Abstract
Streptococcus pneumoniae clinical isolates were recently described that produced capsular polysaccharide with properties of both serotypes 6A and 6B. Their hybrid serological property correlated with mutations affecting the glycosyltransferase WciP, which links rhamnose to ribitol by an α(1-3) linkage for serotypes 6A and 6C and an α(1-4) linkage for serotypes 6B and 6D. The isolates had mutations in the triad residues of WciP that have been correlated with enzyme specificity. The canonical triad residues of WciP are Ala192-Ser195-Arg254 for serotypes 6A and 6C and Ser192-Asn195-Gly254 for serotypes 6B and 6D. To prove that the mutations in the triad residues are responsible for the hybrid serotype, we introduced the previously described Ala192-Cys195-Arg254 triad into a 6A strain and found that the change made WciP bispecific, resulting in 6A and 6B repeat unit expression, although 6B repeat unit production was favored over production of 6A repeat units. Likewise, this triad permitted a 6C strain to express 6C and 6D repeat units. With reported bispecificity in WciN, which adds either glucose or galactose as the second sugar in the serogroup 6 repeat unit, the possibility exists for a strain to simultaneously produce all four serogroup 6 repeat units; however, when genes encoding both bispecific enzymes were introduced into a 6A strain, only 6A, 6B, and 6D repeat units were detected serologically. Nonetheless, this may be the first example of a bacterial polysaccharide with three different repeat units. This strategy of expressing multiple repeat units in a single polymer is a novel approach to broadening vaccine coverage by eliminating the need for multiple polysaccharide sources to cover multiple serogroup members.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a major human pathogen due to its being a leading cause of pneumonia, meningitis, otitis media, and sepsis. The capsular polysaccharide (PS) is an important virulence factor that protects pneumococci from the host innate immune response and greatly enhances their pathogenicity (1, 2). Over 90 capsular serotypes have been defined based on serological properties (3–8). In addition to unique serological properties, each serotype produces a capsular PS with a distinct biochemical structure and has a unique capsule biosynthesis locus (cps) that encodes the enzymes required for capsule synthesis (3). In most serotypes, these enzymes assemble the repeat units on the cytoplasmic leaflet of the membrane through stepwise addition of each sugar, export the completed repeat units to the outer leaflet, and polymerize the repeat units (9).
Serogroup 6 contains four serotypes with unique repeat units—6A, 6B, 6C, and 6D—each with distinct serological, chemical, and genetic features (Fig. 1). The genetic basis for the four serotypes is the allelism of wciN and wciP. Serotypes 6A and 6B encode WciNα, which adds galactose to the repeat unit, whereas serotypes 6C and 6D encode WciNβ, which adds a second glucose (Glc′) (5, 10, 11). wciP allelism is the basis for differentiating serotypes 6A/6C from 6B/6D (6, 12, 13). WciPα links rhamnose to ribitol through an α(1-3) linkage, while WciPβ mediates an α(1-4) linkage (6). The two wciP alleles encode distinct amino acid triads at positions 192, 195, and 254 of the enzyme. Residue 195 correlates most strictly with serotype: a serine in WciPα correlates with serotypes 6A and 6C, but an asparagine in WciPβ correlates with serotypes 6B and 6D (Table 1) (12, 14).
FIG 1.
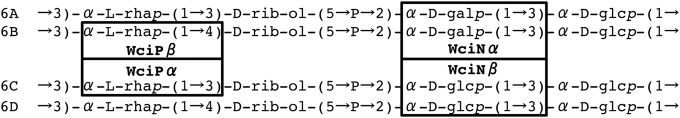
Schematic of the four known serogroup 6 repeat units and linkages mediated by WciP and WciN. P, PO4.
TABLE 1.
Strains used in this studya
| Strain | WciP triad residue at position: |
wciN allele | FCSA resultb | Quellung result | Reference | ||
|---|---|---|---|---|---|---|---|
| 192 | 195 | 254 | |||||
| TIGR6A | A | S | R | α | 6A | 6A | 11 |
| TIGR6B | S | N | G | α | 6B | 6B | 25 |
| TIGR6C | A | S | R | β | 6C | 6C | 11 |
| TIGR6D | S | N | G | β | 6D | 6Dc | 25 |
| PN6AB1d | S | S | G | α | NP | 6A | 17 |
| PN6AB4d | A | C | R | α | NP | 6B | 17 |
| MNZ1126 | S | S | G | α | 6A | 6A | This study |
| MNZ1130 | A | C | R | α | 6X13 | 6B | This study |
| IPZ2002 | A | C | R | β | 6X14 | NP | This study |
| MBO190 | A | C | R | α′e | 6X15 | NP | This study |
Nonencapsulated intermediate strains TIGR6AX, TIGR6AX2, and IPZ2001 are omitted from the table (see Materials and Methods). NP, not performed.
FCSA, flow cytometric serotyping assay.
Positive in Quellung reaction mixtures with both factor serum 6c (recognizes 6B and 6D) and factor serum 6d (recognizes 6C and 6D).
Included for comparative purposes. Strain and associated data were reported in reference 17.
Encodes bispecific WciN (D38N, A150T) (6).
In addition to the serotypes described above, three new serogroup 6 members were recently described. Serotype 6E has a cps locus that differs strikingly from those of known serogroup 6 members and is frequently typed as 6B by the Quellung reaction (15, 16), but its detailed serological and biochemical properties have not yet been reported. Serotypes 6F and 6G are serologically, biochemically, and genetically distinct (6). Serotype 6F capsular PS has repeat units of both serotypes 6A and 6C and expresses serological properties of both serotypes. Genetic studies revealed that 6F encodes a mutant WciNα protein (called WciNα′ here) that can add either glucose or galactose to the repeat unit (6). This bispecific activity of WciNα′ was traced to a single amino acid change, A150T, but the bispecificity was further enhanced by a second mutation, D38N (6). When wciNα′ is present in the serotype 6B cps locus, the mutation creates a new hybrid serotype with serological and biochemical properties of both 6B and 6D, and the hybrid serotype was named 6G (6).
Sheppard et al. described four clinical isolates with serological properties of both serotypes 6A and 6B and showed that the WciP proteins of these 6A/6B hybrid strains contained novel amino acid triads at positions 192, 195, and 254 (17). One isolate had a WciPα protein with A, C, and R at positions 192, 195, and 254, respectively, differing from canonical WciPα, which has residues A, S, and R at these respective positions. The remaining three isolates had a WciPβ protein with S, S, and G at residues 192, 195, and 254, respectively, deviating by a single amino acid from the canonical WciPβ triad of S, N, and G (17). For simplicity, these four WciP variants are herein denoted WciP (ACR), WciP (ASR), WciP (SSG), and WciP (SNG). Although no biochemical or site-directed mutation studies were performed with the four isolates, the correlations suggested that a mutation in residue 195 of WciP may impart the capability to form both α(1-3) and α(1-4) linkages between rhamnose and ribitol in the serogroup 6 repeat unit.
To determine whether either novel triad was sufficient to impart WciP with the ability to form both α(1-3) and α(1-4) linkages, we constructed isogenic strains with a serotype 6A cps locus differing only in the mutations in wciPα by using site-directed mutagenesis and studied the resulting strains serologically and biochemically. In addition, we explored the possibility of engineering a strain producing all four unique serogroup 6 repeat units by introducing the previously described bispecific variant wciNα′ (6) into a strain with bispecific WciP, since such a strain may simplify vaccine formulation while maintaining broad serotype coverage.
MATERIALS AND METHODS
Bacterial strains and other reagents.
Experimental strains are described in Table 1, and genetic intermediate strains are described below (see “Genetic manipulations”). All strains were made in the TIGR4 background (18). For routine cultivation, pneumococci were streaked from frozen glycerol stocks onto blood agar plates (Remel) and grown overnight at 37°C in 5% CO2 before inoculation into liquid culture. For liquid culture growth, pneumococci were grown in Todd-Hewitt broth (BD Biosciences, San Jose, CA) containing 0.5% yeast extract (THY) (for flow cytometry and enzyme-linked immunosorbent assays [ELISAs]; see below) or in chemically defined medium (CDM; JRH Biosciences, Lenexa, KS) (19) supplemented with choline chloride (1 g/liter), sodium bicarbonate (2.5 g/liter), and cysteine-HCl (0.73 g/liter) (for flow cytometry and PS purification; see below) at 37°C in a water bath.
Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO) and used at the following concentrations when appropriate: kanamycin, 100 μg/ml; streptomycin, 300 μg/ml; and spectinomycin, 100 μg/ml. Serotype 6A and 6B capsular PSs were purchased from the Statens Serum Institut (Copenhagen, Denmark). Mouse hybridomas were previously described and are summarized in Table 2 (5, 6, 20, 21).
TABLE 2.
Properties of hybridomas used in this study
| Hybridoma | Isotype | Serotype reactivity |
|||
|---|---|---|---|---|---|
| 6A | 6B | 6C | 6D | ||
| Hyp6AM3 | IgM | + | − | − | − |
| Hyp6AG3 | IgG | + | − | + | − |
| Hyp6BG3 | IgG | − | + | − | − |
| Hyp6BM1 | IgM | − | + | − | − |
| Hyp6CG6 | IgG | − | − | + | − |
| Hyp6DM3 | IgM | − | − | − | + |
Genetic manipulations.
Primers used in this study are listed in Table 3. TIGR6AX (TIGR6A wciN::Janus cassette) (described in reference 11) was transformed with a modified Janus cassette (22) in which the kanamycin resistance gene was replaced with aad9, a spectinomycin resistance gene (obtained from the plasmid pCLT1242 [23]), to generate TIGR6AX2 (TIGR6AX wciP::aad9-rpsL+). Constructs 1 and 2, each encoding different triads of residues at positions 192, 195, and 254, were generated using overlap extension PCR with the primers described in Table 3, as follows. 5′ fragments (wze-wciP′) were amplified from TIGR6A chromosomal DNA by use of primer 5114 and the reverse primer containing the desired mutation(s), and 3′ fragments (′wciP-wzx) were amplified from TIGR6A (construct 2) or TIGR6B (construct 1) chromosomal DNA by use of the forward primer containing the desired mutation(s) and primer 3144. The resulting products were mixed and amplified using internal primers 5113 and 3143, yielding constructs 1 and 2, and transformed into TIGR6AX2 as previously described (11), yielding strains MNZ1126 and MNZ1130, respectively (summarized in Table 1).
TABLE 3.
Primers used in this study
| Primera | Descriptionb | DNA sequence (5′ → 3′)c | Reference |
|---|---|---|---|
| 3143 | wzy10135–10115 | CCTCCCATATAACGAGTGATG | 11 |
| 3144 | wzx12068–12049 | GCGAGCCAAATCGGTAAGTA | 11 |
| 3688 | wciP8886–8852 (S192), construct 1 | ACTATCATGAGACAGTTTTTCTATATATAAATAAT | This study |
| 3691 | wciP8886–8852 (C195), construct 2 | ACAATCATGAGCCAGTTTTTCTATATATAAATAAT | This study |
| 5113 | wchA4893–4870 | GGGAAAAATAAAAAATAGGTCGGG | 11 |
| 5114 | wze4737–4717 | TTAGTGACGGAGGCAGGTGAA | 11 |
| 5689 | wciP8874–8899 (S192), construct 1 | GTCTCATGATAGTTATTTTGCAAAGT | This study |
| 5698 | wciP8872–8899 (C195), construct 2 | CTGGCTCATGATTGTTATTTTGCAAAGT | This study |
Primers beginning with “3” are reverse primers, and primers beginning with “5” are forward primers, with respect to the reference sequence.
Nucleotide sequence numbers correspond to GenBank accession no. CR931638.
Underlined residues indicate divergence from the reference sequence.
To generate IPZ2002, wciNα was replaced with wciNβ in MNZ1130 through allelic exchange with the Janus cassette as previously described (11). To generate MBO190, wciNα′, encoding the bispecific enzyme WciNα′ (D38N, A150T), was introduced into MNZ1130 through allelic exchange with the Janus cassette as described previously (6).
All mutations were confirmed by DNA sequencing by the Heflin Center for Human Genetics, University of Alabama at Birmingham, Birmingham, AL, or Macrogen Company, Seoul, South Korea.
Flow cytometry.
Flow cytometric serotyping assays were performed as previously described (4, 24). In brief, pneumococci were cultured in liquid medium until the optical density at 600 nm (OD600) was ∼0.6 (THY) or 1.2 (CDM). Working samples were generated by mixing cultures with an equal volume of medium, supplementing them with 80% glycerol to a final concentration of 16%, and freezing aliquots at −80°C until use. Frozen bacterial aliquots were washed and resuspended in fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline [PBS] with 3% fetal bovine serum and 0.01% sodium azide). Aliquots of 50 μl (containing ∼5 × 105 CFU) were incubated with 50 μl of hybridoma culture supernatant at 4°C for 30 min. After incubation, bacteria were washed twice and incubated with either (i) goat anti-mouse IgG antibody conjugated with phycoerythrin (Southern Biotech, Birmingham, AL) or (ii) goat anti-mouse IgM antibody conjugated with phycoerythrin-Cy7 (Southern Biotech, Birmingham, AL). After washing, the bacteria were examined in a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA). The data were analyzed with FCS Express, versions 3.0 and 4.0 (De Novo Software, Los Angeles, CA).
Sandwich ELISA.
To investigate whether multiple epitopes were present in a single carbohydrate polymer, a sandwich ELISA was developed using monoclonal antibodies of differing isotypes. ELISA plates were coated with rabbit anti-mouse IgG1 antibody (Zymed) diluted 1:1,000 in PBS with 0.05% Tween 20 (PBST) overnight at 4°C. Plates were washed three times with PBST and incubated with Hyp6AG3 (1:100 in PBST) for 1 h at 37°C. After washing, the plates were blocked with 3% powdered skim milk in PBST. Bacterial lysates, prepared by deoxycholate lysis of THY-grown bacteria as described previously (21), were serially diluted 10-fold in PBST and added to the plates for 1 h at 37°C. After washing with PBST, Hyp6BM1 (1:100 dilution) was added to the wells and incubated for 1 h at 37°C. Detection antibody (anti-mouse IgM–alkaline phosphatase; Sigma) was diluted 1:10,000 in PBST and added to each well prior to incubation at 37°C for 1 h. After washing, plates were incubated with p-nitrophenyl phosphate (Sigma) for 2 h before the optical density at 405 nm was read with a microplate reader (BioTek Instruments Inc., Winooski, VT). (All volumes were 100 μl per well, except for washes.)
Purification of capsular polysaccharide.
Capsular PS was purified from strains MNZ1130 and IPZ2002 by anion-exchange chromatography of mutanolysin-treated cultures as previously described (25), with the modification that elution from DEAE-Sepharose columns was performed with a step gradient of 0 to 1 M NaCl in 0.1 M increments. The capsular PS-containing fractions were identified by capsule-specific ELISA and pooled. Capsular PS in the pool was precipitated with 70% ethanol, dialyzed against H2O, and lyophilized. The lyophilized capsular PSs were dissolved and fractionated by size with Sephacryl S-300 HR. The fractions containing the first capsular PS peak were isolated, pooled, and lyophilized for nuclear magnetic resonance (NMR) analysis.
NMR spectroscopy.
For 1H-NMR analysis, approximately 10 mg of lyophilized capsular PS was dissolved in D2O, and spectra were collected with a 500-MHz NMR spectrometer (INOVA-500) at 25°C. Data were analyzed with an ACD/NMR processor (Advanced Chemistry Development, Inc., Toronto, Canada). Chemical shifts were assigned based on published structures reported elsewhere (6, 13).
Nucleotide sequence accession numbers.
Nucleotide sequences of wciP from MNZ1126 and MNZ1130 were submitted to GenBank under accession no. KJ874435 and KJ874439. A partial cps locus sequence for MBO190, including wciNα′ and wciP, was submitted to GenBank under accession no. KF597302.
RESULTS
WciP (ACR) is sufficient for production of serotype 6A and 6B repeat units in a serotype 6A strain.
To determine whether the previously described mutations in wciP (17) can induce serotype 6A S. pneumoniae to express capsular PS comprised of both serotype 6A and 6B epitopes, we made isogenic TIGR6A-derived variants encoding WciP (ACR) (MNZ1130) or WciP (SSG) (MNZ1126). WciP (ACR) and WciP (SSG) correspond to the mutant WciP enzymes expressed by the previously reported PN6AB4 and PN6AB1 strains, respectively (Table 1) (17). Through standard Quellung serotyping using factor serum 6b (which recognizes serotype 6A) and factor serum 6c (which recognizes serotypes 6B and 6D), MNZ1130 was serotyped as serotype 6B, and MNZ1126 was serotyped as 6A, consistent with the reported Quellung serotypes of isolates PN6AB4 and PN6AB1 (Table 1) (17).
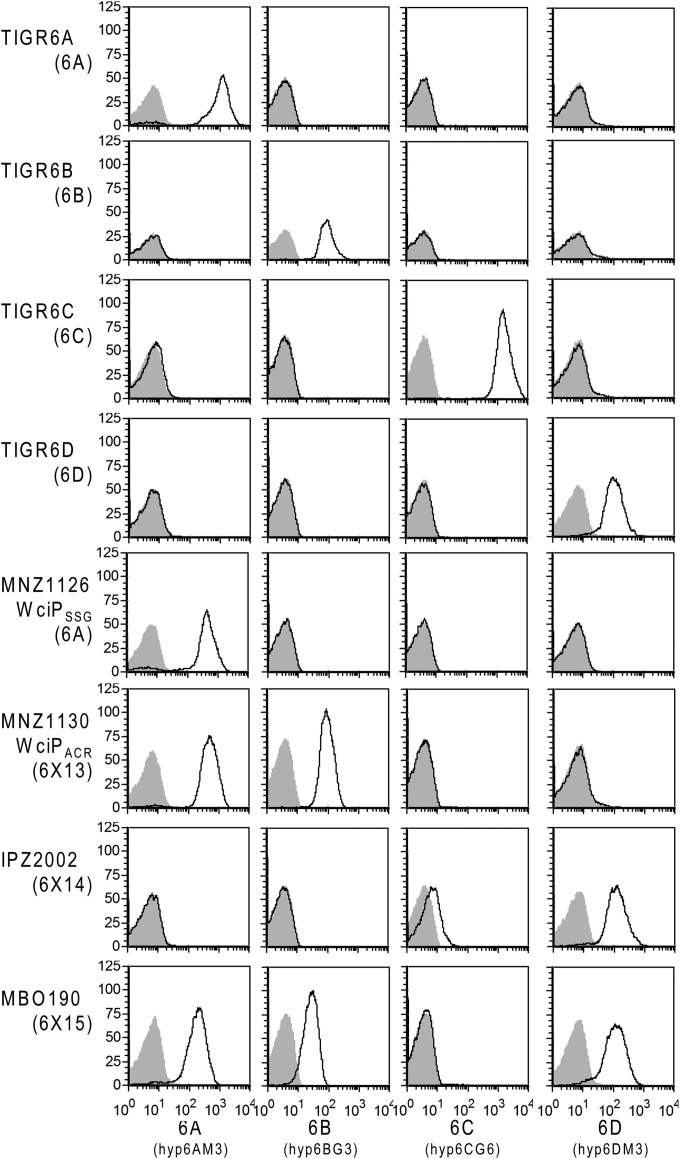
We further serotyped MNZ1126 and MNZ1130 by using monoclonal antibodies. The specificities of the murine hybridomas used to serotype isolates are shown in Fig. 2 (TIGR6A to TIGR6D). MNZ1130 was bound by 6A- and 6B-specific monoclonal antibodies, indicative of expression of both repeat units; however, MNZ1126 was bound by only 6A-specific antibodies (Fig. 2). While an additional mutation (Y91C) in WciP (SSG) of MNZ1126 prevents us from formally concluding that the SSG triad is insufficient for dual repeat unit expression, Y91 is not known to be associated with the functional site. Nonetheless, the ACR triad is sufficient to alter WciP specificity.
FIG 2.
Serological profiles of strains used in this study. The indicated strains (at left) were grown in CDM and then incubated with supernatants from the indicated hybridomas (at bottom), producing antibodies specific for each unique serogroup 6 repeat unit, prior to staining with appropriate secondary antibodies. Similar results were obtained with bacteria grown in THY (data not shown). y axes represent numbers of events at a given fluorescence intensity. Gray shaded areas represent secondary antibody-only controls.
MNZ1130 expresses serotype 6A and 6B repeat units within the same polymer chain.
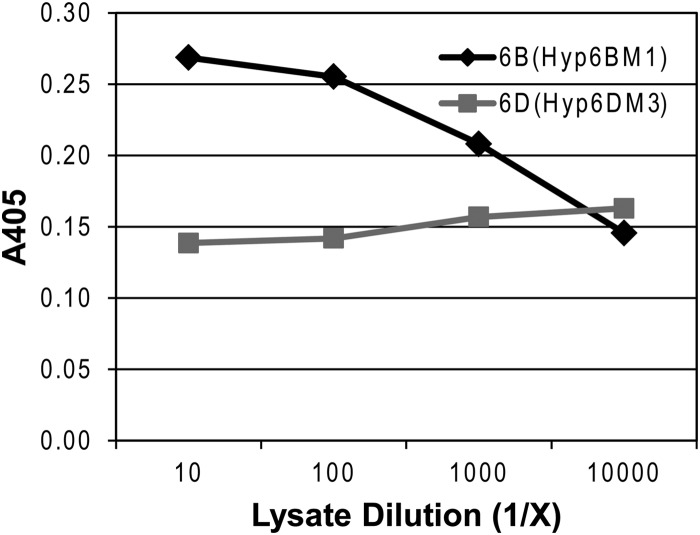
To determine whether capsular PS of MNZ1130 is expressed with 6A and 6B repeat units within a single polymer, as with hybrid capsules composed of 6A/6C repeat units (6F) or 6B/6D repeat units (6G) (6), we developed a sandwich ELISA using monoclonal antibodies of differing isotypes. Serotype 6A repeat units in 6X13 PS (PS expressing both 6A and 6B repeat units) were captured on a microtiter plate by a 6A/6C-specific antibody (Hyp6AG3). Hyp6DM3, an isotype-matched negative-control antibody that is 6D specific (Fig. 2), did not recognize 6X13 PS (Fig. 3), but captured PS was bound by a 6B-specific antibody (Hyp6BM1) in a concentration-dependent manner, indicating that 6X13 contains both serotype 6A and 6B repeat units in a single PS chain (Fig. 3).
FIG 3.
Sandwich ELISA results demonstrating expression of 6A and 6B repeat units in the same polymer. Capsular PS from MNZ1130 (6X13) was captured by a 6A-specific monoclonal antibody (Hyp6AG3) and detected using a 6B-specific monoclonal antibody (Hyp6BM1) as described in Materials and Methods. A 6D-specific isotype-matched antibody (Hyp6DM3) was used as a negative control.
Bispecific WciP (ACR) preferentially forms α(1-4) linkages.
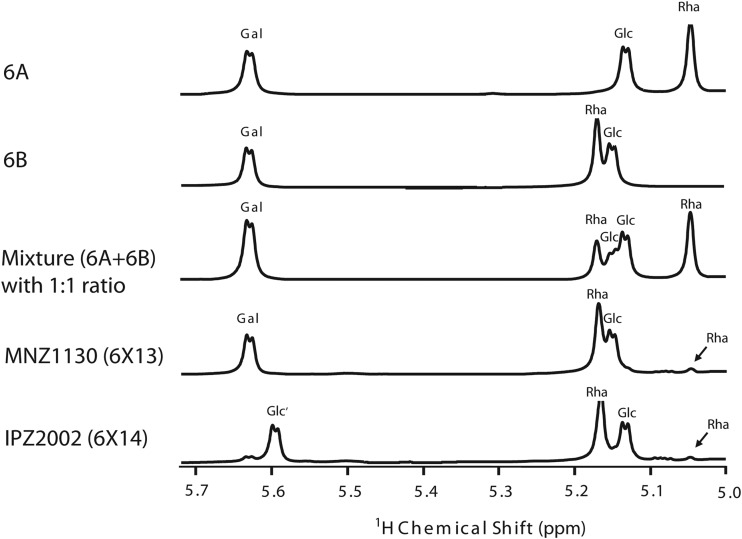
To investigate the chemical structure of 6X13 capsular PS, 1H-NMR was utilized to examine the ratio of 6A and 6B repeat units in MNZ1130. Spectra were consistent with previously described shifts for the anomeric carbons of serogroup 6 repeat units (6, 13). Based on the area under the anomeric peak corresponding to rhamnose for serotype 6A (5.05 ppm) compared to that for serotype 6B (5.17 ppm), 6X13 capsular PS is disproportionately composed of 6B repeat units relative to 6A repeat units (>9:1) (Fig. 4). The strong bias toward 6B repeat units might account for the relatively weak Quellung reaction for MNZ1130 observed with factor serum 6b (serotype 6A specific) relative to that with factor serum 6c (serotype 6B/6D specific), as previously described for isolate PN6AB4 (Table 1 and data not shown) (17).
FIG 4.
One-dimensional 1H-NMR spectra of wciP mutants expressing hybrid repeat units. Gal, galactose; Glc, glucose; Rha, rhamnose; Glc′, the second Glc in serotype 6C/6D repeat units.
Replacement of wciNα with wciNβ in MNZ1130 results in expression of a capsular polysaccharide containing serotype 6C and 6D epitopes.
A serotype 6A strain can be induced to express serotype 6C capsular PS (or a serotype 6B strain to express serotype 6D capsular PS) when the wciNα gene is replaced with wciNβ (4, 11). We replaced wciNα of MNZ1130 with wciNβ from TIGR6C to generate IPZ2002, which should express capsular PS with epitopes for both serotypes 6C and 6D (termed 6X14). Consistent with this prediction, IPZ2002 reacted with 6C- and 6D-specific monoclonal antibodies (Fig. 2). 1H-NMR spectra confirmed that galactose (5.63 ppm) was replaced with glucose (5.60 ppm) in IPZ2002 and that serotype 6C and 6D repeat units were present at a ratio similar to that between serotype 6A and 6B repeat units in MNZ1130 (Fig. 4).
A serogroup 6 strain encoding bispecific WciNα′ and WciP (ACR) expresses a hybrid capsule consisting of serotype 6A, 6B, and 6D repeat units.
Theoretically, combining bispecific wciNα′ and wciP (ACR) alleles should permit a serogroup 6 strain to produce capsular PS containing all four unique serogroup 6 repeat units. To investigate this possibility, we created MBO190, an MNZ1130 derivative encoding the bispecific glucosyl-/galactosyltransferase WciNα′ in addition to WciP (ACR). Staining with monoclonal antibodies clearly demonstrated the presence of 6A, 6B, and 6D repeat units (Fig. 2); however, we were reproducibly unable to detect 6C repeat units serologically.
DISCUSSION
Sheppard et al. reported the existence of atypical serogroup 6 clinical isolates that express antigens associated with both serotypes 6A and 6B and whose encoded WciP proteins differ from the canonical WciP enzymes of serotypes 6A and 6B at residue 195 (17). To investigate these correlations, we produced isogenic mutants and demonstrated that the ACR triad is sufficient for expression of a 6A/6B hybrid, but we were unable to confirm 6A/6B hybrid expression in a strain encoding the reported SSG triad. The residue at position 195 of WciP appears to be a key determinant of WciP specificity, modulating the ability of the enzyme to link rhamnose to ribitol with either an α(1-3) or α(1-4) linkage. Because it was discovered in clinical isolates (17), we propose that this hybrid serotype expressing epitopes for both 6A and 6B (6X13) is a new serotype, which we named “6H.” We also demonstrated that the same bispecificity may be conferred to WciP in serotype 6C strains in vitro, permitting a strain to express 6C and 6D units in the same polymer chain (termed 6X14 here), but such an isolate has yet to be identified in vivo. When it is discovered, we propose that it be designated serotype “6I.”
Genes for at least three bispecific enzymes have been identified within pneumococcal cps loci to date. Two such enzymes, WciN (serogroup 6) and WcrL (serotype 11D), mediate the addition of multiple sugar substrates (6, 26); the third, described here and previously (17), mediates two different linkages of its substrates. These are part of a growing repertoire of described and implicated bispecific transferases in eukaryotes (27, 28) and prokaryotes (6, 26, 29, 30) alike.
It is becoming apparent that bispecific transferases are not as rare as expected and can arise from single nucleotide changes within their respective genes (6, 26, 30; this study). The ability of single nucleotide differences to alter the capsular PS serotype has important ramifications for serotyping by qualitative genetic methods (e.g., PCR) alone. Indeed, small changes in nucleotide sequences have proven troublesome for typing by genetic means; single-base or short changes have caused entire serotype changes in serogroups 6 (6, 17; this study) and 11 (7, 26), and a variant wzy allele in serogroup 19 resulted in mistyping of serotypes 19F and 19A, which vary by the Wzy-mediated linkage, in multiplex PCR serotyping schemes (31–33).
The major limitation of the conjugate vaccine strategy is the difficulty in increasing the number of serotypes covered. Bispecific enzymes present a novel means of simplifying conjugate vaccine production and provide a means for the inclusion of more serotypes in vaccine formulations, without increasing the number of capsular PS sources required. Additional examination of residue 192 in WciP may reveal a point for optimizing WciP toward more equitable linkage formation. For example, D38 of WciN was shown to alter the ratio of 6A to 6C repeat units in serotype 6F while not itself determining the specificity of the enzyme (6). In preliminary studies in mice, we found that immunization with MBO190 (serotype 6X15) elicited antibody responses against serotype 6A, 6B, 6C, and 6D PSs, similar to immunization with TIGR6B (serotype 6B) (data not shown). More realistic studies using appropriate vaccine formulations and vaccination schedules in human volunteers are needed to assess the immunogenicity of hybrid PSs. With carefully applied mutagenesis and optimization, it may be feasible to engineer strains within this and other serogroups producing multiple serotypes of vaccine interest, bypassing reliance upon cross-protection within serogroups and expanding the repertoire of conjugate vaccines.
ACKNOWLEDGMENTS
We thank Charles L. Turnbough, Jr., Department of Microbiology, University of Alabama at Birmingham, for providing pCLT1242, and Jamil S. Saad, Department of Microbiology, University of Alabama at Birmingham, for providing his expertise in evaluating our NMR data. We thank Jisheng Lin, Department of Pathology, University of Alabama at Birmingham, for assistance with animal experiments.
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea, funded by the Ministry of Education (grants NRF-2012R1A1A2005675 and NRF-2013R1A1A2007513), by an Ewha Womans University research grant of 2012, and by National Institute of Allergy and Infectious Diseases grant R56-AI-031473 to M.H.N.
The University of Alabama at Birmingham (UAB) has intellectual property rights on some reagents used in the study, and I.H.P., K.A.G., J.Y., M.B.O., and M.H.N. were UAB employees during some or all of the study period. We have no additional conflicts of interest to disclose.
REFERENCES
- 1.Kim JO, Romero-Steiner S, Sorensen UB, Blom J, Carvalho M, Barnard S, Carlone G, Weiser JN. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect Immun 67:2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun 69:3755–3761. doi: 10.1128/IAI.69.6.3755-3761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratcher PE, Kim KH, Kang JH, Hong JY, Nahm MH. 2010. Identification of natural pneumococcal isolates expressing serotype 6D by genetic, biochemical and serological characterization. Microbiology 156:555–560. doi: 10.1099/mic.0.034116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MC, Nahm MH. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol 45:1225–1233. doi: 10.1128/JCM.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver MB, van der Linden MP, Kuntzel SA, Saad JS, Nahm MH. 2013. Discovery of Streptococcus pneumoniae serotype 6 variants with glycosyltransferases synthesizing two differing repeating units. J Biol Chem 288:25976–25985. doi: 10.1074/jbc.M113.480152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calix JJ, Nahm MH. 2010. A new pneumococcal serotype, 11E, has a variably inactivated wcjE gene. J Infect Dis 202:29–38. doi: 10.1086/653123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calix JJ, Porambo RJ, Brady AM, Larson TR, Yother J, Abeygunwardana C, Nahm MH. 2012. Biochemical, genetic, and serological characterization of two capsule subtypes among Streptococcus pneumoniae serotype 20 strains: discovery of a new pneumococcal serotype. J Biol Chem 287:27885–27894. doi: 10.1074/jbc.M112.380451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yother J. 2011. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 10.Bratcher PE, Nahm MH. 2010. Cross reactivity of current serogroup 6 factor sera from Statens Serum Institut with the recently described pneumococcal serotype 6D. J Clin Microbiol 48:3044–3045. doi: 10.1128/JCM.00839-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park IH, Park S, Hollingshead SK, Nahm MH. 2007. Genetic basis for the new pneumococcal serotype, 6C. Infect Immun 75:4482–4489. doi: 10.1128/IAI.00510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mavroidi A, Godoy D, Aanensen DM, Robinson DA, Hollingshead SK, Spratt BG. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J Bacteriol 186:8181–8192. doi: 10.1128/JB.186.24.8181-8192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai P, Moran J, Pavliak V, Deng C, Khoury N, Marcq O, Ruppen ME. 2012. NMR structural analysis of the capsular polysaccharide from Streptococcus pneumoniae serotype 6C. Carbohydr Res 351:98–107. doi: 10.1016/j.carres.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Pai R, Limor J, Beall B. 2005. Use of pyrosequencing to differentiate Streptococcus pneumoniae serotypes 6A and 6B. J Clin Microbiol 43:4820–4822. doi: 10.1128/JCM.43.9.4820-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko KS, Baek JY, Song JH. 2013. Capsular gene sequences and genotypes of “serotype 6E” Streptococcus pneumoniae isolates. J Clin Microbiol 51:3395–3399. doi: 10.1128/JCM.01645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek JY, Ko KS, Song JH. 2011. Streptococcus pneumoniae serotype 6D cross-reacting with serotype 6A, 6B, and 6C factor sera. J Clin Microbiol 49:765–766. doi: 10.1128/JCM.01933-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheppard CL, Pichon B, George RC, Hall LM. 2010. Streptococcus pneumoniae isolates expressing a capsule with epitopes of both serotypes 6A and 6B. Clin Vaccine Immunol 17:1820–1822. doi: 10.1128/CVI.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 19.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Hwang Y, Nahm MH. 2001. Avidity, potency, and cross-reactivity of monoclonal antibodies to pneumococcal capsular polysaccharide serotype 6B. Infect Immun 69:336–344. doi: 10.1128/IAI.69.1.336-344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Lin J, Kim KH, Benjamin WH Jr, Nahm MH. 2011. Development of an automated and multiplexed serotyping assay for Streptococcus pneumoniae. Clin Vaccine Immunol 18:1900–1907. doi: 10.1128/CVI.05312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong S, McPherson SA, Tan L, Chesnokova ON, Turnbough CL Jr, Pritchard DG. 2008. Anthrose biosynthetic operon of Bacillus anthracis. J Bacteriol 190:2350–2359. doi: 10.1128/JB.01899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calix JJ, Saad JS, Brady AM, Nahm MH. 2012. Structural characterization of Streptococcus pneumoniae serotype 9A capsule polysaccharide reveals role of glycosyl 6-O-acetyltransferase wcjE in serotype 9V capsule biosynthesis and immunogenicity. J Biol Chem 287:13996–14003. doi: 10.1074/jbc.M112.346924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bratcher PE, Park IH, Hollingshead SK, Nahm MH. 2009. Production of a unique pneumococcal capsule serotype belonging to serogroup 6. Microbiology 155:576–583. doi: 10.1099/mic.0.024521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver MB, Jones C, Larson TR, Calix JJ, Zartler ER, Yother J, Nahm MH. 2013. Streptococcus pneumoniae serotype 11D has a bispecific glycosyltransferase and expresses two different capsular polysaccharide repeating units. J Biol Chem 288:21945–21954. doi: 10.1074/jbc.M113.488528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakrishnan B, Qasba PK. 2002. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity. J Biol Chem 277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan B, Qasba PK. 2007. Role of a single amino acid in the evolution of glycans of invertebrates and vertebrates. J Mol Biol 365:570–576. doi: 10.1016/j.jmb.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claus H, Stummeyer K, Batzilla J, Muhlenhoff M, Vogel U. 2009. Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol Microbiol 71:960–971. doi: 10.1111/j.1365-2958.2008.06580.x. [DOI] [PubMed] [Google Scholar]
- 30.Yuki N. 2007. Campylobacter sialyltransferase gene polymorphism directs clinical features of Guillain-Barre syndrome. J Neurochem 103(Suppl 1):S150–S158. doi: 10.1111/j.1471-4159.2007.04707.x. [DOI] [PubMed] [Google Scholar]
- 31.Pimenta FC, Gertz RE Jr, Roundtree A, Yu J, Nahm MH, McDonald RR, Carvalho MG, Beall BW. 2009. Rarely occurring 19A-like cps locus from a serotype 19F pneumococcal isolate indicates continued need of serology-based quality control for PCR-based serotype determinations. J Clin Microbiol 47:2353–2354. doi: 10.1128/JCM.00704-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siira L, Kaijalainen T, Lambertsen L, Nahm MH, Toropainen M, Virolainen A. 2012. From quellung to multiplex PCR, and back when needed, in pneumococcal serotyping. J Clin Microbiol 50:2727–2731. doi: 10.1128/JCM.00689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menezes AP, Reis JN, Ternes YM, Andrade AL, Pimenta FC, Carvalho MG, Beall B. 2013. Update of pneumococcal PCR serotyping assay for detection of a commonly occurring type 19F wzy variant in Brazil. J Clin Microbiol 51:2470–2471. doi: 10.1128/JCM.00743-13. [DOI] [PMC free article] [PubMed] [Google Scholar]