whereas the recombinant viruses carrying the gBsyn3 mutation and either the CL41 or the CL46 mutation produced large syncytial plaques on Vero cells (Fig. 5I and J). Recombinant viruses containing the CL38 mutation (Fig. 5B and H) or the CL49 mutation (Fig. 5E and K) in the gBsyn3 or gKsyn1 genetic background produced significantly smaller syncytial plaques than the gBsyn3 or gKsyn1 viruses specifying the wild-type UL20 gene. The gBsyn3 or gKsyn1 recombinant viruses specifying the 216t mutation produced syncytial plaques similar in size to the gBsyn3 or gKsyn1 viruses (Fig. 5F and L).
FIG. 5.
Plaque phenotypes of recombinant viruses derived from Δ20gKsyn1 (A, B, C, D, E, and F) and Δ20gBsyn3 (G, H, I, J, K, and L) and containing selected UL20 mutations on Vero cells. Confluent cell monolayers were infected with the recombinant viruses containing wild-type UL20 (A and G) or the UL20 mutants CL38 (B and H), CL41 (C and I), CL46 (D and J), CL49 (E and K), or 216t (F and L), and viral plaques were visualized by immunohistochemistry at 24 h.p.i.
Plaque phenotypes of selected UL20 mutations in the context of wild-type gB and gK genes.
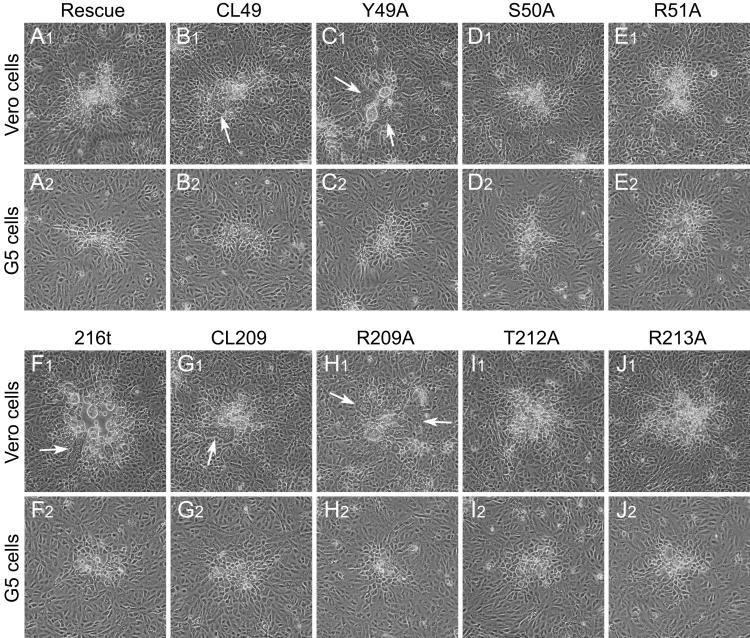
Attempts were made to isolate recombinant viruses specifying each of the 31 engineered UL20 mutations in the Δ20 genetic background specifying wild-type gB and gK genes. As expected, the CL153 mutation and all of the UL20p truncations except 216t failed to yield recombinant viruses on Vero cells. Most other UL20 mutations produced nonsyncytial virus plaques on Vero cells that were similar in size to recombinant viruses produced by rescuing the Δ20 virus with the wild-type UL20 allele in agreement with initial complementation results. Interestingly, recombinant viruses containing the CL49, Y49A, and 216t mutations and which allowed complementation for virus-induced cell fusion but exhibited deficiencies in infectious virus production were isolated (Fig. 3A and B and 4D, F, and I; Table 2). Recombinant viruses containing the CL49, Y49A, 216t, CL209, and R209A mutations produced partially syncytial plaques on Vero cells (Fig. 6B1, C1, F1, G1, and H1) suggesting that specific UL20 domains were involved in virus-induced cell fusion. In contrast, these viruses produced nonsyncytial plaques when plated on the G5 cells (Fig. 6B2, C2, F2, G2, and H2), indicating that the wild-type UL20 gene expressed by G5 cells was dominant over UL20 syncytial mutations. In the amino terminus of UL20p, recombinants expressing the S50A and R51A mutations, which are immediately proximal to the Y49A and contained within the CL49 mutation, did not cause syncytial plaque formation, suggesting that mutation of the Y residue is responsible for the resultant syncytial phenotype (compare Fig. 6B1 and C1 with D1 and E1). In the carboxyl terminus of UL20p, recombinant viruses expressing the CL209 and R209A mutations produced partially syncytial plaques on Vero cells, while those expressing the T212A and R213A did not (compare Fig. 5G1 and H1 with I1 and J1), suggesting that the R residue at position 209 is responsible for the CL209 syncytial phenotype. On average, syncytia produced by recombinant viruses expressing the carboxyl-terminal syncytial mutations 216t, CL209, and R209A were larger than those produced by the amino-terminal syncytial mutations CL49 and Y49A.
FIG. 6.
Domains of UL20p associated with UL20p-induced cell fusion. Plaque phenotypes of recombinant viruses containing selected UL20 mutations are shown on Vero (A1, B1, C1, D1, E1, F1, G1, H1, I1, and J1) and G5 (A2, B2, C2, D2, E2, F2, G2, H2, I2, and J2) cells. Confluent cell monolayers were infected with recombinant viruses containing wild-type UL20 (A1 and A2), or the UL20 mutants CL49 (B1 and B2), Y49A (C1 and C2), S50A (D1 and D2), R51A (E1 and E2), 216t (F1 and F2), CL209 (G1 and G2), R209A (H1 and H2), T212A (I1 and I2), or R213A (J1 and J2) at an MOI of 0.001, and viral plaques were visualized at 30 h p.i. White arrows indicate syncytia.
Ultrastructural characterization of recombinant viruses carrying mutations within the amino and carboxyl termini of UL20p.
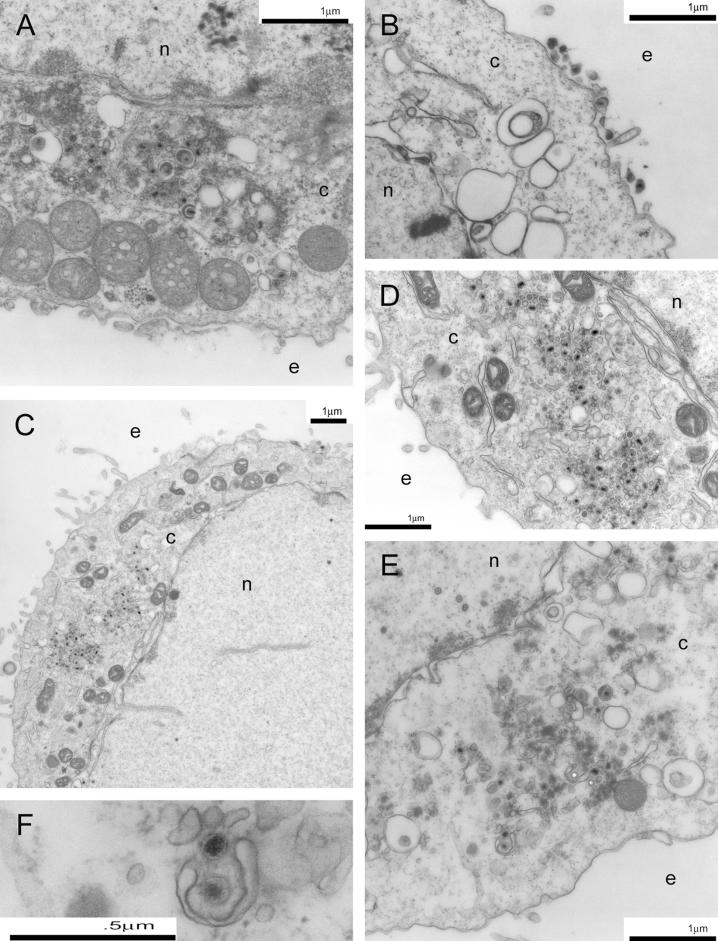
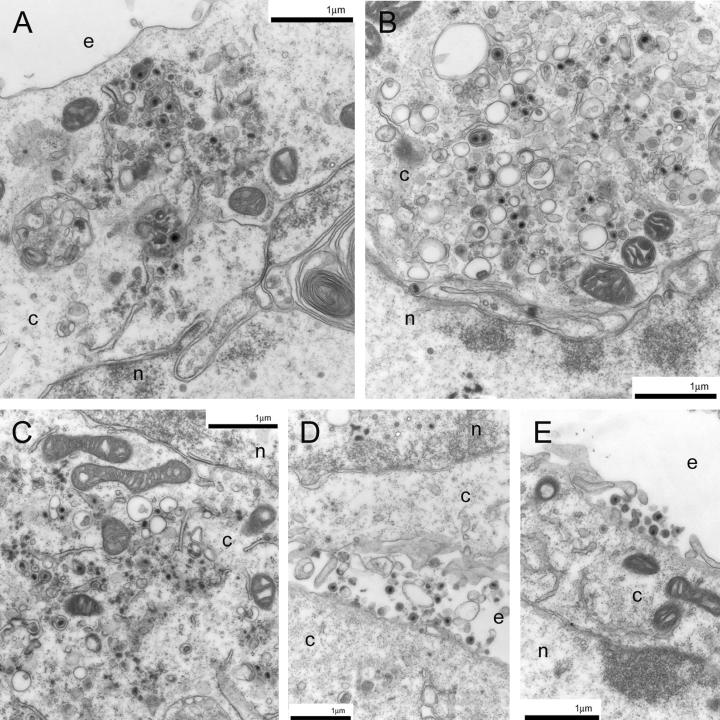
To evaluate the intracellular virion distribution of the recombinant viruses specifying the 216t, CL49, and Y49 UL20 mutations, Vero cells infected with UL20p wild-type and mutant viruses at an MOI of 5 were examined via electron microscopy at 24 h p.i. As previously shown (17), the Δ20 (UL20-null) virus caused accumulation of unenveloped capsids into the cytoplasm in infected Vero cells (Fig. 7A). In contrast, the Δ20-rescued virions were transported to extracellular spaces and exhibited no apparent cytoplasmic defects (Fig. 7B). Importantly, infection with the 216t mutant virus resulted in an accumulation of capsids in the cytoplasm (Fig. 7C, D, E, and F), similar to the UL20-null phenotype. Similarly, infection with the CL49 mutant resulted in the accumulation of capsids as well as a small percentage of enveloped particles within the cytoplasm (Fig. 8A and B). The Y49A mutant virus produced the same ultrastructural phenotype as the CL49 mutant (Fig. 8C). The S50A and R51A mutants exhibited no apparent defects in intracellular morphogenesis, appearing to have an intracellular distribution similar to that of the wild-type KOS virus (Fig. 8D and E).
FIG. 7.
Electron micrographs of Vero cells infected with Δ20DIV5 (A), Δ20-rescue (B), or 216t (C, D, E, and F) viruses. Confluent cell monolayers were infected at an MOI of 5, incubated at 37°C for 24 h, and prepared for TEM. Panel D shows a higher magnification of panel C. Panel F shows a partially enveloped capsid often seen in UL20-null-infected cells. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked.
FIG. 8.
Electron micrographs of Vero cells infected with the CL49 (A and B), Y49Α (C), S50A (D), or R51A (E) viruses. Confluent cell monolayers were infected at an MOI of 5, incubated at 37°C for 24 h, and prepared for TEM. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked.
DISCUSSION
In this study, we constructed and characterized a panel of 31 different mutations within UL20p to delineate domains of UL20p functioning in virion morphogenesis and virus-induced cell fusion. Analysis of the various mutant phenotypes reveals that UL20p contains at least three domains involved in virion morphogenesis and virus-induced cell fusion: the amino and carboxyl termini of UL20 (domains I and V), which are predicted to lie on the cytoplasmic side of membranes, and UL20p domain IV, which is predicted to lie extracellularly. Overall, the data suggest that the role of UL20p in virus-induced cell fusion can be functionally separated from its role in cytoplasmic virion morphogenesis and that certain UL20p domains that function in gB-syn3-mediated virus-induced cell fusion are distinct from those functioning in gKsyn1-mediated virus-induced cell fusion.
The predicted membrane topology of UL20p.
Computer-assisted prediction of the membrane-spanning domains of UL20p indicated that UL20p spanned membranes four times placing both the amino (domain I) and carboxyl terminal (domain V) portions within the cytoplasm of cellular membranes and internal to the virion envelope. This predicted structure includes two extracellular domains: a very small domain of 5 amino acids (domain II) and a second domain of 25 amino acids (domain IV), predicted to be localized in the lumen (extracellular) and external to the virion envelope. Initial epitope tagging results indicate that domain I is located intracellularly, whereas domain IV is located extracellularly, which is in support of this model (data not shown). However, additional work is required in order to fully confirm this UL20p topology model. Domain I is the largest domain (63 amino acids), and it includes stretches of acidic amino acid (D and E) clusters, which could form electrostatic interactions with other proteins. These acidic clusters as well as the amino acid motif YXXφ (YSRL) have been shown to function in endocytosis of alphaherpesvirus envelope proteins from plasma membranes to the TGN (2, 3, 9, 40, 45). The acidic cluster motifs appear to direct TGN localization by binding to a cellular connector protein, PACS-1, which connects the glycoproteins to the AP-1 complex (43), while the YXXφ motif binds adaptor proteins directly (2, 3, 40).
UL20p domains that function in infectious virus production and virus-induced cell fusion.
Complementation experiments of the HSV-1(KOS) UL20-null virus, in the presence or absence of either the gBsyn3 or gKsyn1 mutations, using each mutant UL20 gene revealed that most mutations could be broadly categorized into four major groups based on their differential effects on infectious virus production and virus-induced cell fusion (Table 3). Group I mutations complemented infectious virus production and virus-induced cell fusion. These mutations included CL5, CL11, CL16, CL23, and CL30 in the amino terminus of UL20p (domain I); CL121 (domain III); CL173 and CL177 (domain IV); and CL209 (domain V). Considering that these cluster mutations involved replacement of multiple amino acids by alanine residues, these results indicate that the UL20p regions spanned by these mutations were not crucial for the structure and functions of UL20p.
TABLE 3.
Differential effects of mutations on infectious virus production and cell fusion
| Group | Mutants | Complementation for infectious virion production | Complementation for virus-induced cell fusion
|
|
|---|---|---|---|---|
| gKsyn1 | gBsyn3 | |||
| I | CL5, CL11, CL16, CL23, CL30, CL34, CL121, CL173, CL177, CL209 | Yes | High | High |
| II | CL153, 66t, 149t, 181t, 204t, 211t | No | No | No |
| III | CL49, 216t | No | Low | Low (CL49) High (216t) |
| IV | CL38, CL41, CL46 | Yes | Low | Low (CL38) High (CL41, CL46) |
Group II mutations were unable to complement infectious virus production and virus-induced cell fusion. These mutations included the CL153, 66t, 149t, 181t, 204t, and 211t mutations. The 66t, 149t and 181t mutations specify large carboxyl-terminal UL20p truncations, which are expected to drastically affect the synthesis, transport, and membrane topology of UL20p. Therefore, it is not surprising that these mutations failed to complement both infectious virus production and virus-induced cell fusion. The 204t and 211t mutations encode carboxyl-terminal truncations of 18 and 11 amino acids, respectively, encompassing UL20p domain V. Although, these deletions are rather small and not predicted to affect the membrane incorporation and topology of UL20p (not shown), they may still affect the structure and function of UL20p. Alternatively, this UL20p carboxyl-terminal domain may be involved in protein-protein interactions necessary for UL20p-associated viral functions. The UL20p domain IV is predicted to lie in the lumen or extracellular side of membranes. Based on the fact that the CL173 and CL177 mutations complemented the UL20-null virus, but the CL153 did not, the data suggest that amino acids 150 to 156 are important in infectious virus production. However, none of the single amino acid changes E153A, T154A, S156A, or D158A significantly altered UL20p's ability to complement the UL20-null virus, suggesting that the combined effects of the CL153 mutations may affect the conformation of this domain. Alternatively, this domain may interact with luminal portions of other viral glycoproteins, such as gB or gK, to facilitate infectious virus production and virus-induced cell fusion.
Group III mutations included the CL49 and 216t mutations, located at the amino and carboxyl termini of UL20p, respectively, which complemented for virus-induced cell fusion at a reduced level, but failed to efficiently complement infectious virus production. The carboxyl terminus of UL20p (Domain V) is composed of 18 amino acids and predicted to localize intracellularly. The 216t complementation results suggest that the terminal six amino acids of UL20p are crucial for infectious virus production. This result was further corroborated by the recombinant virus expressing the 216t mutation, which produced an ultrastructural phenotype similar to the UL20-null virus characterized by the accumulation of unenveloped capsids into the cytoplasm. The 216t mutation was able to complement both gBsyn3 and gKsyn1-induced cell fusion, albeit at levels lower than those of the wild-type UL20 gene. However, the recombinant virus carrying the 216t mutation in the Δ20syn1 and Δ20syn3 genetic backgrounds revealed no apparent differences in the size and extent of syncytial plaque formation in comparison to the corresponding Δ20syn1 and Δ20syn3 viruses expressing the wild-type UL20 allele, indicating that the UL20 216t mutation did not adversely affect virus-induced cell fusion.
The CL49 mutant phenotype in the amino terminus of UL20p is of particular interest because alignment of the predicted amino acid sequences of multiple UL20 homologues encoded by alphaherpesviruses revealed conservation of certain amino acid motifs within the UL20p amino terminus (not shown). One such motif is the YXXφ (YSRL) amino acid sequence overlapping the CL49 mutated sequence, which is conserved in HSV-1, HSV-2, and cercopithecine herpesvirus 1 and 2, but not in varicella-zoster virus or pseurodabies virus (not shown). This motif is known to function in the retrieval of cell surface-expressed proteins to the TGN compartment (11, 26, 27, 29, 30, 42). Mutagenesis of the Y residue of a YXXφ (YTKI) motif within gK domain IV, shown to lie in the cytoplasmic side of membranes, produced a gK-null phenotype (16). Similarly, site-directed mutagenesis of the Y49, S50, and R51 residues within this motif revealed that only the Y49A mutation caused failure to complement the UL20-null virus, indicating that this tyrosine residue played an important role in infectious virus production. Tyrosine residues within YXXφ motifs are known to be potentially phosphorylated. Therefore, it is possible that the Y (amino acid position 49) residue of UL20p is phosphorylated and that this phosphorylation may be required for proper intracellular localization and function of UL20p. This result was further supported by the ultrastructural phenotype of the recombinant viruses expressing the CL49 and Y49A mutations, both of which showed an accumulation of unenveloped capsids into the cytoplasm similar to that of the UL20-null virus. However, the CL49 and the Y49A mutations complemented, albeit at reduced levels, both gBsyn3- and gKsyn1-induced cell fusion, suggesting that these mutations only partially inactivated the UL20p functions required for virus-induced cell fusion. This result was further supported by the small but syncytial plaque phenotype of the recombinant viruses expressing the CL49 mutation in either the gKsyn1 or the gBsyn3 genetic background.
Group IV mutations exhibited a variable effect on virus-induced cell fusion but did not drastically reduce infectious virus production. Specifically, the CL38 UL20 mutant gene complemented virus-induced cell fusion caused by the Δ20gBsyn3 and Δ20gKsyn1 viruses at low levels, while the CL41 and CL46 mutant genes complemented Δ20gKsyn1 at low levels but Δ20gBsyn3 at high levels and levels equivalent to the complementation levels produced with the wild-type UL20 gene. The initial complementation results were further supported by the recombinant viruses carrying either the CL41 or the CL46 mutation in the gKsyn1 or gBsyn3 genetic background. Specifically, the gKsyn1 recombinants carrying either the CL41 or CL46 mutations produced small syncytial plaques, while the gBsyn3 recombinants carrying either the CL41 or CL46 mutations produced large syncytial virus plaques indistinguishable to the gBsyn3 recombinant virus carrying the wild-type UL20 allele. These results suggest that the UL20p amino-terminal domain demarcated by the CL38 to CL46 mutations is important in virus-induced cell fusion; however, the specific amino acid requirements within this domain may be different for gBsyn3 versus gKsyn1-induced cell fusion.
UL20p mutations that cause virus-induced cell fusion.
Characteristically, recombinant viruses expressing the CL49, Y49A, CL209, R209A, and 216t UL20p mutations in a wild-type genetic background formed small syncytial plaques on Vero cells. Recombinant viruses specifying the T212A and R213A mutations did not produce syncytial plaques on Vero cells, indicating that the R209A mutation was responsible for the syncytial phenotype of the CL209 mutation. Thus, it is evident that the amino and carboxyl termini of UL20p are directly associated with virus-induced cell fusion in the context of wild-type gB or gK expression. Therefore, the ability of these mutations to complement either gBsyn3 or gKsyn1 virus-induced cell fusion discussed earlier may be in part due to the inherent fusogenic character of the mutated UL20p. Most likely, this is not the case because recombinant viruses carrying the UL20 R209A syncytial mutation as well as either the gBsyn3 or the gKsyn1 mutation did not exhibit increased virus-induced cell fusion in comparison to viruses specifying either the gBsyn3 or gKsyn1 mutation and the wild-type UL20 gene (not shown). Alternatively, UL20p may function as a regulator of virus-induced cell fusion through interactions with other viral proteins such as gB and gK, and disruption of these interactions by specific UL20p mutations may result in the formation of syncytia.
Deletion of the gK gene or lethal gK mutation causes entrapment of enveloped virions within cytoplasmic vesicles presumably originating from the TGN compartment and the accumulation of unenveloped capsids in the cytoplasm (16, 24). Similarly, deletion of the UL20 gene or lethal UL20p mutations cause accumulation of unenveloped capsids into the cytoplasm and a small portion of enveloped capsids within cytoplasmic vesicles (17, 18). These results suggest that UL20p and gK may have interrelated functions with regard to cytoplasmic virion envelopment. The fact that the Y49A, CL49, and 216t mutant viruses produced syncytial plaques, although their ultrastructural phenotypes seemed to be similar to that of the UL20-null virus, suggests that UL20 functions for virus-induced cell fusion may be distinct from those functioning in cytoplasmic virion envelopment. Alternatively, the same domains may function in both cytoplasmic virion envelopment and virus-induced cell fusion; however, virion envelopment may be more susceptible to the negative effects of these mutations than to virus-induced cell fusion.
Overall, our previous findings and the results presented in this paper suggest the existence of functional interactions between gK and UL20p as well as between UL20p and gB. In this regard, it is interesting that syncytial mutations in gB lie in the intracellular carboxyl terminus of gB, allowing the possibility that intracellular domains of UL20p (domains I and V) directly or indirectly interact with the carboxyl terminus of gB altering gB's fusogenic properties.
Acknowledgments
We thank Olga Borkhsenious for her expert technical assistance with electron microscopy. We thank Trisha Olivier and Thaya Guedry for their expert technical assistance.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (AI43000) to K.G.K. J.M.M. was supported by a Louisiana Economic Development Graduate Assistant Fellowship. We acknowledge financial support by the LSU School of Veterinary Medicine to BIOMMED.
REFERENCES
- 1.Aiyar, A., Y. Xiang, and J. Leis. 1996. Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 57:177-191. [DOI] [PubMed] [Google Scholar]
- 2.Alconada, A., U. Bauer, and B. Hoflack. 1996. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 15:6096-6110. [PMC free article] [PubMed] [Google Scholar]
- 3.Alconada, A., U. Bauer, B. Sodeik, and B. Hoflack. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73:377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avitabile, E., G. S. Di, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avitabile, E., P. L. Ward, C. Di Lazzaro, M. R. Torrisi, B. Roizman, and G. Campadelli-Fiume. 1994. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J. Virol. 68:7397-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368-376. [DOI] [PubMed] [Google Scholar]
- 9.Brideau, A. D., T. del Rio, E. J. Wolffe, and L. W. Enquist. 1999. Intracellular trafficking and localization of the pseudorabies virus Us9 type II envelope protein to host and viral membranes. J. Virol. 73:4372-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 11.Canfield, W. M., K. F. Johnson, R. D. Ye, W. Gregory, and S. Kornfeld. 1991. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24-29 of the cytoplasmic tail. J. Biol. Chem. 266:5682-5688. [PubMed] [Google Scholar]
- 12.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36-48. [DOI] [PubMed] [Google Scholar]
- 13.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. Herpes simples virus type 1 (HSV-1) UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bionformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann, K., and W. Stoffel. 1993. TMbase-database of membrane spanning segments. J. Biol. Chem. 377:166. [Google Scholar]
- 21.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 24.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, K. F., and S. Kornfeld. 1992. The cytoplasmic tail of the mannose 6-phosphate/insulin-like growth factor-II receptor has two signals for lysosomal enzyme sorting in the Golgi. J. Cell Biol. 119:249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchhausen, T., J. S. Bonifacino, and H. Riezman. 1997. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr. Opin. Cell Biol. 9:488-495. [DOI] [PubMed] [Google Scholar]
- 28.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72:897-906. [DOI] [PubMed] [Google Scholar]
- 29.Marks, M. S., L. Woodruff, H. Ohno, and J. S. Bonifacino. 1996. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J. Cell Biol. 135:341-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellman, I. 1996. Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12:575-625. [DOI] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100-109. [DOI] [PubMed] [Google Scholar]
- 34.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 35.Ryechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 37.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 38.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press., Boca Raton, Fla.
- 39.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Tirabassi, R. S., and L. W. Enquist. 1999. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J. Virol. 73:2717-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 42.Trowbridge, I. S., J. F. Collawn, and C. R. Hopkins. 1993. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 9:129-161. [DOI] [PubMed] [Google Scholar]
- 43.Wan, L., S. S. Molloy, L. Thomas, G. Liu, Y. Xiang, S. L. Rybak, and G. Thomas. 1998. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94:205-216. [DOI] [PubMed] [Google Scholar]
- 44.Ward, P. L., G. Campadelli-Fiume, E. Avitabile, and B. Roizman. 1994. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J. Virol. 68:7406-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Z., Y. Hao, M. D. Gershon, R. T. Ambron, and A. A. Gershon. 1996. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J. Virol. 70:6563-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]