Abstract
Vesicular stomatitis virus expressing Zaire Ebola virus (EBOV) glycoprotein (VSVΔG/EBOVgp) could be used as a vaccine to meet the 2014 Ebola virus outbreak. To characterize the host response to this vaccine, we used mRNA sequencing to analyze peripheral blood mononuclear cells (PBMCs) from cynomolgus macaques after VSVΔG/EBOVgp immunization and subsequent EBOV challenge. We found a controlled transcriptional response that transitioned to immune regulation as the EBOV was cleared. This observation supports the safety of the vaccine.
TEXT
The 2014 Ebola virus outbreak in West Africa has highlighted the need to deliver a vaccine to those in direct contact with the virus, which includes health care workers and first responders (1). At present, two vaccines are undergoing clinical trials; both have shown effective protection in nonhuman primates (NHPs) and express the Zaire Ebola virus glycoprotein (EBOVgp) on an adenovirus (2) or vesicular stomatitis virus (VSV) (3, 4) backbone. The VSV-based vaccine has the advantage of needing only a single immunization without a separate boost, and it has increased durability. As we have reported, the immunity provided by VSV expressing EBOV glycoprotein (VSVΔG/EBOVgp) vaccine completely abolished the clinical signs of disease and was mediated by IgG antibodies (5). However, the host responses to immunization and subsequent EBOV challenge have not been characterized on a genomic scale; this might aid in assessments of vaccine safety, complementing previous targeted analyses (6–8).
To complement previous measures of viral load and inflammatory mediators in these animals (5), we performed RNA deep sequencing analysis (see Supplemental Methods in the supplemental material) on peripheral blood mononuclear cells (PBMCs) obtained from cynomolgus macaques (CMs) prior to and after immunization with VSV carrying EBOVgp (VSVΔG/EBOVgp) and at multiple points after EBOV challenge. Specifically, three CMs were immunized using a single dose of VSVΔG/EBOVgp 28 days prior (D−28) to EBOV challenge (day 0 [D0]), and PBMCs were collected on D−28, D0, D4, D7, D14, and D35. As a positive control, we also assessed data from animals immunized with a nonprotective vaccine carrying a Marburg virus glycoprotein (VSVΔG/MARVgp) (9). These animals showed the hallmarks of Ebola hemorrhagic fever and succumbed around D7 (7).
To assess the host response to immunization and EBOV challenge, sequenced reads were mapped to both the host and EBOV genomes (GEO accession no. GSE64538). We detected low levels of viral transcription in all three protected animals following challenge, with virus levels peaking at D7 before it was cleared (Table 1), which was consistent with previously measured IgG titers (5). Normalization and differential expression (DE) analysis of the host transcriptome were carried out using the Bioconductor package EdgeR (see Supplemental Methods in the supplemental material). Immunization triggered a restricted transcriptional response, in that the entire time series produced a pool of only 502 DE genes (Fig. 1A) compared to 2,661 in the unprotected animals (see Fig. S1A in the supplemental material). In the protected animals, expression changes that manifested between D−28 and D0 tended to persist throughout the time series; EBOV challenge had a comparatively small effect on gene expression.
TABLE 1.
Numbers of sequenced reads mapping to the Ebola virus genome in each animal immunized with protective (EBOVgp) and nonprotective (MARVgp) vaccines
| Groupa | No. of sequenced reads found on: |
||||
|---|---|---|---|---|---|
| D0 | D4 | D7 | D14 | D35 | |
| EBOVgp 1 | 0 | 0 | 33 | 0 | 0 |
| EBOVgp 2 | 0 | 1 | 55 | 0 | 0 |
| EBOVgp 3 | 0 | 14 | 22 | 15 | 1 |
| MARVgp 1 | 0 | 1,416 | 65,822 | ||
| MARVgp 2 | 0 | 383 | 57,217 | ||
| MARVgp 3 | 0 | 164 | 88,664 | ||
In the protected group, a small number of reads was found, peaking at D7.
FIG 1.
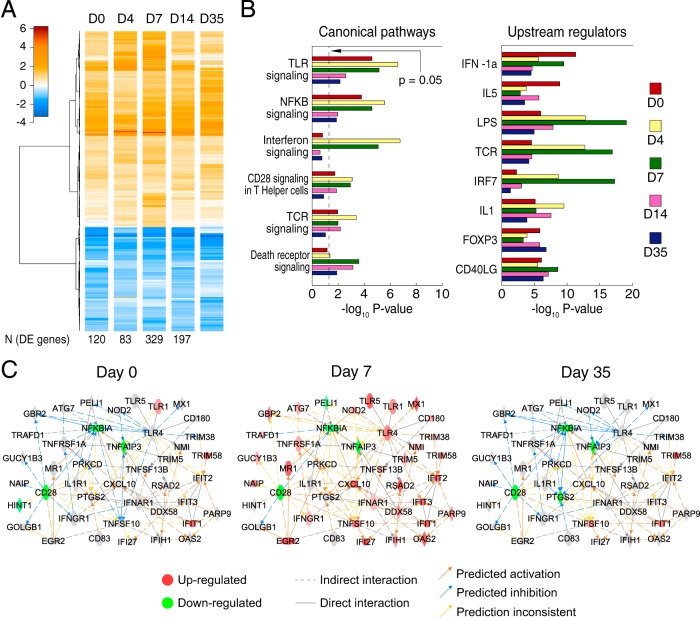
Host response to immunization and Zaire Ebola virus challenge in the protected group. (A) Heatmap showing, at each time point, the average log2 fold change (FC) compared to D−28 of the 502 pooled DE genes from D0 to D35. The differential expression observed at D0 tended to persist throughout the time series, although the number of genes passing the significance threshold varied, peaking at D7. (B) Functional enrichment analysis for canonical pathways and targets of upstream regulators. Both analyses show the activation of interferon responses during the first time points and T-cell activation peaking at D7. LPS, lipopolysaccharide; TCR, T-cell receptor. (C) Constructed network showing interactions between key genes from enriched canonical pathways. The D0 network showed differential expression, suggesting the regulation of immune responses, while the D7 network showed predominantly immune activation. By D35, the network had returned to near that of the D0 state.
To identify the key elements in the host response to immunization and challenge, we used ingenuity pathway analysis (IPA) to perform functional enrichment analysis on the DE genes from each time point (Fig. 1B). The analysis identified biological pathways and predicted transcription factors and other regulators of gene expression (e.g., cytokines) as underlying regulators of the host response to immunization. The unprotected animals showed increasing activation of immune pathways with deteriorating health (see Fig. S1B and Supplemental Results in the supplemental material).
This analysis specifically showed the activation of Toll-like receptor (TLR) signaling and NF-κB activation after immunization (D0), and IPA implicated beta interferon 1a (IFN-β-1a) and interleukin-5 (IL-5) as underlying regulators of this response. TLRs have been implicated in protection against VSV in both in vivo knockout studies (10) and cell culture of dendritic cells (11) and macrophages (12). The TLRs found to be upregulated (TLR1, TLR6, and TLR10) at D0 have not been reported in the context of VSV, but our data suggest that TLR signaling participates in the immunogenicity of VSVΔG/EBOVgp, as it has in other vaccines (13). At D4 and D7, the response to challenge after immunization also included interferon signaling (upregulation of alpha/beta interferon receptor 1 [IFNAR1], gamma interferon receptor 1 [IFNGR1], IFNGR2, interferon-induced protein with tetratricopeptide 1 [IFIT1], and MX dynamin-like GTPase 1 [MX1]), CD28 signaling, and T-cell receptor signaling. The upstream IPA also implicated the T-cell receptor and interferon regulatory factor 7 (IRF7) as regulators of the observed gene expression changes.
In order to identify the interactions between the immunological processes that are activated at different time points, we used IPA to carry out an enrichment analysis of the pool of 502 DE genes in manually curated biological networks (14). This allowed us to include the interactions between DE genes that were not present in canonical signaling pathways. Networks containing key genes from the enriched pathways were merged, and DE genes from each time point were mapped onto this resulting network (Fig. 1C). In this network, TLR4 had the highest number of outgoing interactions (i.e., regulated the highest number of genes). Its downstream-regulated genes (MX1, guanylate binding protein 2 [GBP2], tripartite motif-containing protein 38 [TRIM38], and nucleotide binding oligomerization domain-containing protein 2 [NOD2]) showed predominantly predicted inhibition at D0, activation at D7, and returned to inhibition at D35, indicating that after the initial host response to EBOV challenge, the response resolved and returned close to the baseline state. Immune regulation at D35 was indicated by the activation of a significant number of targets for forkhead box P3 (FOXP3), the marker of regulatory T cells. In contrast, a number of antiviral genes, regulated in the network by DEAD box protein 58 (DDX58) and IFNAR1, showed upregulation or predicted activation throughout the time course (IFIT1, IFI27, tumor necrosis factor [ligand] superfamily, member 13B [TNFSF13B], and radical S-adenosyl methionine domain containing 2 [RSAD2]). Also, the T-cell coreceptor CD28 showed downregulation from D0 throughout the time series, possibly due to the increased frequency of effector memory T cells in response to immunization.
Taken together, these results suggest that in immunized challenged animals, innate immune activation correlates with EBOV replication over time, peaking around D7 after infection, supporting the participation of innate immune responses in IgG-dependent immunity. At D35 after infection, the immune response appears to have resolved; the number of differentially expressed genes decreased, and the immune pathways activated at D7 were downregulated while immune regulators (e.g., FOXP3) were activated. While these results are based on a limited number of animals, they suggest that NHPs immunized by VSVΔG/EBOVgp show restricted immune activation upon EBOV challenge and resolve their immune response after the infection is cleared.
Our observations support the safety of VSVΔG/EBOVgp; the number of DE genes following immunization and EBOV challenge was limited and entailed the early activation of genes associated with antiviral immunity, with subsequent activation of immune regulators. The VSVΔG vector can be redesigned to incorporate glycoproteins from other Ebola viruses; even though such vaccines would need an independent evaluation, our results suggest that this approach is likely to produce safe vaccines.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00733-14.
REFERENCES
- 1.Kanapathipillai R, Henao Restrepo AM, Fast P, Wood D, Dye C, Kieny MP, Moorthy V. 2014. Ebola vaccine—an urgent international priority. N Engl J Med 371:2249–2251. doi: 10.1056/NEJMp1412166. [DOI] [PubMed] [Google Scholar]
- 2.Stanley DA, Honko AN, Asiedu C, Trefry JC, Lau-Kilby AW, Johnson JC, Hensley L, Ammendola V, Abbate A, Grazioli F, Foulds KE, Cheng C, Wang L, Donaldson MM, Colloca S, Folgori A, Roederer M, Nabel GJ, Mascola J, Nicosia A, Cortese R, Koup RA, Sullivan NJ. 2014. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat Med 20:1126–1129. doi: 10.1038/nm.3702. [DOI] [PubMed] [Google Scholar]
- 3.Geisbert TW, Daddario-Dicaprio KM, Geisbert JB, Reed DS, Feldmann F, Grolla A, Stroher U, Fritz EA, Hensley LE, Jones SM, Feldmann H. 2008. Vesicular stomatitis virus-based vaccines protect nonhuman primates against aerosol challenge with Ebola and Marburg viruses. Vaccine 26:6894–6900. doi: 10.1016/j.vaccine.2008.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisbert TW, Geisbert JB, Leung A, Daddario-DiCaprio KM, Hensley LE, Grolla A, Feldmann H. 2009. Single-injection vaccine protects nonhuman primates against infection with Marburg virus and three species of Ebola virus. J Virol 83:7296–7304. doi: 10.1128/JVI.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzi A, Engelmann F, Feldmann F, Haberthur K, Shupert WL, Brining D, Scott DP, Geisbert TW, Kawaoka Y, Katze MG, Feldmann H, Messaoudi I. 2013. Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc Natl Acad Sci U S A 110:1893–1898. doi: 10.1073/pnas.1209591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisbert TW, Daddario-Dicaprio KM, Lewis MG, Geisbert JB, Grolla A, Leung A, Paragas J, Matthias L, Smith MA, Jones SM, Hensley LE, Feldmann H, Jahrling PB. 2008. Vesicular stomatitis virus-based Ebola vaccine is well-tolerated and protects immunocompromised nonhuman primates. PLoS Pathog 4:e1000225. doi: 10.1371/journal.ppat.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mire CE, Miller AD, Carville A, Westmoreland SV, Geisbert JB, Mansfield KG, Feldmann H, Hensley LE, Geisbert TW. 2012. Recombinant vesicular stomatitis virus vaccine vectors expressing filovirus glycoproteins lack neurovirulence in nonhuman primates. PLoS Negl Trop Dis 6:e1567. doi: 10.1371/journal.pntd.0001567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu X, Fernando L, Alimonti JB, Melito PL, Feldmann F, Dick D, Stroher U, Feldmann H, Jones SM. 2009. Mucosal immunization of cynomolgus macaques with the VSVDeltaG/ZEBOVGP vaccine stimulates strong Ebola GP-specific immune responses. PLoS One 4:e5547. doi: 10.1371/journal.pone.0005547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marzi A, Feldmann H, Geisbert TW, Falzarano D. 2011. Vesicular stomatitis-based vaccines for prophylaxis and treatment of filovirus infections. J Bioterror Biodef S1:004. doi: 10.4172/2157-2526.S1-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spanier J, Lienenklaus S, Paijo J, Kessler A, Borst K, Heindorf S, Baker DP, Kroger A, Weiss S, Detje CN, Staeheli P, Kalinke U. 2014. Concomitant TLR/RLH signaling of radioresistant and radiosensitive cells is essential for protection against vesicular stomatitis virus infection. J Immunol 193:3045–3054. doi: 10.4049/jimmunol.1400959. [DOI] [PubMed] [Google Scholar]
- 11.Ahmed M, Mitchell LM, Puckett S, Brzoza-Lewis KL, Lyles DS, Hiltbold EM. 2009. Vesicular stomatitis virus M protein mutant stimulates maturation of Toll-like receptor 7 (TLR7)-positive dendritic cells through TLR-dependent and -independent mechanisms. J Virol 83:2962–2975. doi: 10.1128/JVI.02030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schabbauer G, Luyendyk J, Crozat K, Jiang Z, Mackman N, Bahram S, Georgel P. 2008. TLR4/CD14-mediated PI3K activation is an essential component of interferon-dependent VSV resistance in macrophages. Mol Immunol 45:2790–2796. doi: 10.1016/j.molimm.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins KA, Steffens JT, van Tongeren SA, Wells JB, Bergeron AA, Dickson SP, Dye JM, Salazar AM, Bavari S. 2014. Toll-like receptor agonist augments virus-like particle-mediated protection from Ebola virus with transient immune activation. PLoS One 9:e89735. doi: 10.1371/journal.pone.0089735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer A, Green J, Pollard J Jr, Tugendreich S. 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]