Abstract
Regulatory T (Treg) cells are critical determinants of both immune responses and metabolic control. Here we show that systemic ablation of Treg cells compromised the adaptation of whole-body energy expenditure to cold exposure, correlating with impairment in thermogenic marker gene expression and massive invasion of pro-inflammatory macrophages in brown adipose tissue (BAT). Indeed, BAT harbored a unique sub-set of Treg cells characterized by a unique gene signature. As these Treg cells respond to BAT activation upon cold exposure, this study defines a BAT-specific Treg sub-set with direct implications for the regulation of energy homeostasis in response to environmental stress.
Introduction
Obesity has reached pandemic dimensions with more than 1.5 billion people being affected world-wide [1]. Nutritional overloading of triglyceride storing white adipose tissue (WAT) impairs tissue function and is associated with a status of chronic low-grade inflammation [2]. Upon energy excess, adipocytes release chemokines resulting in adipose tissue infiltration by innate immune cells, e.g. pro-inflammatory macrophages and mast cells, and consequently aberrantly elevated inflammatory signaling [3]. Of note, WAT inflammation represents a causative factor for insulin resistance as a hallmark of obesity-related metabolic dysfunction [4]. Consequently, anti-inflammatory treatment potently improves insulin sensitivity in obesity [5]. In addition to the well-established role of the innate immune system, i.e. macrophage infiltration, in WAT dysfunction and a shift in macrophage polarization from an anti-inflammatory to a more pro-inflammatory status during progressive adiposity, the (aberrant) function of adaptive immune cells is increasingly emerging as a key event in obesity-related metabolic complications [6].
In this respect, regulatory T (Treg) cells represent a diverse sub-population of CD4+ T cells characterized by specific expression of the forkhead-winged helix transcription factor Foxp3 [7]. Treg cells interact with components of both the innate and the adaptive immune system, thereby serving as negative feedback regulators which prevent excess immune responses and ensure self-tolerance [8]. Whereas distinct sub-sets of T lymphocytes, like pro-inflammatory CD4+ T-helper (TH1) cells and cytotoxic T cells, were shown to be upregulated in obese WAT and might also contribute to insulin resistance [9], the number of WAT Treg cells was found to be markedly reduced in obese mice and humans [7,10]. Furthermore, transfer of Treg cells into lymphocyte-deficient obese mice reversed the aberrant glucose metabolism of the animals [11], indicating a key role of Treg cells in controlling WAT inflammation and the associated insulin sensitivity. Importantly, visceral adipose tissue (VAT) Treg cell accumulation, phenotype and function are controlled by the transcription regulator peroxisome proliferator activated receptor gamma (Pparg) [12].
In contrast to the energy-storing WAT, brown adipose tissue (BAT) and inducible brown-in-white (brite) adipocytes are specialized in the dissipation of energy in the form of heat by so-called uncoupling thermogenesis, mediated by the dissociation of mitochondrial respiratory chain electron transport from ATP synthesis via the action of uncoupling protein (UCP)1 [13]. Singular studies indicated that immune cells may also exert important regulatory roles in BAT development and physiology. For instance, alternatively activated anti-inflammatory macrophages (AAM) have been detected in WAT and BAT of mice in response to cold stimulation [14]. Moreover, AAM produced and secreted noradrenaline in an IL-4-dependent manner, thereby increasing thermogenic gene expression in BAT and enhancing energy expenditure [14]. In addition, AAM appear to be centrally involved in WAT browning, i.e. the appearance of brite cells, in response to beta-adrenergic signaling [15]. Recent studies reported that adipose tissue-resident eosinophils induced browning of WAT by stimulating AAM-dependent catecholamine release [16]. Finally, mice lacking mast cells display enhanced energy expenditure, improved glucose homeostasis and elevated expression of UCP1 in BAT [17]. Functional BAT has been detected and implicated in obesity susceptibility in adult humans [18]. Thus, the modulation of BAT-specific immune cell functions may provide future opportunities for BAT-centered systemic control of energy homeostasis and therapeutic targeting of obesity-related metabolic dysfunction. However, the regulatory impact and the molecular nature of BAT-associated Treg cells have not been defined to date.
Methods
2.1 Animals
T reg cells isolation. C57Bl6 female mice (n = 120) were obtained from Charles River Laboratories (CRL) at age 8 weeks and used for isolation of Treg cells from BAT. Mice were housed in a temperature controlled cabinet (Memmert) on a 12h light-dark cycle with unrestricted access to food and water. Mice were acclimatized to 30°C for 2 weeks and afterwards half of them (n = 60) was sacrificed and used for isolation of “warm” Treg cells. The other half (n = 60) was subjected to 12°C cold challenge for 2 days, sacrificed and used for isolation of “cold” Treg cells (details of Treg cells isolation below).
Metabolic phenotyping of Treg-depleted mice. B6N.129(Cg)-Foxp3tm3Ayr (Foxp3DTR; MGI:3698131; Jackson Stock Number: 016958) mice [19] were bred in our specific pathogen free facility at the DKFZ. The mice were housed at ambient temperature 22°C and fed standard chow diet (Kliba Nafag #3437, Provimi Kliba). For metabolic phenotyping of Treg cell-depleted mice, FoxP3DTR female mice were housed individually with unrestricted access to food and water in a control environment on 12h light-dark cycle in the PhenoMaster Cage System (TSE Systems) [20]. In order to deplete Treg cells, mice (n = 10) were intraperitoneally injected with 40 ng per g of body weight diphtheria toxin (DT) (Sigma) diluted in 200 μL PBS on two consecutive days. Vehicle (PBS) injected mice (n = 10) served as control. After the first round of injections, the housing temperature was decreased to 12°C. Oxygen consumption, food intake, locomotor activity and body weight (BW) were recorded continuously for 4 days. Heat production was calculated according to the formula: Heat production = CV*VO2, where CV = 3.941+(1.106*Respiratory Exchange Ratio) wherein numerical values are computed by TSE system. Mice were sacrificed with cervical dislocation in a random-fed state. Serum, brown adipose tissue, inguinal white adipose tissue and spleen were collected, washed in PBS, snap-frozen in liquid nitrogen and stored at -80°C. Animal handling and experimentation was performed in accordance with the European Union directives and the German animal welfare act (Tierschutzgesetz) and approved by local authorities (Regierungspräsidium Karlsruhe).
2.2 Cell sorting
For microarray analysis of Treg cells, stromo-vascular fraction from brown adipose tissue of warm acclimated mice (30°C for 2 weeks), cold treated mice (12°C for 2 days) and spleen was isolated and Treg cells were sorted out using fluorescence-activated cell sorting (FACS). Nonspecific binding of antibodies to cells was blocked by incubation with rat and hamster serum (1.0% (vol/vol) each) and cells were labeled with the following antibodies (clone): CD4 (GK1.5), CD8α (53–6.7), CD19 (6D5), TCRβ (H57–597), CD25 (PC61), CD11b (M1/70) and CD45 (30-F11) (Biolegend). Cells were FACS purified with the FACSAria II instrument (BD Bioscience). Antibody quality was checked and gating was performed using isotype controls. Purity of cell sorts was assured through post-sort analysis and post-sort staining. FlowJo (Treestar) was used for the analysis of flow cytometry data.
2.3 RNA expression profiling of FACS-sorted T cell populations
Conventional T cell (Tconv) (CD4+CD25-) or regulatory T cell (Treg) (CD4+CD25+) populations were FACS purified from the spleen or brown adipose tissue (BAT) from warm (30°C) or cold (12°C) treated mice. RNA was extracted using the RNEasy Plus Micro Kit (Qiagen). The DKFZ Genomics and Proteomics Core Facility amplified and hybridized material to the MouseWG-6 v2.0 Expression BeadChip (Illumina). Microarray data have been deposited in the Gene Expression Omnibus (GEO) with the accession code GSE64909. Microarray confirmation by real-time PCR was not performed for Treg cells due to sample amount limitations.
2.4 Blood and serum parameters
Blood glucose was measured at random-fed state immediately after sacrificing the mice using an automated glucose monitor (One Touch, Lifescan). Commercially available kits were used to quantify serum triglycerides (Sigma) and non-esterified fatty acids (WAKO).
2.5 RNA isolation and real-time RT-PCR
Total RNA was extracted from frozen powderized BAT, scWAT and spleen using Qiazol (Qiagen) and the RNeasy Mini Kit (Qiagen), performing on-column DNAse digest. cDNA was prepared by reverse transcription of 1 μg of total RNA using the First strand cDNA synthesis kit (Fermentas). Real-time RT-PCR was performed in order to measure gene expression of browning (Ucp1, Cidea, Dio2, Pparg, Prdm16) and inflammatory (Cd68, Ccl2, Tnfa, Ifng, Mrc1, Mgl1, Arg1, Il-10, Il-4) markers in BAT and scWAT and to verify Treg cells depletion from BAT and spleen by measuring FoxP3 expression. TaqMan Master Mix (Applied Biosystems) and a gene specific Taqman probes (Applied Biosystems) were used on an ABI StepOnePlus sequence detector (Applied Biosystems). Relative mRNA expression levels were calculated by the delta Ct method using TATA-box binding protein (Tbp) expression as a reference.
2.6 RNA expression profiling of Treg-deficient and -proficient BAT
Total RNA from BAT was isolated as described and subjected to gene expression analysis using GeneChip Mouse Genome 430 2.0 arrays (Affymetrix) performed according to the manufacturer’s recommendations. A Custom CDF Version 14 with Entrez based gene definitions was used to annotate the arrays. The Raw fluorescence intensity values were normalized by quantile normalization. Differential gene expression was analyzed based on loglinear mixed model ANOVA with a commercial software package SAS JMP7 Genomics (version 4) from SAS (SAS Institute). Significance was assumed based on a false positive rate of p = 0.05 with FDR correction. Gene Set Enrichment Analysis (GSEA), was used to determine whether defined lists (or sets) of genes exhibit a statistically significant bias in their distribution within a ranked gene list (http://www.broadinstitute.org/gsea/; [21]. Raw and normalized data are deposited in the Gene Expression Omnibus (GEO) database with an accession number GSE62157.
Real-time PCR was performed on six selected genes, including Prdm16, Dio2, Cd68, Ccl2, Il4, and Infγ to confirm the array results.
2.7 Histology
Brown adipose tissue was fixed in 4% paraformaldehyde for 24 h at room temperature, paraffin-embedded and cut in 3 μm sections. Sections were stained with hematoxylin and eosin (H&E) by standard procedures. For immunohistochemistry, sections were subjected to heat-mediated antigen retrieval in citrate buffer (pH 6) prior to blocking with 1% BSA for 1 h at room temperature. Primary rat monoclonal anti-MAC-2 antibody (Cedarlane Laboratories, #CL8942AP) was diluted 1:3800 in 1% BSA and incubated over night at 4°C. For detection, HPR-conjugated goat anti-rat antibody (Santa Cruz Biotechnology, #sc-2032) diluted 1:500 in 1% BSA was used as a secondary antibody with 3,3’-diaminobenzidine (Sigma) as a chromogenic substrate. Stainings were imaged at 20x magnification (Zeiss). MAC-2 positive signal was quantified as a percentage of total area using Fiji software [22].
2.8 Statistical Analysis
Statistical analysis of experimental data was performed using Prism software (GraphPad) or Excel (Microsoft). The results are shown as means ± SD. Statistical analysis was performed by two-tailed Student’s t-test, for real-time RT-PCR calculated on log10-transformed data values. Results were considered statistically significant if p-values were less than 0.05.
Results
Cold BAT Treg cells display a unique molecular signature
The overall importance of Treg cells for WAT function prompted us to explore the molecular nature of BAT-associated Treg cells. To this end, wild-type C57Bl6 mice were either acclimatized to 30°C or challenged with a 12°C cold exposure for 2 days. After the corresponding acclimatization periods, BAT depots as well as the spleens were collected and Treg cells were sorted out of the corresponding tissue by fluorescence-associated cell sorting (FACS) followed by RNA purification and hybridization to an expression array. Microarray gene expression profiling revealed 430 genes to be more than twofold upregulated in BAT Treg cells compared to splenic Treg cells, with 222 genes significant between three replicates (Fig. 1A). Confirmation of lineage-specific genes in BAT Treg versus BAT Tconv T cells included classic Treg genes such as Foxp3 and cytotoxic T-lymphocyte-associated protein 4 (Ctla4) as over-represented and special AT-rich sequence binding protein 1 (Satb1) and transcription factor 7, T cell specific (Tcf7) as under-represented genes. In addition, chemokine (C-X-C motif) ligand (Cxcl) 1 and 2, and interleukin (IL) 10 were highly over-expressed in BAT Treg cells (Fig. 1B). The top 30 significantly upregulated genes and bottom 10 downregulated genes in BAT Treg cells were then subjected to unsupervised hierarchical clustering (Fig. 1C), which revealed a unique gene expression signature for BAT Treg cells compared to splenic regulatory and conventional T (Tconv) cells as well as BAT Tconv cells. We then compared the BAT Treg gene signature to the VAT-specific gene signature [7], which stratified most of the genes identified in both microarrays to be specific for a general fat Treg gene signature (Fig. 1D, E). However, a specific BAT Treg gene signature could also be discerned, which was independent of the WAT Treg signature (Table 1).
Fig 1. Genotypical comparison of Treg and Tconv cells isolated from brown adipose tissue (BAT) and spleen tissue (SPL) in cold- and warm-conditioned animals generated with an Illumina Mouse Expression Array.
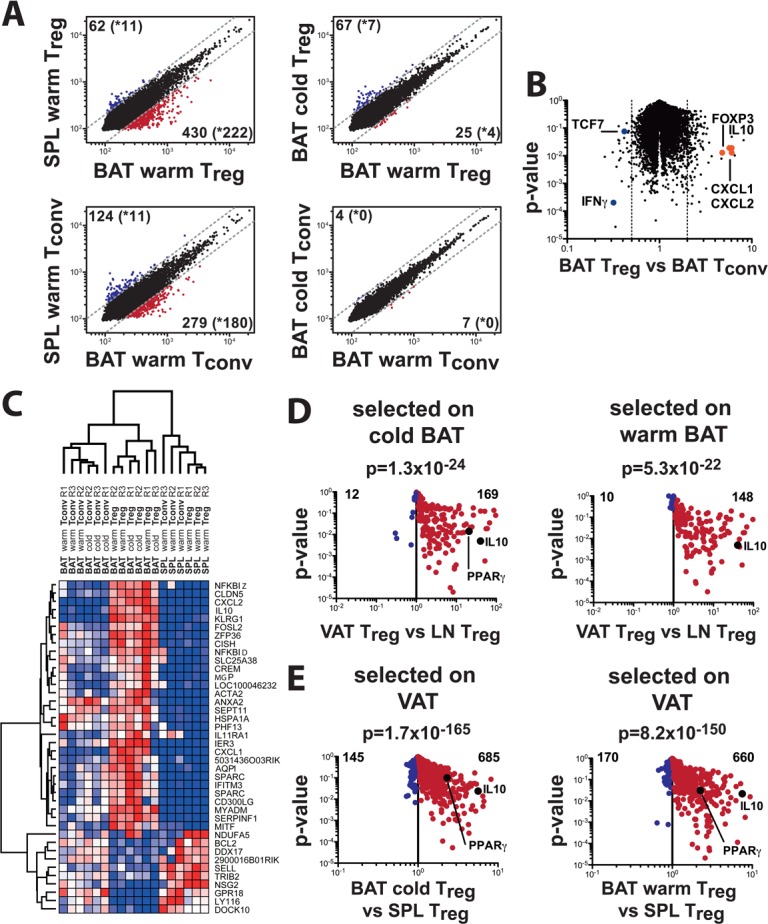
(A) Gene expression profiles comparing Treg (top) or Tconv (bottom) cell populations between spleen and adipose tissue samples isolated from warm-conditioned animals (left) or between cells isolated from cold vs warm-conditioned animals (right). Numbers indicate genes either up- or downregulated more than 2-fold (cut-off: dotted line), with the number of significantly different (p<0.05) genes shown in brackets with an asterisk. (B) Volcano plot comparing gene expression and significance values between Treg and Tconv genes isolated from BAT in warm-conditioned animals. Key up- or downregulated genes in Treg cells are annotated (Foxp3, Il10, Cxcl1/2, Tcf7, Ifng) and serve as quality control to the published consensus Treg-cell signature. (C) Hierarchical clustering of the top-30 upregulated genes and the top-10 downregulated genes in warm-conditioned brown adipose tissue Treg cells versus spleen Treg cells. (D) Comparison of BAT-Treg-specific genes with visceral adipose tissue (VAT)-specific genes. We first determined 430 genes to upregulated in BAT warm-conditioned Treg cells, with 222 genes being significantly altered (p<0.05). We then overlaid BAT Treg-upregulated genes with VAT Treg tissue specific expression gene data. 181 genes were matched between both microarary chips, with 169 genes also upregulated in VAT, and only 12 genes specific for BAT (left). The corresponding analysis of the 516 genes upregulated in cold BAT Treg cells versus warm spleen Treg cells revealed 194 genes to be significantly upregulated. 158 could be matched to VAT Treg-specific genes, of which 148 were VAT-specific, whereas only 10 were specific for BAT. P-values indicate the significance of overrepresentation of BAT Treg-specific genes in the VAT Treg signature. (E) Comparison of VAT-Treg specific genes on BAT warm (left) or BAT cold (right) gene signatures. Of 1839 genes specifically overexpressed in VAT Treg cells, 1059 were statistically significantly (p<0.05) upregulated. Of these 1059 genes, 829 were also detectable in the BAT Treg microarray. When comparing the VAT Treg signature to warm BAT Treg cells, 660 genes were overrepresented in VAT, whereas cold BAT Treg cells show 685 genes to be overrepresented in VAT.
Table 1. Principal biological processes affected by Treg cell depletion in BAT.
| Biological process | GO number | FDR (%) | |
|---|---|---|---|
| Immune processes | Immune response | GO0006955 | <0.001 |
| Inflammatory response | GO0006954 | <0.001 | |
| Innate immune response | GO0045087 | 0.048 | |
| Adaptive immune response | GO0002250 | 0.151 | |
| Chemotaxis | GO0006935 | <0.001 | |
| Leukocyte chemotaxis | GO0030595 | 0.008 | |
| Cell migration | GO0016477 | 13.344 | |
| Cell activation | GO0001775 | 0.057 | |
| Myeloid leukocyte activation | GO0002274 | <0.001 | |
| Leukocyte activation | GO0045321 | 0.097 | |
| T cell activation | GO0042110 | 0.194 | |
| Lymphocyte activation | GO0046649 | 0.500 | |
| Activation of immune response | GO0002253 | 1.459 | |
| Cell proliferation | GO0008283 | 2.271 | |
| Mononuclear cell proliferation | GO0032943 | 0.101 | |
| T cell proliferation | GO0042098 | 0.113 | |
| Lymphocyte proliferation | GO0046651 | 0.125 | |
| Myeloid cell differentiation | GO0030099 | 0.051 | |
| Leukocyte differentiation | GO0002521 | 4.297 | |
| Lymphocyte differentiation | GO0030098 | 5.157 | |
| Cytokines | Cytokine and chemokine mediated signaling pathway | GO0019221 | 0.231 |
| Cytokine production | GO0001816 | 1.637 | |
| Cytokine biosynthetic process | GO0042089 | 3.518 | |
| Cytokine metabolic process | GO0042107 | 4.244 | |
| Metabolism | ATP metabolic process | GO0046034 | 11.567 |
| ATP biosynthetic process | GO0006754 | 8.646 | |
| Phosphate transport | GO0006817 | 0.021 | |
| ATP synthesis coupled proton transport | GO0015986 | 3.864 |
Next, we investigated whether differences exist between the gene expression profiles for cold- and warm-exposed Treg cells. While the majority of genes was expressed at comparable levels in both cell types, 11 genes displayed significant gene expression changes in response to cold challenge (Fig. 1A), including coiled-coil-helix-coiled-coil-helix domain containing 10 (Chchd10), solute carrier protein family 52, member 3 (Slc52a3), protein tyrosine phosphatase, receptor type, S (Ptprs), RFT1 homolog (S. cerevisiae) (Rft1), dishevelled associated activator of morphogenesis 2 (Daam2), zinc finger protein 668 (Zfp668), stromal interaction molecule 1 (Stim1), family with sequence similarity 65, member B (Fam65b), RAS-like, family 11, member B (Rasl11b), centromere protein L (Cenpl), and polymerase (RNA) I polypeptide A (Polr1a) suggesting that this gene cluster represents a cold-specific Treg cell signature in BAT.
Cold activation of BAT requires Treg cells for proper thermogenic gene expression
To next test the impact of Treg cells on systemic energy expenditure under cold conditions, we performed deep metabolic phenotyping of Treg-proficient and Treg-deficient mice. Due to the lack of fat-specific Treg targeting systems to date, we employed transgenic mice expressing the diphtheria toxin (DT) receptor under the control of Foxp3 gene regulatory elements [19]. Given the development of systemic immune dysfunction two weeks after DT administration in these animals [19], we restricted our analysis to the adaptation of systemic metabolism to an acute cold challenge and analyzed 48 h after Treg depletion. Therefore, control littermates and Treg-depleted mice were challenged with an acute 4°C cold exposure after adaptation to room temperature (22°C). In line with previous studies [7], administration of DT to these animals resulted in the rapid systemic depletion of Foxp3-expressing cells (Fig. 2A).
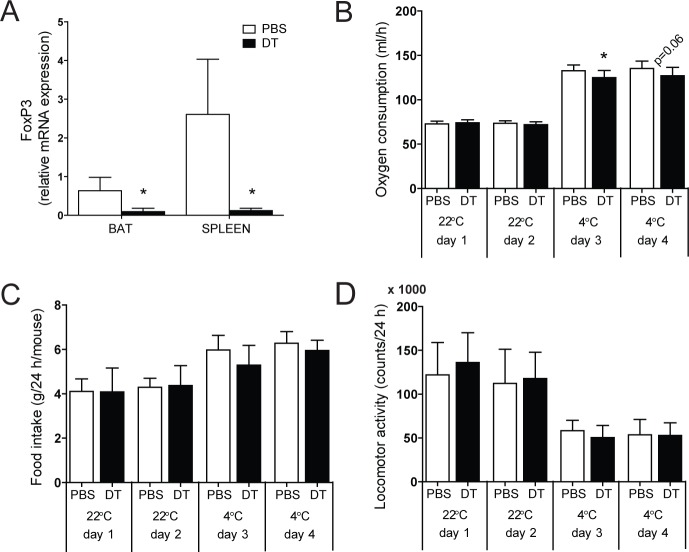
Fig 2. Treg cell depletion and metabolic phenotyping.
(A) Real-time RT-PCR analysis of FoxP3 expression in brown adipose tissue (BAT) and spleen in Treg cell-proficient (PBS) and Treg cell-deficient (DT) mice. (B) Oxygen consumption, (C) food intake and (D) locomotor activity during 4 consecutive days at 22°C and 4°C. Injections of vehicle (PBS) or diphtheria toxin (DT) were performed at day 2 and day 3. Values are mean ± SD (n = 9–10); *P<0.05 (Student’s t-test).
While Treg deficiency did not exert any effects on respiratory activity under room temperature conditions as demonstrated by unchanged oxygen consumption rates (Fig. 2B), depletion of Treg cells significantly reduced oxygen consumption at cold conditions (Fig. 2B), while leaving food intake and locomotor activity unchanged (Fig. 2C, D).
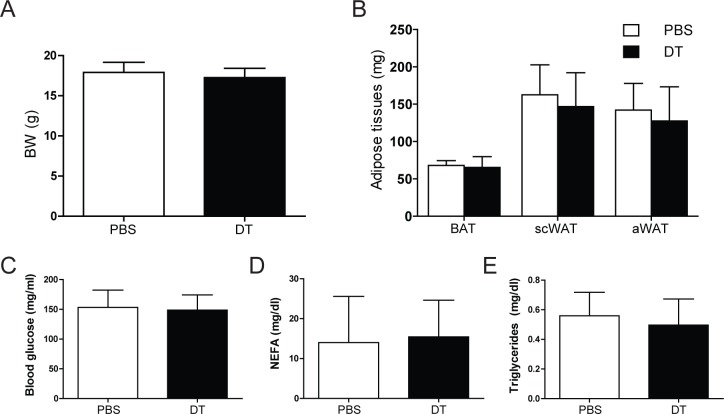
Of note, diphtheria toxin administration had no effect on oxygen consumption in wild-type animals (data not shonw), demonstrating the specificity of the observed effects. Also, body and adipose tissue weights and other metabolic parameters, including blood glucose, serum non-esterified fatty acids (NEFA) and triglycerides, remained unaffected by Treg cell-depletion (Fig. 3A-E).
Fig 3. Physiological parameters.
(A) Body weight (BW) and (B) adipose tissues weights of Treg cell-proficient (PBS) and Treg cell-deficient (DT) mice after cold exposure. BAT, brown adipose tissue; scWAT, subcutaneous white adipose tissue, aWAT, abdominal white adipose tissue. (C) Blood glucose, (D) serum non-estherified fatty acids (NEFA) and (E) serum triglycerides in PBS and DT mice. Values are mean ± SD (n = 9–10); *P<0.05 (Student’s t-test).
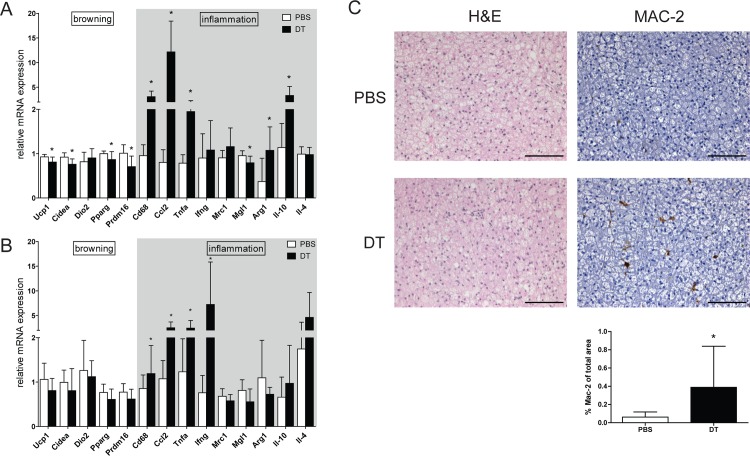
These data supported the hypothesis that Treg cells may exert regulatory functions in BAT activation in response to cold stimulation. Consistent with impaired BAT activity, heat production per animal was decreased (S1 Fig.). Also, selective gene expression profiling revealed that particularly genes in the thermogenic gene program were downregulated in cold-exposed BAT upon Treg cell deficiency (Fig. 4A), without affecting browning markers in subcutaneous WAT (Fig. 4B). Pro-inflammatory markers, including chemokine (C-C motif) ligand 2 (Ccl2) and tumor necrosis factor alpha (Tnfa), were found to be induced upon the lack of Treg cell function. Along with these observations, Treg-deficient BAT displayed a substantial increase in macrophage infiltration (Fig. 4C).
Fig 4. Inflammatory status of adipose tissue.
Real-time RT-PCR analysis of (A) brown adipose tissue (BAT) and (B) subcutaneous white adipose tissue of Treg cell-proficient (PBS) and Treg cell-deficient (DT) mice after cold exposure. Ucp1, uncoupling protein 1; Cidea,cell death-inducing DNA fragmentation factor, alpha subunit-like effector A; Dio2,deiodinase, iodothyronine, type II; Pparg, peroxisome proliferator-activated receptor gamma; Prdm16, PR domain containing 16; Cd68, Cd68 antigen; Ccl2,chemokine (C-C motif) ligand 2; Tnfa, tumor necrosis factor alpha; Ifng, interferon, gamma; Mrc1, mannose receptor, C type 1; Mgl1, macrophage galactose-type C-type lectin 1; Arg1,arginase 1; Il-10, interleukin 10; Il-4, interleukin 4. Data are mean ± SD (n = 9–10); *p<0.05 (Student’s t-test). (C) Representative hematoxylin and eosin (H&E) staining (left) and immunohistochemical anti-MAC-2 staining (right; brown color) in BAT from PBS and DT mice. Scale bar 100 μm. Quantification of MAC-2 positive area (panel below MAC-2 staining) as a percentage of total area.
Treg cells control distinct inflammatory and metabolic pathways in BAT
The data thus far indicated that suppressive Treg cell function plays an important role in the execution of proper BAT function with implications for systemic energy expenditure, prompting us to analyze Treg-dependent molecular networks in BAT in more detail. To this end, we performed microarray analysis of cold-exposed Treg-deficient and -proficient BAT. Hierarchical clustering analysis showed a clear difference in transcription pattern between respective groups (S2 Fig.). It revealed that the absence of Treg cells led to the marked upregulation of genes involved in immune processes. Most prominent among 107 significantly upregulated genes in Treg-deficient BAT were CC chemokine family members, namely Ccl2, -3, -6, -7 and -9. In parallel, expression of the macrophage marker Cd68 was upregulated. Increased infiltration of macrophages in BAT was indeed confirmed by immunohistochemical staining and its quantification for an additional macrophage marker MAC-2 (Fig. 4C). Moreover, high transcript levels of members of the Toll-like receptor family (Tlr7, -8, -13) were observed, suggesting activation of innate immune system in Treg cell-deficient BAT. At the same time, a glycolytic enzyme aldolase A (Aldoa) and a glucose/fructose transporter solute carrier family 2 (facilitated glucose transporter), member 5 (Slc2a5) were downregulated in Treg-deficient BAT, which might indicate impaired glucose utilization. Manual inspection of the gene expression data for gene members of glycolytic pathway indeed demonstrated that expression of pyruvate dehydrogenase kinase, isoenzyme 1 (Pdk1), glucose-6-phosphatase, catalytic (G6pc), phosphoglycerate kinase 1 (Pgk1), hexokinase 1 (Hk) and phosphoglucomutase 3 (Pgm3) were decreased as well, although did not reach the stringent criteria for statistical significance.
Subsequent pathway analysis using Gene Set Enrichment Analysis (GSEA) software confirmed the hierarchical clustering and showed that the absence of Treg cells led to substantial upregulation of immune processes, in particular of pro-inflammatory pathways involving cell differentiation, proliferation, trafficking and activation (Table 1). Furthermore, at the same time ATP metabolic processes were upregulated in Treg-deficient BAT (Table 1). Thus, Treg cell ablation altered the activity in particular of pro-inflammatory pathways while at the same time controlling ATP synthesis-related genes and glycolysis overall suggesting that BAT-associated Treg cells indeed impose a critical regulatory input into BAT macrophage activity as well as energy production.
Discussion
In this study, we characterized the gene expression profile of BAT Treg cells under two different temperature conditions and investigated the role of Treg cells in BAT activation after cold challenge.
Recently, a unique population of Treg cells with a distinct gene expression signature has been characterized in VAT [23] and its contribution to proper white adipose function has been described [11]. Given the opposing functions of white and brown adipose tissues, this prompted us to investigate Treg cell population characteristics in BAT under different temperature conditions. The gene expression profile of BAT Treg cells clearly differentiated them from spleen Treg cells and from conventional BAT and spleen T cells. Direct comparison to the visceral Treg cell expression profile revealed a close similarity of BAT and VAT Treg cells, but also a BAT specific Treg signature. Furthermore, we compared the Treg cell expression profile from warm and cold temperature condition and identified 11 unique differentially expressed genes. The high correlation of BAT- and VAT-specific Treg genes indicate common adipose tissue factors that induce the phenotype of fat resident Treg cells. Indeed, Pparg, the central transcription factor involved in VAT Treg cell accumulation and function [12], is also over-expressed in BAT Treg cells, however, much less pronounced. The identification of a VAT independent but BAT-specific Treg cell signature suggests that additional factors shape BAT Treg cell phenotype and function.
Lack of Treg cells [7] or their phenotypic change [24] in WAT upon high-fat feeding is associated with VAT dysfunction. In agreement with these findings, metabolic phenotyping after systemic Treg cell depletion also indicated impaired BAT function, demonstrated by decreased oxygen consumption in Treg depleted mice and prevention of the activation of thermogenic genes. Decreased thermogenesis is often translated into enhanced adiposity [25]; however, BW and fat pad weight after cold stimulation remained unchanged in Treg-proficient and Treg-deficient mice due to increased food intake, which most likely compensated for increased energy expenditure. Decreased thermogenesis in Treg-deficient mice did not affect adiposity, given only the short period of observation.
Treg cell depletion potentiates tissue inflammation and influences number and activation state of macrophage and dendritic cells [19]. It has been shown that sustaining adaptive thermogenesis requires alternative activation of BAT-resident macrophages in cold-challenged mice [14], which was precluded upon Treg cell depletion. Indeed, pro-inflammatory Tnfa and several members of CC chemokine family were among the most upregulated genes in cold-challenged Treg cell depleted BAT. Therefore, impaired BAT function might be at least partly attributed to inability of alternative macrophage activation due to the loss of Treg cell suppressive function and the markedly increased inflammatory state of the tissue, since inflammation can suppress induction of Ucp1 expression [26,27].
Cold exposure is associated with lipolysis stimulation and increased glucose uptake in BAT [28–30], where released fatty acids and glucose represent a metabolic fuel. In our setting, glucose transporter together with glycolytic pathway activation in Treg-deficient mice remained downregulated compared to control mice, indicating impaired fuel utilization. Moreover, ATP metabolic processes were upregulated in Treg-deficient BAT, suggesting that high inflammatory status of the tissue prevented heat production.
Our study defines the molecular signature of BAT-specific Treg cells and also implies a functional role of Treg cells in BAT function during cold exposure. Given the therapeutic potential of BAT activation for conditions of metabolic dysfunction, the modulatory role of BAT-associated Treg cells may now provide clues for novel BAT-based therapies in obesity and type 2 diabetes.
Supporting Information
Treg cell-proficient (PBS) and Treg cell-deficient (DT) mice heat production during 4 consecutive days at 22°C and 4°C. Injections of vehicle (PBS) or diphtheria toxin (DT) were performed at day 2 and day 3. Values are mean ± SD (n = 9–10); *P<0.05.
(TIF)
Heat map showing all FDR-significant genes and genes with fold-change higher than twofold with nominal p-value below 0.01.
(TIF)
Genes over-expressed in warm-conditioned BAT Treg cells compared to spleen Treg cells from the same mice were identified (2-fold up-regulated). These genes were then compared with visceral adipose tissue (VAT) Treg cells and selected for not-different expressed in visceral Treg vs lymph node Treg cells (less than 1.2-fold change). These 28 genes are listed with their differential expression values color-coded to indicate significance (p<0.05, red font color) and expression change between BAT and SPL tissue (two-fold induction, green font color). Genes with more than two-fold induction only in Treg BAT cells, but not in Tconv BAT cells, comprise a Treg-specific BAT gene signature.
(DOCX)
Acknowledgments
We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Expression BeadChips services, and the DKFZ Flow Cytometry Core Facility for expert service and support.
Data Availability
All relevant data are within the paper and its Supporting Information files. For RNA expression profiling of brown adipose tissue, raw and normalized data are deposited in the Gene Expression Omnibus (GEO) database with an accession number GSE-62157.
Funding Statement
This work was funded by a grant of the Deutsche Forschungsgemeinschaft (HE-3260/8-1) and the EU-FP7 Collaborative Project DIABAT (HEALTH -F2-2011-278373) to S. H., Helmholtz Association of German Research Centers (HZ-NG-505 to M. F.), and the German-Israeli-Helmholtz Research School in Cancer Biology (to M. D.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet 2011; 377: 557–567. 10.1016/S0140-6736(10)62037-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hotamisligil GS Inflammation and metabolic disorders. Nature 2006; 444: 860–867. [DOI] [PubMed] [Google Scholar]
- 3. Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003; 112: 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shoelson SE, Herrero L, Naaz A Obesity, inflammation, and insulin resistance. Gastroenterology 2007; 132: 2169–2180. [DOI] [PubMed] [Google Scholar]
- 5. Yuan M, Konstantopoulos N, Lee J, Hansen L, Li ZW, Karin M, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science 2001; 293: 1673–1677. [DOI] [PubMed] [Google Scholar]
- 6. Eheim A, Medrikova D, Herzig S Immune cells and metabolic dysfunction. Semin Immunopathol 2014; 36: 13–25. 10.1007/s00281-013-0403-7 [DOI] [PubMed] [Google Scholar]
- 7. Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med 2009; 15: 930–939. 10.1038/nm.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakaguchi S, Takahashi S, Sasaki T, Kumagai T, Nagata K Progression of alcoholic and non-alcoholic steatohepatitis: common metabolic aspects of innate immune system and oxidative stress. Drug Metab Pharmacokinet 2011; 26: 30–46. [DOI] [PubMed] [Google Scholar]
- 9. Osborn O, Olefsky JM The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med 2012; 18: 363–374. 10.1038/nm.2627 [DOI] [PubMed] [Google Scholar]
- 10. Deiuliis J, Shah Z, Shah N, Needleman B, Mikami D, Narula V, et al. Visceral adipose inflammation in obesity is associated with critical alterations in tregulatory cell numbers. PLoS One 2011; 6: e16376 10.1371/journal.pone.0016376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med 2009; 15: 921–929. 10.1038/nm.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012; 486: 549–553. 10.1038/nature11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diaz MB, Herzig S, Vegiopoulos A Thermogenic adipocytes: From cells to physiology and medicine. Metabolism 2014; 63: 1238–1249. 10.1016/j.metabol.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 14. Nguyen KD, Qiu Y, Cui X, Goh YP, Mwangi J, David T, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature 2011; 480: 104–108. 10.1038/nature10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee YH, Thacker RI, Hall BE, Kong R, Granneman JG Exploring the activated adipogenic niche: interactions of macrophages and adipocyte progenitors. Cell Cycle 2014; 13: 184–190. 10.4161/cc.27647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qiu Y, Nguyen KD, Odegaard JI, Cui X, Tian X, Locksley RM, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 2014; 157: 1292–1308. 10.1016/j.cell.2014.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J, Divoux A, Sun J, Zhang J, Clement K, Glickman JN, et al. Genetic deficiency and pharmacological stabilization of mast cells reduce diet-induced obesity and diabetes in mice. Nat Med 2009; 15: 940–945. 10.1038/nm.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009; 360: 1500–1508. 10.1056/NEJMoa0808718 [DOI] [PubMed] [Google Scholar]
- 19. Kim JM, Rasmussen JP, Rudensky AY Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol 2007; 8: 191–197. [DOI] [PubMed] [Google Scholar]
- 20. Rohm M, Sommerfeld A, Strzoda D, Jones A, Sijmonsma TP, Rudofsky G, et al. Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metab 2013; 17: 575–585. 10.1016/j.cmet.2013.02.010 [DOI] [PubMed] [Google Scholar]
- 21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A 2010; 107: 5919–5924. 10.1073/pnas.1002006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han JM, Patterson SJ, Speck M, Ehses JA, Levings MK Insulin inhibits IL-10-mediated regulatory T cell function: implications for obesity. J Immunol 2014; 192: 623–629. 10.4049/jimmunol.1302181 [DOI] [PubMed] [Google Scholar]
- 25. Vegiopoulos A, Muller-Decker K, Strzoda D, Schmitt I, Chichelnitskiy E, Ostertag A, et al. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science 2010; 328: 1158–1161. 10.1126/science.1186034 [DOI] [PubMed] [Google Scholar]
- 26. Sakamoto T, Takahashi N, Sawaragi Y, Naknukool S, Yu R, Goto T, et al. Inflammation induced by RAW macrophages suppresses UCP1 mRNA induction via ERK activation in 10T1/2 adipocytes. Am J Physiol Cell Physiol 2013; 304: C729–738. 10.1152/ajpcell.00312.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valladares A, Roncero C, Benito M, Porras A TNF-alpha inhibits UCP-1 expression in brown adipocytes via ERKs. Opposite effect of p38MAPK. FEBS Lett 2001; 493: 6–11. [DOI] [PubMed] [Google Scholar]
- 28. Klingenspor M Cold-induced recruitment of brown adipose tissue thermogenesis. Exp Physiol 2003; 88: 141–148. [DOI] [PubMed] [Google Scholar]
- 29. Shimizu Y, Nikami H, Saito M Sympathetic activation of glucose utilization in brown adipose tissue in rats. J Biochem 1991; 110: 688–692. [DOI] [PubMed] [Google Scholar]
- 30. Marette A, Bukowiecki LJ Noradrenaline stimulates glucose transport in rat brown adipocytes by activating thermogenesis. Evidence that fatty acid activation of mitochondrial respiration enhances glucose transport. Biochem J 1991; 277 (Pt 1): 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Treg cell-proficient (PBS) and Treg cell-deficient (DT) mice heat production during 4 consecutive days at 22°C and 4°C. Injections of vehicle (PBS) or diphtheria toxin (DT) were performed at day 2 and day 3. Values are mean ± SD (n = 9–10); *P<0.05.
(TIF)
Heat map showing all FDR-significant genes and genes with fold-change higher than twofold with nominal p-value below 0.01.
(TIF)
Genes over-expressed in warm-conditioned BAT Treg cells compared to spleen Treg cells from the same mice were identified (2-fold up-regulated). These genes were then compared with visceral adipose tissue (VAT) Treg cells and selected for not-different expressed in visceral Treg vs lymph node Treg cells (less than 1.2-fold change). These 28 genes are listed with their differential expression values color-coded to indicate significance (p<0.05, red font color) and expression change between BAT and SPL tissue (two-fold induction, green font color). Genes with more than two-fold induction only in Treg BAT cells, but not in Tconv BAT cells, comprise a Treg-specific BAT gene signature.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. For RNA expression profiling of brown adipose tissue, raw and normalized data are deposited in the Gene Expression Omnibus (GEO) database with an accession number GSE-62157.