Abstract
We report the isolation of 24 novel genotypes of TT viruses from a surgically removed spleen of a patient with Hodgkin's disease. The sequence analysis of our 24 isolates revealed the remarkable heterogeneity of TT virus isolates not only from the same patient but also from the same biopsy material. These isolates belong to four phylogenetic groups of TT viruses. Nucleotide sequence analyses revealed five distinct genotypes (tth3, tth4, tth5, tth6, and tth7). The limited variation in sequence identity of the other isolates defines the latter as variants of four of these genotypes. A group of 6 isolates (the tth7 group) revealed a reorganization of open reading frame 1 (ORF1) leading to one larger and a varying number of smaller ORFs. The nucleotide difference of the full-length genomes was less than 1%. A variation of 69 to 97% in amino acids of a second group of 8 isolates (the tth3 group) was restricted to the hypervariable region of ORF1, indicating the existence of a quasi-species. These isolates differed by less than 2% in the remainder of their nucleotide sequences. An alignment of these isolates with 79 previously reported TT virus genotypes permits the proposal of TT virus genera and species within the family Anelloviridae in analogy to a previous proposal for the papillomaviruses (family Papillomaviridae).
TT viruses (TTV) are ubiquitous in nature and have been demonstrated in more than 90% of serum samples from healthy individuals where they persist over time (14, 16, 27, 41). Viral particles have been purified from feces (17, 34) and, in addition, are excreted in saliva, breast milk, and bile juice (9, 38, 47, 49). Transplacental transmission from mother to child has been controversial, but postnatal transmission has been confirmed repeatedly (15, 20, 24, 30, 33, 37, 48, 51, 52, 57). Replicative intermediate forms of the viral genome have been isolated from bone marrow cells (42) and demonstrated in a number of other organs (39, 44). Peripheral blood mononuclear cells act as a reservoir for TTV (41), but the highest viral load is found in the granulocytes (54). Viral transcription naturally occurs in bone marrow cells and not in peripheral blood mononuclear cells (40), although transcription in vitro has been achieved by DNA transfection into stimulated peripheral blood monocytes (29) and in a monkey cell line (19).
A large number of full-length or near full-length genomes of TTV has been isolated from humans and primates. They have also been identified in farm animals (23), although the genomes of recent isolates from pigs, dogs, and cats proved to be considerably smaller in size than the TTV found in humans and primates (43). The highly conserved noncoding region constitutes about one-third of the genome. The coding region consists of a large open reading frame (ORF), ORF1, coding for the viral capsid protein as well as the smaller ORF2, ORF3, and ORF4. Additional mRNAs result from splicing events (19, 40). Many different genotypes have been isolated, and an extremely wide range of DNA sequence divergence has been demonstrated (45). The steadily increasing number of new genotypes and the high sequence variability point to the need for a uniform and defined classification of this virus family.
Several attempts have been made to link TTV to the etiology of a specific disease. TTV were originally isolated from a patient suffering from posttransfusion hepatitis (35), but a direct role in the pathogenesis of this disease or in hepatocellular carcinoma has not been established (31, 32, 46). Several studies have failed to demonstrate a direct association of TTV with lymphomas and myelomas (2, 8, 60). Conditions of immune suppression may, however, lead to increased virus levels in the blood (39, 60). Our laboratory recently reported the presence of TTV DNA in specific human cancers, most notably in cancers of the gastrointestinal tract, lung, and breast and in a number of multiple myelomas (6). PCR amplification of fragments of the genome was used to detect a series of newly identified TTV-related sequences.
The present study describes the isolation of 24 full-length genomes and three 2-kb fragments from the spleen of a patient with Hodgkin's disease. These isolates belong to four different genotypic groups of TTV. The sequences of 8 clones from one TTV type were nearly identical to each other over the length of the complete genome, except for a high variation between clones in the highly variable region. In a second set, 6 isolates of the same TTV type varied in as little as 1% of nucleotides over the full genome length, but this variation induced stop codons within the ORF1, generating smaller individual ORFs in this region. We propose a more-defined classification system resulting from comparisons of our TTV types to 79 previously reported TTV genomes. The genomic diversity of the TTV justify the classification of these viruses into a family, Anelloviridae, consisting of genera and species.
MATERIALS AND METHODS
Tumor and DNA extraction.
The spleen from a German patient with Hodgkin's disease was collected 30 years ago and stored frozen. At that time, a histological differentiation of the Hodgkin type was not available. DNA was extracted after proteinase K digestion with phenol and chloroform-isoamyl alcohol.
PCR amplification of short fragments.
Total cellular DNA (100 ng) was used for amplification by using the B set of primers as previously described by Leary et al. (23). Amplicons (243 bp in size) were eluted, cloned, and sequenced as previously described (6).
PCR amplification of full-length genomes with inverted primers.
Total cellular DNA (650 ng) was used as a template for long PCR amplification with inverted consensus primers designed on short amplicons obtained with the B set primers. The inverted primers tbb188.1a (forward) 5′-GAA AGT GAG TGG GGG CCA GAC TTC GC-3′ and tbb188.1s (reverse) 5′-CGA AAC GCG CAA GCG TTT CGG GTGG-3′ were used, and amplification was performed in the presence of LA Taq polymerase in GC-I buffer under conditions described by the manufacturer (TaKaRa, Gennevilliers, France). To perform a hot-start PCR, enzyme was added to the reaction mixture after preheating the reaction mixture at 94°C for 3 min followed by 40 cycles of denaturation for 50 s at 95°C, annealing for 45 s at 68°C, and elongation for 8 min at 72°C. The final elongation step was performed at 72°C for 5 min. Amplicons were purified after gel electrophoresis by using the High Pure PCR product purification kit (Roche, Mannheim, Germany) and cloned into the pMOSBlue vector according to the manufacturer's instructions (pMOSBlue blunt end cloning kit; Amersham). The inserts of the recombinant clones were subjected to restriction fragment length polymorphism by using 6 restriction enzymes (EcoRI, PstI, XbaI, HindIII, NdeI, and EcoRV) to distinguish between different isolates. Sequencing of both strands of each of the 29 inserts was performed on an ABI model sequencer with Big Dye terminator chemistry (Perkin Elmer Applied Biosystems Division).
PCR amplification of overlapping short fragment to complete the genome.
Primers were selected in regions overlapping by at least 200 nucleotides (nt) with the long fragment. The specific primers, amplification conditions, and resulting fragment lengths for each individual TTV type are summarized in Table 1. Amplification with the respective primer sets was performed on total cellular DNA (100 ng) by using AmpliTaq Gold polymerase (Applied Biosystems).
TABLE 1.
Primer used to amplify overlapping short fragments tth3, tth4, tth5, tth6, and tth7
| Fragment | Primer | Primer sequence (5′-3′) | Annealing temp (°C) and time (s) | Product size (bp) |
|---|---|---|---|---|
| tth3 | af1h3 | TGG GCA CCA AAA CCC CAA GAG AAA GTC AG | 70, 30 | 606 |
| air1h3 | TTG TGA CCC CTC CCC AGT CAG GTG ACT TGT G | |||
| tth4 | af1h4 | AAA CAG AGT GTA CCT TTT CCC CCC CGA C | 70, 30 | 776 |
| ar1h4 | TTG ACT TCC GGG TTA TAC GGC AAC CCT C | |||
| tth5 | af1h5 | ATT GTT AGA GTA GGC CCC GAG CAG TG | 65, 90 | |
| ar1h5 | TTT TGA GTA GGT GTG GCT GAT GGT G | |||
| if1h5 (nested) | CGA GGA ATA CAC TGC CTG TAA ATA CTG | 594 | ||
| ir1h5 (nested) | GTG GCT GAT GGT GAC CTT TGA ACT C | |||
| tth6 | af1h6 | TAT CTG TTC CCA GAA CGC TTA CCA C | 60, 30 | |
| arir1h6 | GCT TAC TTA AAA TGG CGG CCA TGA C | |||
| if1h6 (seminested) | ACA TAC CCA GAA ACC CAG GCT TCA G | 65, 30 | 495 | |
| tth7 | af1h7 | ATG CCT ACA GTA GGT CCC AGG CAG TG | 60, 30 | |
| ar1h7 | GTG GCT GAT GGT GAC CTT TGA ACT C | |||
| if1h7 (nested) | AGA AAG TCA GCC CCA GCT CCT AGC AG | 620 | ||
| ir1h7 (nested) | TGA CCT TTG AAC TCA CGC CAC CGT C |
PCR amplification to verify stop codons in the 5′ half of ORF1 of tth7.
Additional amplicons (837 bp) overlapping the 5′ end of ORF1 were generated to verify whether the premature stop codons present in two cloned full-length sequences could be ascribed to PCR artifacts. The primers used were h7g2f 5′-TTC ACA TTA CTG CAC TTG CTG-3′ (forward) and h7g2r 5′-AAC TTG TTC CTT TTG TGC CTG-3′ (reverse) in a first round of 45 cycles of denaturation for 40 s at 94°C and annealing for 40 s at 55°C, followed by elongation for 1.5 min at 72°C. A nested reaction followed with the primers h7g43f (forward) 5′-CCC TCA CAG GTT GAT TCG AGA C-3′ and h7g43r (reverse) 5′-GGT GAC TTA TGC ATC CTT CTG-3′ for 45 cycles with the same conditions as described previously, except that annealing was performed at 64°C for 40 s. ProofStart DNA polymerase (QIAGEN) was used according to the manufacturer's instructions. Amplification was performed on the total cellular DNA from the biopsy sample (400 ng) as well as the cloned long fragments initially generated by PCR amplification. The length of the first product was 1,082 bp, and the length was 837 bp after the second nested amplification.
Transformation into two different strains of bacteria to verify whether stop codons resulted from mutations induced in bacteria.
One of the short clones was transformed into the bacterial strain XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)]} of Escherichia coli (catalog no. 200249; Stratagene) as well as into TOP10F′ bacterial strain {F′ [lacIq Tn10 (Tetr)] mcrA (mrr-hsdRMS-mcrBC) V80 lacZ M15 lacX74 deoR recA1 araD139 (ara-leu)7697 galU galK rpsL endA1 nupG} (catalog no. C3030-03; Invitrogen). DNA from single colonies of each strain was isolated, and DNA preparations from a total of 107 clones were sequenced. The sequences of these cloned plasmids were all identical to that of the original clone used for transformation.
Sequence analyses.
Sequences were compared to TTV sequences available in all databanks by using the HUSAR software package (50).
The genomic sequences of the full-length genomes of 103 TTV were submitted to phylogenetic analysis. The alignment was performed with the ClustalW program (56) by using a gap creation penalty of 10 and a gap extension penalty of 5. Distance matrix calculation was carried out by using the Distances program of the GCG Wisconsin package, version 10.2 (Accelrys), with the Kimura distance correction method. The phylogenetic tree from the distance matrix was constructed by using the Growtree program (GCG Wisconsin package, version 10.2), with neighbor-joining analysis. The tree was displayed by using the Treeview program of the University of Glasgow. The translated ORFs 1a were analyzed for homologous proteins and functional domains by using ProtSweep (4).
Nucleotide sequence accession number.
Nucleotide sequences were deposited in the EMBL Nucleotide Sequence Database, and the accession numbers are as follows: tth6, AJ620212; tth10, AJ620213; tth11g2, AJ620214; tth18, AJ620215; tth20, AJ620216; tth 21, AJ620217; tth3, AJ620218; tth9, AJ620219; tth16, AJ620220; tth17, AJ620221; tth25, AJ620222; tth26, AJ620223; tth27, AJ620224; tth31, AJ620225; tth4, AJ620226; tth5, AJ620227; tth14, AJ620228; tth29, AJ620229; tth7, AJ620230; tth8, AJ620231; tth13, AJ620232; tth19, AJ620233; tth22g4, AJ620234; tth23, AJ620235; N57.72, AJ620236; N58.46, AJ620237; N58.22, AJ620238; N58.15, AJ620239; N58.11, AJ620240; N58.5, AJ620241; L2T1.57, AJ620242.
RESULTS
Initial amplification of a short conserved region of the TTV genome led to the identification of 15 individual TTV sequences present in the spleen of a patient with Hodgkin's disease. These isolates were divided into 6 groups according to sequence homology to facilitate the selection of primers for amplifying the remaining part of the full-length genome of each type (data not shown). Amplification of the tumor DNA with one of these primer sets resulted in the isolation of 29 fragments larger than 3 kb and 2 fragments of 2 kb in size. A total number of 128 clones were grouped after restriction fragment length polymorphism analyses. Of these, 30 clones were sequenced and arranged into 5 groups according to sequence homology. The tth3 group consisted of 12 isolates, 2 of which were identical to two other clones (tth16 to tth12 and tth28 to tth26). Two fragments (tth1 and tth31) represented only partial TTV genomes. Summaries of all isolates and their DNA sequence homologies are presented in Tables 2, 3, 4, and 5.
TABLE 2.
Pairwise percent homology comparisons of full-length nucleotide sequences of tth clones
| Clone | % Homology with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| tth16 | tth17 | tth25 | tth26 | tth27 | tth3 | tth31 | tth9 | |
| tth16 | 100.0 | 97.6 | 97.3 | 97.3 | 97.3 | 97.5 | 99.4 | 98.2 |
| tth17 | 100.0 | 98.9 | 98.8 | 98.6 | 98.7 | 97.7 | 98.6 | |
| tth25 | 100.0 | 99.5 | 99.4 | 98.3 | 97.4 | 98.3 | ||
| tth26 | 100.0 | 99.3 | 98.3 | 97.4 | 98.2 | |||
| tth27 | 100.0 | 98.2 | 97.3 | 98.1 | ||||
| tth3 | 100.0 | 97.6 | 98.3 | |||||
| tth31 | 100.0 | 98.2 | ||||||
| tth9 | 100.0 | |||||||
TABLE 3.
Pairwise percent homology comparisons of full-length nucleotide sequences of tth clones
| Clone | % Homology with:
|
||
|---|---|---|---|
| tth14 | tth29 | tth5 | |
| tth14 | 100.0 | 99.7 | 99.7 |
| tth29 | 100.0 | 99.7 | |
| tth5 | 100.0 | ||
TABLE 4.
Pairwise percent homology comparisons of full-length nucleotide sequences of tth clones
| Clone | % Homology with:
|
|||||
|---|---|---|---|---|---|---|
| tth10 | tth11g2 | tth18 | tth20 | tth21 | tth6 | |
| tth10 | 100.0 | 99.4 | 99.6 | 98.0 | 99.4 | 98.2 |
| tth11g2 | 100.0 | 99.3 | 97.8 | 99.3 | 98.0 | |
| tth18 | 100.0 | 97.9 | 99.3 | 98.1 | ||
| tth20 | 100.0 | 97.9 | 99.2 | |||
| tth21 | 100.0 | 98.2 | ||||
| tth6 | 100.0 | |||||
TABLE 5.
Pairwise percent homology comparisons of full-length nucleotide sequences of tth clones
| Clone | % Homology with:
|
|||||
|---|---|---|---|---|---|---|
| tth13 | tth19 | tth22g4 | tth23 | tth7 | tth8 | |
| tth13 | 100.0 | 99.1 | 99.1 | 99.2 | 99.1 | 99.3 |
| tth19 | 100.0 | 99.3 | 99.5 | 99.5 | 99.6 | |
| tth22g4 | 100.0 | 99.4 | 99.4 | 99.5 | ||
| tth23 | 100.0 | 99.6 | 99.7 | |||
| tth7 | 100.0 | 99.7 | ||||
| tth8 | 100.0 | |||||
The nearly full-length genomes of 79 TTV isolates were aligned to the 24 isolates described here. These included all isolates of at least 3,200 bp. The genome of isolate tth4 falls into group 3 of the TTV (45) and is most closely related to senv-e (55), sharing 70% identity. In addition, tth4 shares 60 to 70% identity with SANBAN (11), tjn02 (58), tchn-g2 (26), ttvsan-ir1031, and ttvsan-s039 (53) but less than 60% identity with all other tth and TTV isolates. Isolate tth5 shares 99% identity with isolates tth14 and tth29. It also shares 91% identity to jt33f (45) and 92% identity to l01 (25). The isolates tth7, tth8, tth13, tth19, tth22g4, and tth23 shared 99% identity with each other and 71% identity with the next closest jt33f, l01, and tth5. The group of tth6, tth10, tth11g2, tth18, tth20, and tth21 shared 97 to 99% identity with each other and 66% identity with the closest related KAV (10). The tth3 group comprises 8 full-length isolates, namely tth3, tth9, tth16, tth17, tth25, tth26, tth27, and tth31. The isolates tth16 and tth31 share 99% identity, and tth25, tth26, and tth27 share 99% identity with each other, forming 2 subgroups which, with tth3, tth9, tth17, share 97 to 98% identity with each other. The closest known TTV is P1C1 (13), sharing 85% identity. The next related is HEL32/6a (18), sharing 70% identity. These TTV are members of the group 1 of TTV (45).
The new isolates all constituted a genome organization characteristic of TTV (1, 45). By comparing the full-length sequences between isolates of the tth7 group on the one hand and the tth3 group on the other, two distinct observations were made. The 8 full-length isolates of tth3 group may be considered variants of one type, as measured by a sequence difference of less than 2% between isolates. The only region in which a high nucleotide sequence variability (80 to 99%) (Table 6 and Fig. 1) was observed was restricted to the highly variable region (HVR) within the ORF1 (36), whereas only single nucleotide differences were present in the remaining part of the genome. This HVR, spanning nt 1425 to 1693 of our isolates, varied not only in nucleotide sequence identity but also substantially in amino acid homology (between 69 and 97%). Several of the amino acid substitutions were nonconservative, which suggests a possible role of these changes in immune modulation of the ORF1 protein. It is obviously not possible to determine which isolate represented the prototype responsible for the initial infection or which isolates developed over time within this individual patient.
TABLE 6.
Pairwise percent identity comparisons of nucleotide and amino acid sequences of the HVR of tth3 clonesa
| Clone | % Identity with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| tth16 | tth17 | tth25 | tth26 | tth27 | tth3 | tth31 | tth9 | |
| tth16 | 82.7 | 81.4 | 80.7 | 81.7 | 82.1 | 99.0 | 83.7 | |
| tth17 | 69.1 | 93.0 | 91.7 | 91.4 | 91.7 | 83.1 | 94.7 | |
| tth25 | 70.0 | 84.5 | 98.3 | 98.3 | 88.4 | 81.7 | 92.4 | |
| tth26 | 70.0 | 82.7 | 97.3 | 97.0 | 88.0 | 81.1 | 91.4 | |
| tth27 | 71.8 | 80.9 | 96.4 | 94.5 | 87.7 | 82.1 | 91.4 | |
| tth3 | 72.7 | 81.8 | 78.2 | 79.1 | 78.2 | 82.4 | 91.0 | |
| tth31 | 97.3 | 70.0 | 70.9 | 70.9 | 72.7 | 73.6 | 84.1 | |
| tth9 | 70.9 | 86.4 | 84.5 | 84.5 | 84.5 | 83.6 | 71.8 | |
Comparisons of nucleotide sequences are shown above the diagonal, and comparisons of amino acid sequences are shown below the diagonal.
FIG. 1.
Nucleotide and amino acid sequences of the HVRs (HVR1, HVR2, and HVR3) of the variants of tth3. Dashes indicate nucleotides or amino acids identical to those of the consensus sequence.
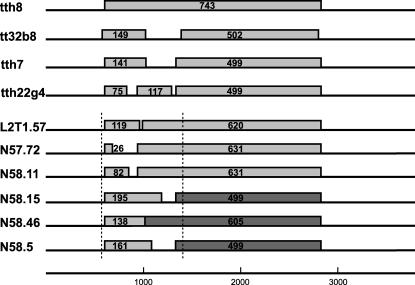
The second observation was the presence of premature stop codons within ORF1 in the isolates tth7 and tth22g4, falling into group 5 (45) of TTV. Instead of the complete ORF1 constituting 743 amino acids (aa), one nucleotide change in the tth7 sequence resulted in a premature stop codon at position nt 1022, coding for a shortened ORF1 (ORF1a, 141 aa, 423 bp). The second ORF1b of the tth7 (nt 1331 to 2828) leads to a putative protein of 499 aa (1,497 bp) in length (Fig. 2). The ORF1 of isolate tth22g4 was interrupted by 2 stop codons, resulting in 3 putative ORFs: ORF1a (75 aa, nt 599 to 824, 225 bp), ORF1b (117 aa, nt 935 to 1286, 351 bp), and ORF1c (499 aa, nt 1331 to 2828, 1,497 bp). An alternative start codon (21) for ORF1 of isolate tth13 was GTG at nt 599, resulting in a putative protein of 743 aa. Single nucleotide changes occurred scattered throughout the rest of the genomes not leading to any obvious modifications. To verify whether the stop codons of isolates tth7, tth22g4, and tth13 resulted as PCR artifacts during amplification of the long fragments, we generated primers to amplify the region spanning the 5′ end of ORF1 (nt 445 to 1506 of tth8). The original tumor DNA was used as a template, and an amplicon of 837 bp in size was generated by nested PCR with a low-error-rate proofreading enzyme. After cloning of the amplicon, the inserts of 244 resulting clones were sequenced on both strands, and the sequences were compared to the tth8 isolate (with intact ORF1). Of these, 158 clones were identical to each other, but all varied from tth8 at nt 794 of the complete genome sequence (G in tth8 and A in the amplicon, leading to an amino acid modification of glycine [GGA] to arginine [AGA]). Seven additional clones contained premature stop codons, resulting in putative shorter ORF1 proteins. Two of these were identical to each other. The remaining 6 clones were fragment L2T1.57 with 2 ORFs, ORF1a (357 bp, 119 aa) and ORF1b (1,068 bp, 620 aa), fragment N57.72 with ORF1a (78 bp, 26 aa) and ORF1b (1,893 bp, 631 aa), fragment N58.5 with ORF1a (483 bp, 161 aa) and ORF1b (1,497 bp, 499 aa), fragment N58.11 with ORF1a (346 bp, 82 aa) and ORF1b (1,893 bp, 631 aa), fragment N58.15 with ORF1a (585 bp, 195 aa) and ORF1b (1,497 bp, 499 aa), and fragment N58.46 with ORF1a (414 bp, 138 aa) and ORF1b (1,815 bp, 605 aa). By comparison to all other isolates, we assumed the 3′ region of the amplicons to result in the putative larger proteins as indicated (Fig. 2). In addition, 79 clones harbored scattered single nucleotide differences which did not influence the ORF structure. As the bacterial strains used in amplifying the plasmids may have induced such mutations into the plasmids, we transformed one clone with two different bacteria strains (TOP10F′ and XL1-Blue) and sequenced the resulting clones from each strain (107 in total). All obtained sequences were identical to the input clone.
FIG. 2.
Additional ORFs within ORF1 amplified from the spleen of a patient with Hodgkin's disease. The number of amino acids in each of the respective ORFs is indicated.
Databank searches with ProtSweep against Swissprot, TREMBL, and EMBL without expressed sequence tags and sequence tagged sites revealed no significant hits for any of the resulting putative proteins, except for the ORF1a of N58.46 which shared some homology to the UL31-like protein of herpes simplex virus (accession number AAG14224).
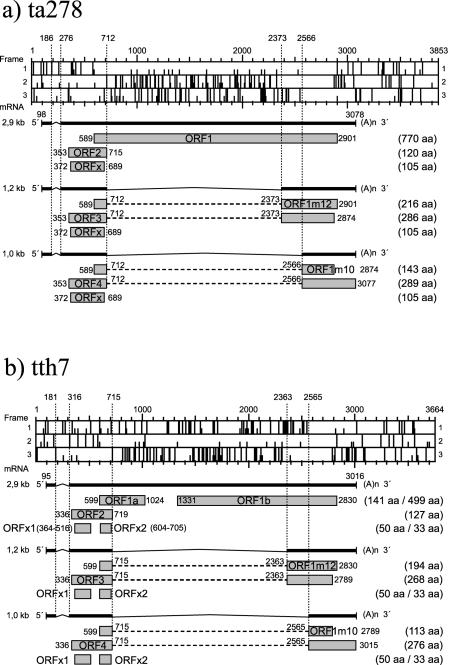
Replicative forms of TTV have been reported in bone marrow cells (42). Three mRNA species (2.9, 1.2, and 1.0 kb) have, in addition, been reported either in bone marrow cells (40) or in COS1 cells transfected with a TTV construct (19). We have analyzed the TTV genomes of our isolates and that of TA278 (accession no. AB017610) for additional initiation codons according to the Kozak rules (22). The use of the start codon at nt 372 in the third reading frame of the TA278 sequence would result in an additional ORFx (stop codon nt 689), coding for a putative protein of 105 aa in length (Fig. 3a). This ORFx is not present in all TTV types. Donor and acceptor splice sites used for the transcription of the 1.2- and 1.0-kb mRNA species may also lead to additional ORFs. The splice sites for the 1.2-kb mRNA may result in an additional protein coded for on the second reading frame of TA278 (Fig. 3a). The splicing site at nt 712 linking with the acceptor site at nt 2373 will result in an ORF1m12 mRNA coding for a putative protein of 216 aa (ORF1m12, nt 589 to 712 linked to nt 2373 to 2898). Similarly, the 1.0-kb mRNA splice sites located in reading frame 2 at nt 712 and linked to nt 2566 will create the ORF1m10 mRNA coding for a putative protein of 143 aa in length (ORF1m10, nt 589 to 712 linked to nt 2566 to 2872). These two additional ORFs can be generated in all known TTV genomes. The corresponding putative proteins expressed by transcription of the tth7 genome will be smaller in size, i.e., 195 and 98 aa, respectively. Additional studies have to be performed to determine whether these mRNA species are present in cells infected in vivo with TTV.
FIG. 3.
Putative ORFs for TTV ta278 (a) and tth7 (b).
DISCUSSION
The present study describes the isolation of 24 full-length TTV from a single spleen biopsy sample from a patient with Hodgkin's disease. They were grouped into 5 different TTV genotypes, based on the sequence analyses of the complete genomes. A group of 6 different isolates constituted the tth6 group, 8 isolates constituted the tth3 group, 3 isolates constituted the tth5 group, and 6 isolates constituted the tth7 group. The isolate tth4 represented another TTV genotype.
It became very evident from sequence comparisons between our isolates, tth3, tth9, tth16, tth17, tth25, tth26, tth27, and tth31, that the sequence divergence restricted to the HVR, a small region located in the ORF1, does not influence the definition of a genotype. It is, however, of great interest that the sequence identity in this HVR varied between 80 and 99% between the isolates, leading to a modification in the amino acid composition. Only single and scattered insignificant nucleotide changes were present in the remaining part of the genomes. This demonstrates the circulation of TTV as quasi-species (36). The modification in the structure of the putative capsid protein points to an interesting possible mechanism by which these persisting virus infections may escape immune surveillance.
The modifications observed in the tth7 group of TTV isolates described here may be of significance for a pathogenic role of these viruses. It is interesting that this modification of ORF1 into separate smaller ORFs was restricted to this one group of isolates, although the genomes of all of the other TTV isolates described here were generated in exactly the same way. Additional experiments also verified that these modifications had originated within the patient and did not result from any experimental artifacts. It is also of interest that the ORF1 of certain species contains as many as 14 individual start sites, which when used, will all result in proteins larger than 100 aa. The possible existence of interrupted ORF1s has previously been reported (7, 21, 26). Other known TTV with an interrupted ORF1 are l03 (AF371370) (25), senv-b (AX025677) (55), tchn-a (AF345526), tchn-c1 (AF345523), tchn-d2 (AF345525) (26), us32 (AF122921) (7), and tt32b8 (AX781361) (6; unpublished data). The smaller ORFs resulting from the premature stop codons are summarized in Table 7. Our laboratory has previously described the identification of TTV in several malignant tumors (6). The complete TTV genome from a case of lung cancer, isolate tt32b8, also revealed an interrupted ORF1, resulting in smaller ORFs in this region. In vitro functional studies with these newly created proteins may assist in defining a possible role for TTV in the etiology of some malignancies.
TABLE 7.
Nucleotide and amino acids of additional smaller ORFs within ORF1 of various TTV isolates
| Isolate | ORF1a
|
ORF1b
|
||||||
|---|---|---|---|---|---|---|---|---|
| Position (nt) of:
|
Length in:
|
Position (nt) of:
|
Length in:
|
|||||
| Start codon | Stop codon | bp | aa | Start codon | Stop codon | bp | aa | |
| Full-length TTV isolates | ||||||||
| tth7 | 599 | 1022 | 423 | 141 | 1331 | 2828 | 1,497 | 499 |
| tth22g4a | 599 | 824 | 225 | 75 | 935 | 1286 | 351 | 117 |
| 103 | 490 | 1015 | 525 | 175 | 1068 | 2730 | 1,662 | 554 |
| senv-b | 592 | 772 | 180 | 60 | 832 | 2869 | 2,037 | 679 |
| tchn-a | 502 | 685 | 183 | 61 | 767 | 2834 | 2,067 | 689 |
| tchn-c1 | 478 | 1060 | 582 | 194 | 1050 | 2766 | 1,716 | 572 |
| tchnd2 | 479 | 1757 | 1,278 | 426 | 1869 | 2709 | 840 | 280 |
| tt32b8 | 416 | 863 | 447 | 149 | 1139 | 2645 | 1,506 | 502 |
| Partial ORF1 isolates | ||||||||
| L2T1.57 | 599 | 956 | 357 | 119 | 986 | 2828b | 1,860 | 620 |
| N57.72 | 599 | 677 | 78 | 26 | 935 | 2828b | 1,893 | 631 |
| N58.11 | 599 | 845 | 246 | 82 | 935 | 2828b | 1,893 | 631 |
| N58.15 | 599 | 1184 | 585 | 195 | 1331 | 2828b | 1,497 | 499 |
| N58.46 | 599 | 1014 | 414 | 138 | 1013 | 2828b | 1,815 | 605 |
| N58.5 | 599 | 1082 | 483 | 161 | 1331 | 2828b | 1,497 | 499 |
ORF1 is divided into 3 ORFs in the tth22g4 sequence. The start codon of ORF1c is at nt 1331, and the stop codon is at nt 2828 (total length, 1,497 bp or 499 aa).
Assumed stop codon of ORF1b by comparison to all other isolates.
Multiple reports have dealt with the isolation and characterization of a large number of TTV, and many authors have attempted to classify these viruses into groups and/or genotypes (1, 12). The justification for different genera and species has, in addition, been suggested (45, 55). The use of different regions of the genome for establishing phylogeny has frequently been questioned and debated (3, 39, 45, 55). Recombination between TT isolates has been discussed (28, 59), although according to our experience this may represent PCR artifacts (generating genomes or partial genomes by PCR amplification on DNA templates harboring several closely related TTV genomes) (unpublished data). The TT-like mini viruses have been lumped together with the TTV into the proposed genus Anellovirus, based on a similar genome organization and similar modes of transmission and infection (12). Not only are the genomes of the TT-like mini viruses about 30% smaller than those of TTV but the viral particles of these two groups differ accordingly in size (53). This development is very analogous to that previously experienced in the papillomavirus field (5). The family Papillomaviridae consists of more than 100 isolates with a large divergence in genome sequence, very similar to that seen for TTV. The rules currently applied to the classification of viruses (e.g., pathology and serology) do not apply for any of these groups because too little is known about the biological functions of each individual isolate. In addition, the absence of suitable in vitro systems for culturing the individual virus types indicates that the desired information about these characteristics may not be obtained. The ubiquitous nature of TTV and the rapid isolation of novel types may create substantial taxonomic confusion.
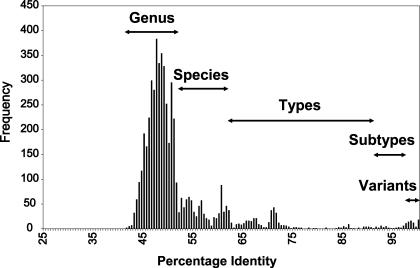
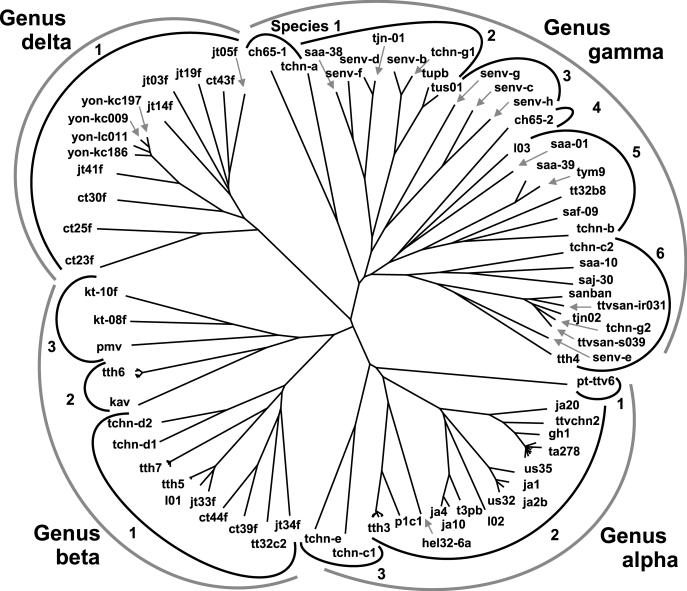
We have compared the overlapping 3,200 bp (nt 136 to 3264 of TTV type TA278) of 79 full-length TTV genomes and the 24 TTV described here and determined the percentage of nucleotide sequence identity between all isolates (data available upon request). The frequency distribution of the percent identities of the TTV is presented in Fig. 4. A phylogenetic tree has been compiled of these sequences by using neighbor-joining analysis (Fig. 5). Based on this information and the above-mentioned reasons, we propose the classification of TTV as a separate family, Anelloviridae, including TTV groups with an identity of less than 50% constituting individual genera and TTV genomes with sequence identity greater than 50% and less than 60% belonging to the same genus. A species will be defined as a group of TTV types sharing greater than 60% and less than 90% sequence homology to each other. A TTV type is defined by a complete viral genome sharing less than 90% homology with the closest related known TTV type. Subtypes may be defined as sequences sharing more than 90% homology, and those sharing homologies of 96% and higher may be defined as variants of each other. Applying this, the 5 previously named groups (45) will be reduced to 4 genera (α, β, γ, and δ), each harboring a number of species (Table 8 and Fig. 5). The isolates in groups 2 and 5 (45) share more than 50% and less than 60% sequence identity and will therefore be combined into one genus, β. A number of hitherto known TTV types become variants of a prototype when applying the proposed criteria. We propose a priority according to the date of submission to a databank for selection of a specific type as the prototype. The TTV types which will be affected by this classification are summarized in Table 9.
FIG. 4.
Frequency distribution of pairwise identity percentages from nucleotide sequence comparisons (3,200 nt) of 103 TTV isolates.
FIG. 5.
Phylogenetic tree containing sequences of 103 TTV. The outermost semicircular symbols identify TTV genera, and the number at each inner semicircular symbol refers to the TTV species.
TABLE 8.
Proposed classification of TTV
| Genus | Species | Type (accession no.) |
|---|---|---|
| Alpha | 1 | pt-ttv6 (AB041957) |
| 2 | ta278 (AB017610) | |
| ttvchn2 (AF129887) | ||
| ja20 (AF122914) | ||
| ja1 (AF122916) | ||
| 102 (AY026466) | ||
| ja4 (AF122917) | ||
| hel32-6a (AY034068) | ||
| p1c1 (AF298585) | ||
| tth3 (AJ620218) | ||
| 3 | tchn-c1 (AF345523) | |
| tchn-c (AF345522) | ||
| Beta | 1 | jt34f (AB064607) |
| tt32c2 (AX781379) | ||
| ct39f (AB064604) | ||
| ct44f (AB064605) | ||
| 101 (AY026465) | ||
| tth7 (AJ620212) | ||
| tchn-d1 (AF345524) | ||
| tchn-d2 (AF345525) | ||
| 2 | kav (AF435014) | |
| tth6 (AJ620212) | ||
| 3 | pmv (AF261761) | |
| kt-08f (AB054647) | ||
| kt-10f (AB054648) | ||
| Gamma | 1 | ch65-1 (AB037926) |
| tchn-a (AF345526) | ||
| 2 | saa-38 (AB060593) | |
| senv-f (AX025822) | ||
| senv-d (AX025730) | ||
| senv-b (AX025677) | ||
| tus01 (AB017610) | ||
| 3 | senv-g (AX025830) | |
| senv-c (AX025718) | ||
| senv-h (AX025838) | ||
| 4 | ch65-2 (AB049607) | |
| 5 | 103 (AF371370) | |
| saa-01 (AB060597) | ||
| saa39 (AB060592) | ||
| tt32b8 (AX781362) | ||
| saf-09 (AB060596) | ||
| tchn-b (AF348409) | ||
| 6 | tchn-c2 (AF345527) | |
| saa-10 (AB060594) | ||
| saj-30 (AB060595) | ||
| sanban (AB025946) | ||
| ttvsan-ir031 (AB038619) | ||
| tjn02 (AB028669) | ||
| ttvsan-s039 (AB038620) | ||
| senv-e (AX025761) | ||
| tth4 (AJ620226) | ||
| Delta | 1 | ct23f (AB064595) |
| ct25f (AB064596) | ||
| ct30f (AB064597) | ||
| jt41f (AB064603) | ||
| ttvyon-kc009 (AB038621) | ||
| jt14f (AB064601) | ||
| jt03f (AB064599) | ||
| jt19f (AB064602) | ||
| ct43f (AB064598) | ||
| jt05f (AB064600) |
TABLE 9.
Definition of prototypes with subtypes and variants
| Genus | Species | Prototype (accession no.) | Subtype (accession no.) | Variant(s) (accession no.) |
|---|---|---|---|---|
| Alpha | 2 | ta278 (AB017610) | gh1 (AF122913) | ja9 (AF122915), bdh1 (AF116842), trm1 (AB026345), tk16 (AB026346), tp1-3 (AB026347), ttvchn1 (AF079173), twh (AF351132) |
| ja1 (AF122916) | ja2b (AF122918), us32 (AF122921) | |||
| us35 (AF122920) | ||||
| ja4 (AF122917) | t3pb (AF247138) | ja10 (AF122919) | ||
| tth3 (AJ620218) | tth9 (AJ620219), tth16 (AJ620220), tth17 (AJ620221), tth25 (AJ620222), tth26 (AJ620223), tth27 (AJ620224), tth31 (AJ620225) | |||
| Beta | 1 | 101 (AY026465) | jt33f (AB064606) | |
| tth5 (AJ620227) | tth14 (AJ620228), tth29 (AJ620229) | |||
| tth7 (AJ620230) | tth8 (AJ620231), tth13 (AJ620232) | |||
| tth19 (AJ620233), tth22g4 (AJ620234), tth23 (AJ620235) | ||||
| 2 | tth6 (AJ620212) | tth10 (AJ620213), tth11g2 (AJ620214), tth18 (AJ620215), tth20 (AJ620216), tth21 (AJ620217) | ||
| Gamma | 2 | senv-d (AX025730) | tjn01 (AB028668) | |
| senv-b (AX025677) | tchn-g1 (AF345521) | |||
| tus01 (AB017613) | tupb (AF247137) | |||
| 5 | tym9 (AB050448) | saa-39 (AB060592) | ||
| 6 | ttvsan-s039 (AB038620) | tchn-g2 (AF345529) | ||
| Delta | 1 | ttvyon-kc009 (AB038621) | ttvyon-kc197 (AB038624) | |
| ttvyon-kc186 (AB038623) | ||||
| ttvyon-lc011 (AB038622) |
The demonstration of the in vivo modification within the HVR of a single TTV type opens a possibility for further investigation into the role these changes may play in immune evasion and the persistence of these viruses in its host. A second mechanism by which the TTV genome is modified within its host, and which needs further investigation, is the induction of single nucleotide mutations in ORF1 leading to stop codons. The smaller proteins resulting from the usage of the additional start codons in this ORF may have different additional functions which may play an important role in the pathogenesis of these viruses.
Acknowledgments
We thank Corinna Whitley, Elsbeth Schneider, Sabine Serick, Imke Grewe, Karin Gunst, Sonja Stephan, Helene Rahn, Christine Nitsch, Martin Dietrich, and Romana Cop for excellent technical assistance.
REFERENCES
- 1.Bendinelli, M., M. Pistello, F. Maggi, C. Fornai, G. Freer, and M. L. Vatteroni. 2001. Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin. Microbiol. Rev. 14:98-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cacoub, P., E. Rosenthal, V. Gerolami, P. Hausfater, P. Ghillani, Y. Sterkers, V. Thibault, H. Khiri, J. C. Piette, and P. Halfon. 2003. Transfusion-associated TT virus co-infection in patients with hepatitis C virus is associated with type II mixed cryoglobulinemia but not with B-cell non-Hodgkin lymphoma. Clin. Microbiol. Infect. 9:39-44. [DOI] [PubMed] [Google Scholar]
- 3.Cong, M., B. Nichols, X. Dou, J. E. Spelbring, K. Krawczynski, H. A. Fields, and Y. E. Khudyakov. 2000. Related TT viruses in chimpanzees. Virology 274:343-355. [DOI] [PubMed] [Google Scholar]
- 4.del Val, C., A. Mehrle, M. Falkenhahn, M. Seiler, K.-H. Glatting, A. Poustka, S. Suhai, and S. Wiemann. 2004. High-throughput protein analysis integrating bioinformatics and experimental assays. Nucleic Acids Res. 32:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Villiers, E.-M. 2001. Taxonomic classification of papillomaviruses. Pap. Rep. 12:57-63. [Google Scholar]
- 6.de Villiers, E.-M., R. Schmidt, H. Delius, and H. zur Hausen. 2002. Heterogeneity of TT virus related sequences isolated from human tumour biopsy specimens. J. Mol. Med. 80:44-50. [DOI] [PubMed] [Google Scholar]
- 7.Erker, J. C., T. P. Leary, S. M. Desai, M. Chalmers, and I. K. Mushahwar. 1999. Analyses of TT virus full-length genomic sequences. J. Gen. Virol. 80:1743-1750. [DOI] [PubMed] [Google Scholar]
- 8.Garbuglia, A. R., T. Iezzi, M. R. Capobianchi, P. Pignoloni, A. Pulsoni, J. Sourdis, E. Pescarmona, D. Vitolo, and F. Mandelli. 2003. Detection of TT virus in lymph node biopsies of B-cell lymphoma and Hodgkin′s disease, and its association with EBV infection. Int. J. Immunopathol. Pharmacol. 16:109-118. [DOI] [PubMed] [Google Scholar]
- 9.Hayakawa, E., Y. Edamoto, D. Xin, H. T. Tran, Y. Iwaki, Y. Sato, T. Sata, and K. Abe. 2001. Detection of TT virus in human bile juice. Jpn. J. Infect. Dis. 54:127-128. [PubMed] [Google Scholar]
- 10.Heller, F., R. Zachoval, A. Koelzer, H. Nitschko, and G. G. Froesner. 2001. Isolate KAV: a new genotype of the TT-virus family. Biochem. Biophys. Res. Commun. 289:937-941. [DOI] [PubMed] [Google Scholar]
- 11.Hijikata, M., K. Takahashi, and S. Mishiro. 1999. Complete circular DNA genome of a TT virus variant (isolate name SANBAN) and 44 partial ORFs sequences implicating a great degree of diversity beyond genotypes. Virology 260:17-22. [DOI] [PubMed] [Google Scholar]
- 12.Hino, S. 2002. TTV, a new human virus with single stranded circular DNA genome. Rev. Med. Virol. 12:151-158. [DOI] [PubMed] [Google Scholar]
- 13.Hohne, M., T. Berg, A. R. Muller, and E. Schreier. 1998. Detection of sequences of TT virus, a novel DNA virus in German patients. J. Gen. Virol. 79:2761-2764. [DOI] [PubMed] [Google Scholar]
- 14.Irving, W. L., J. K. Ball, S. Berridge, R. Curran, A. M. Grabowska, C. L. Jameson, K. R. Neal, S. D. Ryder, and B. J. Thomson. 1999. TT virus infection in patients with hepatitis C: frequency, persistence and sequence heterogeneity. J. Infect. Dis. 180:27-34. [DOI] [PubMed] [Google Scholar]
- 15.Iso, K., Y. Suzuki, and M. Takayama. 2001. Mother-to-infant transmission of TT virus in Japan. Int. J. Gynaecol. Obstet. 75:11-19. [DOI] [PubMed] [Google Scholar]
- 16.Itoh, K., M. Takahashi, M. Ukita, T. Nishizawa, and H. Okamoto. 1999. Influence of primers on the detection of TT virus DNA by polymerase chain reaction. J. Infect. Dis. 180:1750-1751. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, Y., M. Takahashi, M. Fukuda, T. Shibayama, T. Ishikawa, F. Tsuda, T. Tanaka, T. Nishizawa, and H. Okamoto. 2000. Visualization of TT virus particles recovered from the sera and feces of infected humans. Biochem. Biophys. Res. Commun. 279:718-724. [DOI] [PubMed] [Google Scholar]
- 18.Kakkola, L., K. Hedman, H. Vanrobaeys, L. Hedman, and M. Soderlund-Venermo. 2002. Cloning and sequencing of TT virus genotype 6 and expression of antigenic open reading frame 2 proteins. J. Gen. Virol. 83:979-990. [DOI] [PubMed] [Google Scholar]
- 19.Kamahora, T., S. Hino, and H. Miyata. 2000. Three spliced mRNAs of TT virus transcribed from a plasmid containing the entire genome in COS1 cells. J. Virol. 74:9980-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazi, A., H. Miyata, K. Kurokawa, M. A. Khan, T. Kamahora, S. Katamine, and S. Hino. 2000. High frequency of postnatal transmission of TT virus in infancy. Arch. Virol. 145:535-540. [DOI] [PubMed] [Google Scholar]
- 21.Khudyakov, Y. E., M. Cong, B. Nichols, D. Reed, X. Dou, S. O. Viazov, J. Chang, M. W. Fried, I. Williams, W. Bower, S. Lambert, M. Purdy, M. Roggendorf, and H. A. Fields. 2000. Sequence heterogeneity of TT virus and closely related viruses. J. Virol. 74:2990-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak, M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leary, T. P., J. C. Erker, M. L. Chalmers, S. M. Desai, and I. K. Mushahwar. 1999. Improved detection systems for TT virus reveal high prevalence in humans, non-human primates and farm animals. J. Gen. Virol. 80:2491-2499. [DOI] [PubMed] [Google Scholar]
- 24.Lin, H. H., J. H. Kao, P. I. Lee, and D. S. Chen. 2002. Early acquisition of TT virus in infants: possible minor role of maternal transmission. J. Med. Virol. 66:285-290. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Z., K. Luo, R. Zheng, J. Hu, and H. He. 2002. Novel TTV variant isolated in an epidemic of hepatitis of unknown etiology. J. Med. Virol. 67:113-117. [DOI] [PubMed] [Google Scholar]
- 26.Luo, K., H. He, Z. Liu, D. Liu, H. Xiao, X. Jiang, W. Liang, and L. Zhang. 2002. Novel variants related to TT virus distributed widely in China. J. Med. Virol. 67:118-126. [DOI] [PubMed] [Google Scholar]
- 27.Maggi, F., M. Pistello, M. Vatteroni, S. Presciuttini, S. Marchi, P. Isola, C. Fornai, S. Fagnani, E. Andreoli, G. Antonelli, and M. Bendinelli. 2001. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J. Virol. 75:11999-12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manni, F., A. Rotola, E. Caselli, G. Bertorelle, and D. Di Luca. 2002. Detecting recombination in TT virus: a phylogenetic approach. J. Mol. Evol. 55:563-572. [DOI] [PubMed] [Google Scholar]
- 29.Mariscal, L., J. Lopez-Alcotocho, E. Rodriguez-Inigo, N. Ortiz-Movilla, S. de Lucas, J. Bartolome, and V. Careno. 2002. TT virus replicates in stimulated but not in nonstimulated peripheral blood mononuclear cells. Virology 301:121-129. [DOI] [PubMed] [Google Scholar]
- 30.Matsubara, H., K. Michtaka, N. Horiike, T. Kihana, M. Yano, T. Mori, and M. Onji. 2001. Existence of TT virus DNA and TT-like mini virus DNA in infant cord blood: mother-to-neonatal transmission. Hepatol. Res. 21:280-287. [DOI] [PubMed] [Google Scholar]
- 31.Moriyama, M., W. Longren, Z. Zi-Yi, S. Oshiro, H. Matsumura, H. Aoki, T. Shimizu, K. Nakai, H. Yamagami, M. Kaneko, A. Shioda, S. Ichijima, K. Iwaguchi, H. Iwasaki, N. Tanaka, and Y. Arakawa. 2003. TT virus infection does not affect the clinical profiles of patient with hepatitis B and C in Yanbian City, China. Intervirology 46:214-221. [DOI] [PubMed] [Google Scholar]
- 32.Moriyama, M., H. Matsumura, T. Shimizu, A. Shioda, M. Kaneko, K. Miyazawa, H. Miyata, N. Tanaka, T. Uchida, and Y. Arakawa. 2001. Histopathologic impact of TT virus infection on the liver of type C chronic hepatitis and liver cirrhosis in Japan. J. Med. Virol. 64:74-81. [DOI] [PubMed] [Google Scholar]
- 33.Morrica, A., F. Maggi, M. L. Vatteroni, C. Fornai, M. Pistello, P. Ciccorossi, E. Grassi, A. Gennzzani, and M. Bendinelli. 2000. TT virus: evidence for transplacental transmission. J. Infect. Dis. 181:803-804. [DOI] [PubMed] [Google Scholar]
- 34.Mushahwar, I. K., J. C. Erker, A. S. Muerhoff, T. P. Leary, J. N. Simons, L. G. Birkenmeyer, M. L. Chalmers, T. J. Pilot-Matias, and S. M. Dexai. 1999. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc. Natl. Acad. Sci. USA 96:3177-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshikawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 36.Nishizawa, T., H. Okamoto, F. Tsuda, T. Aikawa, Y. Sugai, K. Konishi, Y. Akahane, M. Ukita, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1999. Quasispecies of TT virus (TTV) with sequence divergence in hypervariable regions of the capsid protein in chronic TTV infection. J. Virol. 73:9604-9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohto, H., N. Ujie, C. Takeuchi, A. Sato, A. Hayashi, H. Ishiko, T. Nishizawa, and H. Okamoto. 2002. TT virus infection during childhood. Transfusion 42:892-898. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, H., Y. Akahane, M. Ukita, M. Fukuda, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1998. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G-hepatitis. J. Med. Virol. 56:128-132. [PubMed] [Google Scholar]
- 39.Okamoto, H., T. Nishizawa, M. Takahashi, S. Asabe, F. Tsuda, and A. Yoshikawa. 2001. Heterogenous distribution of TT virus of distinct genotypes in multiple tissues from infected humans. Virology 288:358-368. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto, H., T. Nishizawa, A. Tawara, M. Takahashi, J. Kishimoto, T. Sai, and Y. Sugai. 2000. TT virus mRNAs detected in the bone marrow cells from an infected individual. Biochem. Biophys. Res. Commun. 279:700-707. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto, H., M. Takahashi, N. Kato, M. Fukuda, A. Tawara, S. Fukuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 2000. Sequestration of TT virus of restricted genotypes in peripheral blood mononuclear cells. J. Virol. 74:10236-10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto, H., M. Takahashi, T. Nishizawa, A. Tawara, Y. Sugai, T. Sai, T. Tanaka, and F. Tsuda. 2000. Replicative forms of TT virus DNA in bone marrow cells. Biochem. Biophys. Res. Commun. 270:657-662. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto, H., M. Takahashi, T. Nishizawa, A. Tawara, K. Fukai, U. Muramatsu, Y. Naito, and A. Yoshikawa. 2002. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 83:1291-1297. [DOI] [PubMed] [Google Scholar]
- 44.Okamoto, H., M. Ukita, T. Nishizawa, J. Kishimoto, Y. Hoshi, H. Mizuo, T. Tanaka, and M. Miykawa. 2000. Circular double-stranded forms of TT virus DNA in the liver. J. Virol. 74:5161-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng, Y. H., T. Nishizawa, M. Takahashi, T. Ishikawa, A. Yoshikawa, and H. Okamoto. 2002. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch. Virol. 147:21-41. [DOI] [PubMed] [Google Scholar]
- 46.Pineau, P., M. Meddeb, R. Raselli, L. X. Qin, B. Terris, Z. Y. Tang, P. Tiolais, V. Mazzaferro, and A. Dejean. 2000. Effect of TT virus infection on hepatocellular carcinoma development: results of a Euro-Asian survey. J. Infect. Dis. 181:1138-1142. [DOI] [PubMed] [Google Scholar]
- 47.Ross, R. S., S. Viazov, V. Runde, U. W. Schaefer, and M. Roggendorf. 1999. Detection of TT virus in specimens other than blood. J. Clin. Virol. 13:181-184. [DOI] [PubMed] [Google Scholar]
- 48.Saback, F. L., S. A. Gomes, V. S. de Paula, R. R. da Silva, L. L. Lewis-Ximenez, and C. Niel. 1999. Age-specific prevalence and transmission of TT virus. J. Med. Virol. 59:318-322. [DOI] [PubMed] [Google Scholar]
- 49.Schroter, M., S. Polywka, B. Zollner, P. Schafer, R. Laufs, and H. H. Feucht. 2000. Detection of TT virus DNA and GB virus type C/hepatitis G virus RNA in serum and breast milk: determination of mother-to-child transmission. J. Clin. Microbiol. 38:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senger, M., T. Flores, K.-H. Glatting, P. Ernst, A. Hotz-Wagenblatt, and S. Suhai. 1998. W2H: WWW interface to the GCG sequence analysis package. Bioinformatics 14:452-457. [DOI] [PubMed] [Google Scholar]
- 51.Simmonds, P., L. E. Prescott, C. Logue, F. Davidson, A. E. Thomas, and C. A. Ludlam. 1999. TT virus-part of the normal flora? J. Infect. Dis. 180:1748-1749. [DOI] [PubMed] [Google Scholar]
- 52.Sugiyama, K., K. Goto, T. Ando, F. Mizutani, K. Terabe, and T. Yokoyama. 2001. Highly diverse TT population in infants and their mothers. Virus Res. 73:183-188. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi, K., M. Hijikata, E. I. Samokhvalov, and S. Mishiro. 2000. Full or near full-length nucleotide sequences of TT virus variants (types SANBAN and YONBAN) and the TT virus-like mini virus. Intervirology 43:119-123. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi, M., S. Asabe, Y. Gotanda, J. Kishimoto, F. Tsuda, and H. Okamoto. 2002. TT virus is distributed in various leukocyte subpopulations at distinct levels, with the highest viral load in granulocytes. Biochem. Biophys. Res. Commun. 290:242-248. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka, Y., D. Primi, R. Y. H. Wang, T. Umemura, A. E. T. Yeo, M. Mizokami, H. J. Alter, and J. W. Shih. 2001. Genomic and molecular evolutionary analysis of a newly identified infectious agent (SEN virus) and its relationship to the TT virus family. J. Infect. Dis. 183:359-367. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toyoda, H., M. Naruse, S. Yokozaki, K. Morita, I. Nakano, A. Itakura, M. K. Okamura, Y. Fukuda, and T. Hayakawa. 1999. Prevalence of infection with TT virus (TTV), a novel DNA virus, in healthy Japanese subjects, newborn infants, cord blood and breast milk. J. Infect. 38:198-199. [DOI] [PubMed] [Google Scholar]
- 58.Ukita, M., H. Okamoto, T. Nishizawa, A. Tawara, M. Takahashi, H. Iizuka, Y. Miyakawa, and M. Mayumi. 2000. The entire nucleotide sequences of two distinct TT virus (TTV) isolates (TJN01 and TJN02) remotely related to the original TTV isolates. Arch. Virol. 145:1543-1559. [DOI] [PubMed] [Google Scholar]
- 59.Worobey, M. 2000. Extensive homologous recombination among widely divergent TT viruses. J. Virol. 74:7666-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, S., W. Yeo, M. Tang, C. Liu, X. Lin, W. M. Ho, P. Hui, and P. J. Johnson. 2002. Frequent detection of the replicative form of TT virus DNA in peripheral blood mononuclear cells and bone marrow cells in cancer patients. J. Med. Virol. 66:428-434. [DOI] [PubMed] [Google Scholar]